Abstract
Systemic or hepatic inflammation is caused by intraperitoneal application of lipopolysaccharide (LPS). In this study, we investigated anti-inflammatory and antioxidant properties of combination of ginsenoside-Rg2 (G-Rg2) and -Rh1 (G-Rh1) on liver function under LPS challenging. We first confirmed that G-Rg2 and -Rh1 at 100 μg/ml did not show cytotoxicity in HepG2 cells. G-Rg2 and -Rh1 treatment significantly inhibited activation of STAT3 and TAK1, and inflammatory factors including iNOS, TNF-α, and IL-1β in peritoneal macrophages. In HepG2 cells, G-Rg2 and -Rh1 treatment inhibited activation of STAT3 and TAK1/c-Jun N-terminal kinase, and down-regulated nuclear translocation of NF-κB transcription factor. In addition, LPS-induced mitochondrial dysfunction was restored by treatment with G-Rg2 and -Rh1. Interestingly, pretreatment with G-Rg2 and -Rh1 effectively inhibited mitochondrial damage-mediated ROS production induced by LPS stimulation, and alterations of Nrf2 nuclear translocation and ARE promotor activity were involved in G-Rg2 and -Rh1 effects on balancing ROS levels. In liver tissues of LPS-treated mice, G-Rg2 and -Rh1 treatment protected liver damages and increased Nrf2 expression while reducing CD45 expression. Taken together, G-Rg2 and -Rh1 exerts a protective effect on liver function by increasing antioxidant through Nrf2 and anti-inflammatory activities through STAT3/TAK1 and NF-κB signaling pathways in liver cells and macrophages.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The liver plays a key role in the regulation of metabolism, homeostasis and immunological activities (Nesseler et al. 2012). However, these vital functions are usually affected by inflammatory factors, oxidative stress and pathogens and lipopolysaccharide (LPS), a major component of the outer membrane of gram-negative bacteria, is the most characterized pathogen that triggers systemic inflammatory response, leading to the dysfunction of multiple organs, particularly liver (Cecconi et al. 2018; Lee et al. 2019). This may result from immune cell infiltration in liver as well as the overproduction and prolonged secretion of inflammatory mediators and oxidative stress (Li et al. 2016).
Under the stimulation of LPS, various signaling pathways including nuclear factor kappa-light-chain-enhancer of activated B (NF-κB), mitogen-activated protein kinases (MAPKs) and Janus kinase/signal transducers and activators of transcription (JAK/STAT) are activated, resulting in the upregulation of a variety of pro-inflammatory mediators and cytokines, such as inducible nitric oxide synthase (iNOS), tumor necrosis factor alpha (TNF-α) and interleukin-1 beta (IL-1β), which play an important role in inflammatory process (Lee et al. 2017; Huynh et al. 2020a). Additionally, transforming growth factor-β activated kinase-1 (TAK1), a member of the mitogen-activated 3 kinase family, is also activated by a wide range of inflammatory stimuli such as inflammatory cytokines and toll-like receptor (TLR) ligands (Mihaly et al. 2014), which leads to the activation of NF-κB (Irie et al. 2000). Furthermore, activation of MAPKs lead to reduction in antioxidant enzymes (Bak et al. 2012).
Reactive oxygen species (ROS) accumulation induced by LPS leads to the state of oxidative stress, which also promotes the inflammatory progression (Lugrin et al. 2014). Nuclear factor-erythroid 2-related factor 2 (Nrf2), which is a transcription factor that plays a pivotal role in regulating the expression of antioxidant genes, such as heme oxygenase 1, NAD(P)H quinone oxidoreductase 1, superoxide dismutase (SOD), glutathione peroxidase (GPx) and catalase (Taguchi and Kensler 2020). Notably, this transcription factor has been demonstrated to contribute to the suppression of inflammatory conditions by regulating gene expression via the antioxidant response element (ARE) (Ahmed et al. 2017). The upregulation of antioxidant genes mediated Nrf2 has been revealed to inhibit the expression of pro-inflammatory cytokines (Ahmed et al. 2017). Therefore, targeting the Nrf2 signaling not only controls oxidative stress, but also ameliorates the inflammatory process, which is a potential therapeutic to protect liver function during sepsis condition.
Ginsenosides (G), the major active components of Panax ginseng Mayer (P. ginseng), which have been demonstrated to exhibit various pharmacological effects, including antioxidant, anticancer, anti-inflammatory effects and beneficial effects on cardiovascular diseases and immunodeficiency (Baatar et al. 2018; Huynh et al. 2020a; Jin et al. 2020). Particularly, ginsenoside-Rg2 (G-Rg2) and -Rh1 (G-Rh1) are classified as minor ginsenosides, which can be converted from the major ginsenosides Re and Rg1 (Quan et al. 2012; Kim 2018). In our previous study, G-Rg2 and -Rh1 has been revealed to exert the anti-inflammatory effect in LPS-stimulated macrophages and the protective effects on liver and kidney injury through the inhibition of TLR4-STAT1 signaling pathway (Huynh et al. 2020a). However, whether the beneficial effects of this combination on the regulation of oxidative stress to protect liver function from inflammatory responses still remains to be elucidated. In this study, we further investigated the effects of G-Rg2 and -Rh1 on maintaining the redox balance to protect liver function during sepsis shock induced by LPS and the potential mechanisms in liver cells and macrophages.
Materials and methods
Materials
Primary antibodies for phospho-NF-κB p65, STAT3, phospho-STAT3, phospho-TAK1, extracellular signal-regulated kinase 1/2 (ERK1/2), phospho-ERK1/2, c-Jun N-terminal kinase (JNK), phospho-JNK, p38, phospho-p38, Nrf2 and histone deacetylase 1 (HDAC1) were purchased from Cell Signaling Technology, Inc. (Danvers, MA, USA). Antibodies for TAK1, TNF-α, IL-1β and CD45 were purchased from Santa Cruz Biotechnology Inc. (Dallas, TX, USA). Anti-iNOS antibody was purchased from Cayman chemical, Inc. (Ann Arbor, MI, USA). Anti-α-tubulin antibody, anti-IgG antibody, LPS, dexamethasone (Dex), 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolylcarbocyanine iodide (JC-1), thioglycolate broth, 2′,7′-dichlorodihydrofluorescein diacetate (DCF-DA), hematoxylin and eosin (H&E) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Mounting medium with 4′,6-diamidino-2-phenylindole (DAPI) was purchased from Vector Laboratories, Inc. (Burlingame, CA, USA). Phosphate buffered saline (PBS) and reverse transcription 5X master mix were purchased from Elpisbio (Daejeon, Korea). Bovine serum albumin (BSA) was purchased from Bioworld (Dublin, OH, USA). Tri-RNA reagent was purchased from Favorgen (Pingtung, China). Dulbecco’s modified Eagle’s medium (DMEM) and fetal bovine serum (FBS) were purchased from Gibco (Carlsbad, CA, USA). G-Rg2 and G-Rh1 (purity 99%) were purchased from Ace EMzyme (Anseong-si, Kyeonggi-do, Korea). In G-Rg2 and -Rh1, G-Rg2 and G-Rh1 were prepared by mixing in a 1:1 ratio, e.g., 10 μg/mL of the combination contained 5 μg/mL Rg2 and 5 μg/mL Rh1, and so forth.
Cell culture
HepG2 cell line was purchased from Korea Cell Line Bank (KCLB, #88065, Seoul, Korea). The cells were maintained by DMEM including 10% FBS, 100 U/mL penicillin and streptomycin at 37◦C in humidified atmosphere 5% CO2 (HERAcell 150i, hermo Electron Corp., Waltham, MA, USA).
Proliferation assay
Proliferation of HepG2 cells was determined using a Muse®Ki67 Proliferation Kit (Merck Millipore, Billerica, MA, USA). HepG2 cells were treated with various concentrations of G-Rg2 and -Rh1 for 24 h. After treatment, cells were collected by trypsinization, fixed, permeabilized and labelled with Muse® human Ki-67-PE or Muse® human IgG1-PE antibody. Afterwards, the cells were detected by Muse® Cell Analyzer and the data was analyzed with Muse® 1.5 analysis software.
Measurement of reactive oxygen species
Intercellular ROS was measured by using DCF-DA. HepG2 cells were seeded into 96-well plates at the 1 × 104 cells/well overnight. After starvation for 4 h, HepG2 cells were pretreated with various concentrations of G-Rg2 and -Rh1 for 3 h prior to 500 ng/mL LPS treatment for 24 h. Next, cells were incubated with 10 µM DCF-DA solution for 30 min at 37 °C. After labeling with DCF-DA, residual DCF-DA was removed and cells were washed with PBS and added 100 µl PBS for measuring fluorescence intensity of DCF at an excitation wavelength 485 nm and emission wavelength 535 nm using the fluorescence microplate reader (TECAN, Männedorf, Switzerland).
Western blot analysis
The cells were seeded into 12-well plates. After the starvation with FBS-free medium for 4 h, cells were pretreated with various concentrations of G-Rg2 and -Rh1 followed by LPS treatment depending on different experiments. After treatment, cells were lysed using 2X SDS lysis buffer (1 M Tris–HCl (pH 7.4), 25% Glycerol, 10% SDS, 5% 2-mercaptoethnol and 1% bromphenol blue). Cell lysates were boiled at 95 °C for 10 min and afterward resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto a nitrocellulose membrane. Then, the membranes were blocked for 1 h at room temperature (RT) using 5% skim milk, incubated with primary antibodies at 4 °C overnight, and then incubated with secondary antibodies for 90 min at RT. After being washed 5 times with PBS, proteins on membranes were visualized by using the enhanced chemiluminescence detection reagents (Amersham Pharmacia Biotech, Piscataway, NJ, USA).
Real-time quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
After treatment, total RNA was extracted from cells using Tri-RNA reagent according to the manufacturer’s instructions (Favorgen, Pingtung, China). Total RNA (1 µg) was reverse transcribed into single-stranded cDNA using the reverse transcription 5X master mix. The mRNA expression of the genes was quantified with iQ™ SYBR green supermix (Bio-rad Inc., Hércules, CA, USA) using a CFX Connect™ (Bio-rad Inc.). The 2−∆∆Ct method was used to calculate the fold-change in genes expression (GAPDH was used as the internal control). The following primers and probes were used: TNF-α (mouse), forward-5′-GCCTCTTCTCATTCCTGCTTG-3′ and reverse-5′-CTGATGAGAGGGAGGCCATT-3′; iNOS (mouse), forward-5′-TTGGAGCGAGTTGTGGATTTG-3′ and reverse-5′-GTAGGTGAGGGCTTGGCTGA-3′; IL-1β (mouse), forward-5′-AACCTGCTGGTGTGTGACGTTC-3′ and reverse-5′- CAGCACGAGGCTTTTTTGTTGT-3′; GAPDH (mouse), forward-5′-AACGTCATCCCAGAGCTGAA-3′ and reverse-5′-CTGCTTCACCACCTTCTTGA-3′; SOD (human), forward-5′-TGGAGATAATACAGCAGGCT-3′ and reverse-5′-AGTCACATTGCCCAAGTCTC-3′; GPx-1 (human), forward-5′-AGAAGTGCGAGGTGAACGGT-3′ and reverse-5′-CCCACCAGGAACTTCTCAAA-3′; β-actin (human), forward-5′-TCCACCTTCCAGCAGATGTG-3′ and reverse-5′-GCATTTGCGGTGGACGAT-3’.
Mitochondrial membrane potential assay
JC-1 is a cationic dye, expressing the fluorescence emission in two types, including red (J aggregates) and green (J monomers). The decrease in red intensity and increase in green intensity exhibit the loss of mitochondrial membrane potential. For this experiment, HepG2 cells were seeded on coverslips in 12-well plates overnight at 37 °C and 5% CO2. After reaching 80% confluence and starvation with FBS-free medium for 4 h, HepG2 cells were treated with G-Rg2 and -Rh1 (10, 25, 50 µg/mL) for 3 h followed by stimulation with LPS (1 µg/mL) for 12 h. 2 µM Dex was used as a positive control. Then, the cells were proceeded to incubation with 10 µg/mL JC-1 for 20 min at 37 °C and 5% CO2. The cells were then observed under a laser scanning confocal spectral microscope (K1-Fluo, Nanoscope systems, Daejeon, Korea).
Luciferase reporter gene assay
Luciferase reporter gene assay was performed as described previously (Huynh et al. 2020b). Briefly, HepG2 cells seeded in 12-well plates were co-transfected with pARE and pRL-TK (normalization control) reporter gene. Then, the cells were pretreated with different concentrations of G-Rg2 and -Rh1 for 3 h followed by treatment with or without LPS (500 ng/mL) for 12 h. ARE promoter activity was measured with dual-luciferase kit (E1960, Promega) using a Glomax 20/20 luminometer (Promega BioSystems Sunnyvale, Inc., USA).
Preparation of cytosolic and nuclear fractions
The HepG2 cells were pretreated with G-Rg2 and -Rh1 (25, 50 µg/mL) for 3 h followed by LPS (500 ng/mL) or H2O2 (200 µM) for 24 h. Nuclear proteins were prepared as previously described (Huynh et al. 2020a). The nuclear extracts were subjected to western blot analysis.
Immunofluorescence staining
Immunofluorescence staining was performed as described previously (Hai et al. 2019). HepG2 cells were seeded on coverslips in 12-well plates overnight at 37 °C and 5% CO2. After treatment, the cells were fixed with 4% paraformaldehyde for 15 min at RT, washed 3 times using PBS, then permeated with 0.2% Triton X-100 for 10 min, and further washed 2 times with PBS. After that, the cells were blocked with 3% BSA for 30 min at RT before proceeding incubation with first antibody for 2 h at RT. The processed cells were incubated with fluorescent secondary antibodies for 1 h at RT followed by DAPI staining for nuclear detection. Subsequently, the cells were observed using a laser scanning confocal spectral microscope (K1-Fluo, Nanoscope systems, Daejeon, Korea). Image J software was used for quantitative analysis.
Animal experiments
ICR mice (male) were obtained from Samtako (Osan, Korea). The mice were acclimated for a week before the experiments and fed with standard mice feed under the 23 ± 2 °C, 12:12 dark to light conditions. All animal studies were performed with the approval from Institutional Animal Care and Use committee of Chungnam National University (201912-CNU-191). Mice were randomly divided into 3 groups (group 1: control group, group 2: 10 mg/kg LPS, group 3: 20 mg/kg G-Rg2 and -Rh1 + 10 mg/kg LPS for 24 h, n = 5 in each group). Mice were administered G-Rg2 and -Rh1 at a dosage of 20 mg/kg for 24 h by intraperitoneal injection except groups 1 and 2. After 24 h, the mice of groups 2 and 3 were intraperitoneal injected with 10 mg/kg LPS for 24 h. Then, the mice were sacrificed, and tissues were collected. Liver tissues were fixed with 4% paraformaldehyde and embedded in paraffin for histological analysis.
Isolation of peritoneal macrophages
ICR mice were intra-peritoneal injected with 4% thioglycolate broth as described previously (Huynh et al. 2020a). After 48 h, peritoneal macrophages in peritoneal cavity were collected using DMEM containing 10% FBS, 100 U/mL penicillin and streptomycin. After brief centrifuge, the pellets afterward were collected, suspended and seeded into 6-well plates overnight at 37 °C under 5% CO2 in a humidified incubator before treatment.
Histological analysis
3 μm thick sections of liver tissues were cut using automated rotary microtome and were stained with H&E stain to evaluate pathological changes in tissue. The sections were examined using the light microscope (Olympus IX71, Tokyo, Japan).
Immunohistochemistry (IHC) staining
IHC staining was performed as described previously (Huynh et al. 2020a). Briefly, 3 μm thick sections of liver tissue were deparaffinized and rehydrated prior to incubation with 1% hydrogen peroxide for 10 min. To avoid non-specific binding, sections were blocked with 5% BSA in 30 min. Then, the sections were incubated with primary antibodies at RT for 2 h followed by incubation in biotinylated secondary antibody for 1 h, the avidin–biotin complex (ABC) and the 3,3′-diaminobenzidine (DAB) chromogens reaction, respectively. To counterstain nuclear, hematoxylin was used. Next, sections were proceeded to dehydration before mounting. The sections were analyzed under the light microscope (Olympus IX71, Tokyo, Japan).
Statistical analysis
All experiments were performed independently at least 3 times. All data was presented using GraphPad Prism 5 (version 5.02, GraphPad Software Inc., San Diego, CA, USA) for statistical analysis. One-way analysis of variance (ANOVA) followed by a Bonferroni multiple comparison was used. All results were expressed as the mean ± SEM. A p value < 0.05 was considered significant.
Results
Effects of G-Rg2 and -Rh1 on cell proliferation and STAT3 and TAK1 signaling pathways in HepG2 cells
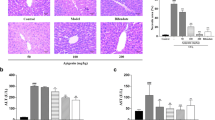
To choose appropriate concentrations of ginsenoside for treatment condition, we first investigated cytotoxicity of G-Rg2 and -Rh1 in HepG2 cells via Ki67 proliferation assay. The cells were treated with various concentrations of G-Rg2 and -Rh1 (0, 25, 50, and 100 µg/mL) for 24 h. Ki67 is a nuclear protein which is absent in phage G0 and presenting in phages G1, S, G2 and mitosis, indicating for proliferating cells (Scholzen and Gerdes 2000). As shown in Fig. 1a and b, there were no significant changes in cell proliferation and cytotoxicity by treatment with G-Rg2 and -Rh1 up to 100 µg/mL in HepG2 cell line. Therefore, we chose the concentration of G-Rg2 and -Rh1 at 50 µg/mL for following experiments. Next, we investigated effects of G-Rg2 and -Rh1 on key signaling pathways of inflammation and oxidative stress including STAT3 and TAK1 and its downstream targets, MAPKs in HepG2 cells. We found that ginsenoside treatment inhibited phosphorylation of STAT3, TAK1 and JNK in a dose-dependent manner (Fig. 1c). However, there were no remarkable changes in phosphorylation of p38 and ERK1/2 (Fig. 1c). We further examined above proteins under LPS stimulation in HepG2 cells. As shown in Fig. 1d and e, a dose-dependent downregulations of LPS-induced phosphorylation against STAT3, TAK1 and JNK were observed in ginsenoside-treated groups.
Cytotoxicity on HepG2 cells demonstrated by cell proliferation and inhibitory effect of ginsenoside Rg2 and Rh1 combination (G-Rg2 and -Rh1) on STAT3 and TAK1 signaling pathways. a Histogram of Ki67 expression in HepG2 cells treated with different concentrations of G-Rg2 and -Rh1 for 24 h. Ki67-positive cells were assessed using Muse® Ki67 proliferation assay, indicating for proliferating cells. b Quantified bar graph indicates Ki67 expression in ginsenoside-treated groups compared to the vehicle group. Data shown are representative of two independent experiments. c-d Expression of phosphorylated and total protein was analyzed by western blotting. HepG2 were treated with various concentrations G-Rg2 and -Rh1 for 30 min followed by treatment with or without lipopolysaccharide (LPS) (500 ng/mL) for 30 min. e Quantitative data of the relative protein expression levels of the phosphorylated form to the total form. Data represents the mean ± SEM of two independent experiments. ***p < 0.001 versus control, ##p < 0.01 and ###p < 0.01 compared to LPS-treated group
Effect of G-Rg2 and -Rh1 on LPS-induced inflammation in peritoneal macrophages
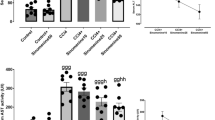
Since peritoneal macrophages are involved in systemic inflammation, we further investigated anti-inflammatory effects of G-Rg2 and -Rh1 on these cells stimulated with LPS. Peritoneal macrophages were pretreated with various concentrations of G-Rg2 and -Rh1 for 2 h and then treated with or without LPS (200 ng/mL) for 3 h. The phosphorylation of STAT3 and TAK1 was significantly increased in LPS-stimulated peritoneal macrophages, whereas G-Rg2 and -Rh1 pretreatment markedly reduced the LPS-induced phosphorylation of STAT3 and TAK1 in a dose-dependent manner (Fig. 2a, b). Furthermore, effect of ginsenoside on major inflammatory transcriptional factor, NF-κB activation was examined and we found that LPS-stimulated NF-κB phosphorylation was strongly suppressed by the ginsenoside treatment (Fig. 2c). We next examined inflammatory mediators, including iNOS, TNF-α and IL-1β, using western blot analysis (Fig. 2d) and qRT-PCR (Fig. 2e-g). Consistently, ginsenoside treatment exhibited the significant inhibition in LPS-triggered production of iNOS, TNF-α and IL-1β in terms of both protein expressions (Fig. 2d) and mRNA levels (Fig. 2e–g). Taken together, these results implied that inflammation in peritoneal macrophages induced by LPS treatment significantly inhibited by G-Rg2 and -Rh1, which in turn contributes liver inflammation.
Inhibitory effect of G-Rg2 and -Rh1 on LPS-induced inflammation in peritoneal macrophages. Peritoneal macrophages were pretreated with various concentrations of G-Rg2 and -Rh1 for 2 h and then treated with or without LPS (200 ng/mL) for indicated times in each figure (a-g). a Inflammatory makers (phosphorylated and total forms of STAT3 and TAK1) were evaluated by western blot analysis. b Quantitative data of the relative protein expression levels of the phosphorylated form to the total form. c Phosphorylated form of NF-κB p65 was determined by western blot analysis. d Expression of inflammatory mediators including iNOS, TNF-α, and cytokine IL-1β determined by western blot analysis. e–g Cellular mRNAs of iNOS, TNF-α and IL-1β were analyzed by quantitative reverse transcription-polymerase chain reaction. GAPDH was used as the housekeeping gene. Each value of the ratio of mRNA represents the mean ± SEM of two independent experiments. *p < 0.05, ***p < 0.001 versus control group, #p < 0.05, ##p < 0.01 compared to LPS-treated group
Effects of G-Rg2 and -Rh1 on NF-κB inflammatory signaling pathway in HepG2 cells
Since TAK1 and STAT3 activated inflammatory signaling pathway is associated with activation of NF-κB, leading to the subsequent inflammation (Irie et al. 2000; Liu et al. 2018), the effects of G-Rg2 and -Rh1 on NF-κB p65 activity were elucidated by immunofluorescence staining in HepG2 cells. LPS treatment significantly increased NF-κB p65 in both cytosol and nuclear compared with untreated group, whereas it was effectively suppressed by G-Rg2 and -Rh1 treatment (Fig. 3a, b).
Effects of G-Rg2 and -Rh1 on NF-κB inflammatory signaling pathway in HepG2 cells. a Immunofluorescence analysis was performed after fixing and immunostaining with NF-kB p65 and α-tubulin. DAPI was co-stained for visualizing nucleus. Images are shown using a laser scanning confocal spectral microscope. Scale bar indicates 30 μm. b Nuclear NF-κB p65 was quantified in the bar graph using Image J software. Data are represented as mean ± SEM (n = 6). **p < 0.01 versus vehicle group, #p < 0.05 and ##p < 0.01 compared to LPS-treated group
Effect of G-Rg2 and -Rh1 on LPS-induced mitochondrial dysfunction in HepG2 cells
Mitochondrial dysfunction is involved in excessive levels of ROS, leading to liver disorders (Hassanein and Frederick 2004). Thus, we further investigated the protective effect of G-Rg2 and -Rh1 on mitochondria under LPS stimulation. HepG2 cells were pretreated with different concentrations of G-Rg2 and -Rh1 for 3 h followed by stimulation with 1 µg/mL LPS for 12 h. Dex (2 µM) was used as a positive control. We found that LPS treatment resulted in the remarkable shift of fluorescence emission from red (aggregates) to green (monomers) in the comparison with control (p < 0.001), which indicated for a decline in mitochondrial membrane potential (Fig. 4a, b). Interestingly, the treatment with G-Rg2 and -Rh1 significantly rescued the loss of mitochondrial membrane potential trigged by LPS in a dose-dependent manner (Fig. 4a, b). These results suggest that the combination of G-Rg2 and -Rh1 possesses the positive effect in preserving mitochondrial function impaired by LPS in HepG2 cells.
G-Rg2 and -Rh1 protects HepG2 cells against LPS-induced mitochondrial dysfunction. HepG2 cells were treated with different concentrations of G-Rg2 and -Rh1 for 3 h followed by stimulation with LPS (1 µg/mL) for 12 h. a Mitochondrial membrane potential was determined with JC-1 staining using confocal microscopy. The fluorescence emission turns from red (JC-1 aggregates) to green (JC-1 monomers) exhibits for mitochondrial membrane damages. Dexamethasone at 2 µM (Dex) was used as a positive control. b Quantitative analysis of red/green fluorescent intensity using Image J software. Data are represented as mean ± SEM (n = 6). ***p < 0.001 compared with control sample, #p < 0.05 and ##p < 0.01 compared to LPS-treated group
Effect of G-Rg2 and -Rh1 treatment on ROS production and Nrf2/ARE anti-oxidant pathway under LPS stimulation in HepG2 cells
ROS are associated with inflammation formation, which in turn leads to liver injury (Jaeschke 2000). On the other hand, under physiological conditions, antioxidant systems play a pivotal role in maintain redox status balance by scavenging excessive ROS (Nguyen et al. 2009). To investigate anti-oxidant effects of G-Rg2 and -Rh1, we first explored effect of G-Rg2 and -Rh1 on LPS-induced ROS production using DCF-DA assay in HepG2 cells. We found that LPS-treated group significantly increases ROS level compared with the control (p < 0.001), while pretreatment with G-Rg2 and -Rh1 at 10, 25 and 50 µg/mL effectively inhibited LPS-induced ROS generation (Fig. 5a). Next, to clarify changes in antioxidant systems, we evaluated ARE promotor activity and Nrf2 expression. The results of ARE-luciferase assay showed that G-Rg2 and -Rh1 dose-dependently elevated ARE promotor activity in HepG2 cells (Fig. 5b) as well as recovered LPS-decreased ARE activity in the cells (Fig. 5c). Western blot analysis and immunofluorescence staining also demonstrated that G-Rg2 and -Rh1 restored Nrf2 nuclear translocation diminished by LPS (500 ng/mL) or H2O2 (200 µM)-induced ROS stimulation (Fig. 5d–g). Since Nrf2 is a transcription factor that regulates the expression of antioxidant genes, we next investigated the effects of G-Rg2 and -Rh1 on mRNA expression of antioxidant genes including SOD and GPx-1. We found that G-Rg2 and -Rh1 treatment significantly recovered LPS-induced suppression of SOD and GPx-1 (Fig. 5h, i). It suggests that G-Rg2 and -Rh1 enhances antioxidant genes via up-regulating ARE-Nrf2 transcriptional activity. Collectively, our findings demonstrate that G-Rg2 and -Rh1 exerts potential effects in balancing ROS levels by enhancing Nrf2/ARE antioxidant defense.
Effect of G-Rg2 and -Rh1 treatment on ROS production and Nrf2/ARE anti-oxidant pathway under LPS stimulation in HepG2. a Measurement of ROS using DCF-DA assay in HepG2 cells pretreated with various concentrations of G-Rg2 and -Rh1 for 3 h prior to 500 ng/mL LPS treatment for 24 h. b-c HepG2 cells co-transfected with pARE and pRL-TK (normalization control) reporter gene 24 h, were pretreated with G-Rg2 and -Rh1 for 3 h and then treated with or without LPS (500 ng/mL) for 12 h. ARE promoter activity was measured by luciferase assay. Data are represented as mean ± SEM of three experiments. d HepG2 cells were pretreated with G-Rg2 and -Rh1 for 3 h prior to 500 ng/mL LPS treatment for 24 h. Nuclear proteins extracted were subjected to western blotting. Histone deacetylase 1 (HDAC1) was used as loading control for nuclear proteins. e HepG2 cells were pretreated with G-Rg2 and -Rh1 for 3 h followed by 200 µM H2O2 treatment for 24 h. Nuclear proteins extracted were subjected to western blotting. HDAC1 was used as loading control for nuclear proteins. f Nrf2 translocation examined using immunofluorescence analysis. HepG2 cells were pretreated with G-Rg2 and -Rh1 (25 µg/mL) for 3 h prior to LPS (1 µg/mL) treatment for 12 h. Immunofluorescence analysis was performed after fixing and immunostaining with Nrf2 and α-tubulin. DAPI was co-stained for visualizing nucleus. Images were shown using a laser scanning confocal spectral microscope. Scale bar indicates 30 μm. g Nuclear Nrf2 was quantified in the bar graph using Image J software. Data are represented as mean ± SEM (n = 6). (H-I) HepG2 cells were treated with various concentrations of G-Rg2 and -Rh1 (10, 25 µg/mL) for 3 h followed by LPS (1 µg/mL) treatment for 12 h. Cellular mRNA levels of SOD and GPx from HepG2 cells was analyzed by qRT-PCR reaction. β-actin was used as the housekeeping gene. Each value of the ratio of mRNA represents the mean ± SEM of two independent experiments. *p < 0.05, **p < 0.01 and ***p < 0.001 versus control group, #p < 0.05, ##p < 0.01 and ###p < 0.01 compared to LPS-treated group
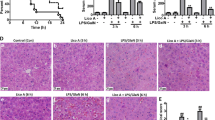
Protective effect of G-Rg2 and -Rh1 in LPS-induced liver damage
We used the acute peritonitis mouse model following LPS (10 mg/kg) injection intraperitoneally as indicated in Fig. 6a. H&E data showed that LPS treatment significantly induced liver injury compared to the control, showing through increased appearances of immune cell infiltration and vacuolization. Intriguingly, these damages were dramatically suppressed in the G-Rg2 and -Rh1-treated group (Fig. 6b, top panel and Fig. 6c). Since inflammation results in liver damages in these animal models, IHC staining for CD45 and Nrf2 was conducted to determine anti-inflammatory and antioxidant effects of G-Rg2 and -Rh1. As seen in Fig. 6b (middle panel) and 6E, the increment of intensity of CD45 in the LPS group markedly decreased by G-Rg2 and -Rh1 treatment compared to control group. In addition, G-Rg2 and -Rh1 treated group altered LPS-inhibited Nrf2 expression compared to liver tissue of control group (Fig. 6b, bottom panel and 6d). Altogether, G-Rg2 and -Rh1 administration are able to inhibit LPS-induced liver injury.
Protective effect of G-Rg2 and -Rh1 in LPS-induced liver damage. a Animal experimental scheme. b-c Liver tissue was collected from mice applied intraperitoneal injection of G-Rg2 and -Rh1 (20 mg/kg) for 24 h followed by with or without LPS (10 mg/kg) treatment 24 h. b H&E and IHC staining of Nrf2 and CD45 in the liver sections. Scale bar indicates 100 µm. c Quantified bar graph indicates tubular injury score of liver tissues treated with control, LPS, and G-Rg2 and Rh1. d-e Quantified bar graph indicates CD45 (d) and Nrf2 (e) expression in liver tissues. ***p < 0.001 versus control group, ###p < 0.001 compared to LPS-treated group
Discussion
Excessive accumulation of ROS and inflammatory responses play a vital role in liver dysfunction. Various natural compounds have been reported to exert protective effect on liver due to their anti-inflammatory and anti-oxidant effects (Qi et al. 2017; Jiang et al. 2018; Yang et al. 2018). Furthermore, combination of natural compounds is a potential strategy that can facilitate a high efficiency with a lower dosage to reduce drug toxicity.
In previous studies, G-Rg2 has been found to have antidepressant-like and anti-adipogenesis effects and reduce hepatic glucose production in HepG2 cells (Yuan et al. 2012; Ren et al. 2017; Liu et al. 2019). We also previously demonstrated that G-Rg2 and -Rh1 displayed synergistically the anti-inflammatory effect on LPS-stimulated macrophages as well as kidney and liver injury through suppressing the expression of pro-inflammatory cytokines and mediators via the inhibition of TLR4/STAT1 signaling pathway. In this study, we extended the investigation of this combination effect on liver function. Subsequently, the results revealed that G-Rg2 and -Rh1 repressed inflammatory response not only by inhibiting the inflammatory pathway, but also by upregulating the Nrf2 anti-oxidant signaling pathways in macrophages and liver cells, which consequently exerted the protective effect on liver function under the stimulation of LPS.
Previous studies showed that TAK1 activation is associated with LPS-induced NF-κB activation in macrophages, leading to the subsequent inflammation (Irie et al. 2000). In the current study, LPS treatment induced TAK1 activation and increased protein and gene expressions of pro-inflammatory cytokines (TNF-α and IL-1β) and iNOS associated with activation of NF-κB transcription factor. However, G-Rg2 and -Rh1 suppressed all these effects, and thereby, suppressed the inflammatory responses in LPS-treated HepG2 cells and peritoneal macrophages (Figs. 1, 2, 3). These results suggest that one of the mechanisms by which G-Rg2 and -Rh1 alleviate inflammation in LPS-treated cells is to reduce the transcriptional activity of NF-κB through suppressing the activation of TAK1.
In addition to TAK1 activation, STAT3, a member of the STAT family, plays a vital role in inflammation (Walker and Frank 2012; Villarino et al. 2015). LPS-induced JAK/STAT3 activation leads to the activation of the downstream NF-κB involved in transcription of pro-inflammatory genes (Liu et al. 2018). In our study, as shown in Fig. 2a, G-Rg2 and -Rh1 inhibited the phosphorylation of STAT3 in peritoneal macrophages. In addition to the inhibitory effect on STAT1 activity as we demonstrated in previous study, G-Rg2 and -Rh1 also suppressed LPS-induced STAT3 activation, which is possibly a potential mechanism to protect liver from LPS-induced systemic inflammation.
Since oxidative stress is highly linked to inflammatory process, we examined the effects of the combination on ROS levels in HepG2 stimulated with LPS. Interestingly, G-Rg2 and -Rh1 reduced LPS-stimulated ROS accumulation and restored LPS-impaired mitochondrial function (Fig. 4, 5a). Redox homeostasis is closely linked to mitochondrial function (Qi et al. 2017). Notably, antioxidant treatment has been shown to recover impaired mitochondrial function in a sepsis model (Zapelini et al. 2008). Consistently, G-Rg2 and -Rh1 was found to increase ARE promoter activities and protein expression of Nrf2 in a dose-dependent manner (Fig. 5). These results indicate that in addition to the anti-inflammatory effect, G-Rg2 and -Rh1 the combination also exerted the antioxidant activity, which may further enhance the inhibitory effect of the combination on inflammation progression.
Excessive ROS production is closely associated with cell death, which contributes to the progression of inflammation (Jaeschke 2000). In the present study, G-Rg2 and -Rh1 reduced LPS-increased ROS generation, and thereby supported liver cell survival, decreasing the inflammatory responses in the cells. Consistent to the in vitro results, H&E and IHC staining of liver tissues showed G-Rg2 and -Rh1 treatment had a recovery effect on LPS-induced pathological changes and immune cell infiltration accompanied by Nrf2 expression (Figs. 6, 7). Altogether, these findings indicate that maintaining the balance of redox status in liver cells plays an important role in protecting the liver from the inflammatory stimuli, and the compounds with anti-oxidant properties serve as potential candidates for this issue.
A schematic diagram representing the signaling pathways involved in antioxidant and anti-inflammatory effects of G-Rg2 and -Rh1 in HepG2 cells. The combination of these ginsonosides impaired the binding of LPS to TLR4, leading to downregulation of the TLR4/TAK1 signaling pathway. In general, the phosphorylation of TAK1 results in activation and nuclear translocation of NF-κB, which in turn produces the transcription of inflammatory cytokines and mediators such as TNF-α, IL-1β and iNOS. In addition to TAK1 activation, LPS up-regulates the phosphorylation of STAT3, leading to increase inflammation through nuclear translocation of NF-κB. TAK1 activation also induces phosphorylation of JNK, causing reduction of nuclear accumulation of Nrf2 and the activity of ARE promotor. As a result, the productions of antioxidant enzymes such as NQO-1, GPx-1, SOD are diminished, resulting in an increase of intracellular ROS levels. In this study, G-Rg2 and -Rh1 not only decreased inflammatory cytokines and mediators, but also effectively inhibited mitochondrial damage-induced ROS production. LPS lipopolysaccharide, TLR4 toll-like receptor 4, ROS reactive oxygen species, TAK1 transforming growth factor β-activated kinase 1, NF-κB nuclear factor-kappa B, STAT3 signal transducer and activator of transcription 3, TNF-α tumor necrosis factor-alpha, iNOS inducible nitric oxide synthase, IL-1β interleukin-1 beta, JNK c-Jun N-terminal kinase, Nrf2 nuclear factor-erythroid 2-related factor 2, ARE antioxidant response element, NQO-1 NAD(P)H quinone oxidoreductase-1, SOD superoxide dismutase, GPx-1 glutathione peroxidase-1, P phosphorylation
In conclusion, our study illustrated that G-Rg2 and -Rh1 exerted a protective effect on liver function under LPS challenging by increasing anti-inflammatory effect and antioxidant activity through the STAT3/TAK1-mediated NF-κB and Nrf2-ARE signaling pathways in macrophages and liver cells.
References
Ahmed SM, Luo L, Namani A, Wang XJ, Tang X (2017) Nrf2 signaling pathway: pivotal roles in inflammation. Biochim Biophys Acta Mol Basis Dis 1863:585–597. https://doi.org/10.1016/j.bbadis.2016.11.005
Baatar D, Siddiqi MZ, Im WT, Ul Khaliq N, Hwang SG (2018) Anti-inflammatory effect of ginsenoside Rh2-Mix on lipopolysaccharide-stimulated RAW 264.7 murine macrophage cells. J Med Food 21:951–960. https://doi.org/10.1089/jmf.2018.4180
Bak MJ, Jun M, Jeong WS (2012) Antioxidant and hepatoprotective effects of the red ginseng essential oil in H(2)O(2)-treated hepG2 cells and CCl(4)-treated mice. Int J Mol Sci 13:2314–2330. https://doi.org/10.3390/ijms13022314
Cecconi M, Evans L, Levy M, Rhodes A (2018) Sepsis and septic shock. Lancet 392:75–87. https://doi.org/10.1016/S0140-6736(18)30696-2
Hai Y, Wang X, Song P, Li JY, Zhao LH, Xie F, Tan XM, Xie QJ, Yu L, Li Y, Wu ZR, Li HY (2019) Realgar transforming solution-induced differentiation of NB4 cell by the degradation of PML/RARalpha partially through the ubiquitin-proteasome pathway. Arch Pharm Res 42:684–694. https://doi.org/10.1007/s12272-019-01170-9
Hassanein T, Frederick T (2004) Mitochondrial dysfunction in liver disease and organ transplantation. Mitochondrion 4:609–620. https://doi.org/10.1016/j.mito.2004.07.015
Huynh DTN, Baek N, Sim S, Myung CS, Heo KS (2020a) Minor ginsenoside Rg2 and Rh1 attenuates LPS- via downregulating induced acute liver and kidney damages activation of TLR4-STAT1 and inflammatory cytokine production in macrophages. Int J Mol Sci 21:6656. https://doi.org/10.3390/ijms21186656
Huynh DTN, Jin Y, Myung CS, Heo KS (2020b) Inhibition of p90RSK is critical to abolish Angiotensin II-induced rat aortic smooth muscle cell proliferation and migration. Biochem Biophys Res Commun 523:267–273. https://doi.org/10.1016/j.bbrc.2019.12.053
Irie T, Muta T, Takeshige K (2000) TAK1 mediates an activation signal from toll-like receptor(s) to nuclear factor-kappaB in lipopolysaccharide-stimulated macrophages. FEBS Lett 467:160–164. https://doi.org/10.1016/s0014-5793(00)01146-7
Jaeschke H (2000) Reactive oxygen and mechanisms of inflammatory liver injury. J Gastroenterol Hepatol 15:718–724. https://doi.org/10.1046/j.1440-1746.2000.02207.x
Jiang Z, Meng Y, Bo L, Wang C, Bian J, Deng X (2018) Sophocarpine attenuates LPS-induced liver injury and improves survival of mice through suppressing oxidative stress, inflammation, and apoptosis. Mediat Inflamm 2018:5871431. https://doi.org/10.1155/2018/5871431
Jin Y, Huynh DTN, Nguyen TLL, Jeon H, Heo KS (2020) Therapeutic effects of ginsenosides on breast cancer growth and metastasis. Arch Pharm Res 43:773–787. https://doi.org/10.1007/s12272-020-01265-8
Kim DH (2018) Gut microbiota-mediated pharmacokinetics of ginseng saponins. J Ginseng Res 42:255–263. https://doi.org/10.1016/j.jgr.2017.04.011
Lee J, Park JS, Roh YS (2019) Molecular insights into the role of mitochondria in non-alcoholic fatty liver disease. Arch Pharm Res 42:935–946. https://doi.org/10.1007/s12272-019-01178-1
Lee SB, Lee WS, Shin JS, Jang DS, Lee KT (2017) Xanthotoxin suppresses LPS-induced expression of iNOS, COX-2, TNF-alpha, and IL-6 via AP-1, NF-kappaB, and JAK-STAT inactivation in RAW 264.7 macrophages. Int Immunopharmacol 49:21–29. https://doi.org/10.1016/j.intimp.2017.05.021
Li S, Hong M, Tan HY, Wang N, Feng Y (2016) Insights into the role and interdependence of oxidative stress and inflammation in liver diseases. Oxid Med Cell Longev 2016:4234061. https://doi.org/10.1155/2016/4234061
Liu H, Liu M, Jin Z, Yaqoob S, Zheng M, Cai D, Liu J, Guo S (2019) Ginsenoside Rg2 inhibits adipogenesis in 3T3-L1 preadipocytes and suppresses obesity in high-fat-diet-induced obese mice through the AMPK pathway. Food Funct 10:3603–3614. https://doi.org/10.1039/c9fo00027e
Liu X, Yin S, Chen Y, Wu Y, Zheng W, Dong H, Bai Y, Qin Y, Li J, Feng S, Zhao P (2018) LPSinduced proinflammatory cytokine expression in human airway epithelial cells and macrophages via NFkappaB, STAT3 or AP1 activation. Mol Med Rep 17:5484–5491. https://doi.org/10.3892/mmr.2018.8542
Lugrin J, Rosenblatt-Velin N, Parapanov R, Liaudet L (2014) The role of oxidative stress during inflammatory processes. Biol Chem 395:203–230. https://doi.org/10.1515/hsz-2013-0241
Mihaly SR, Ninomiya-Tsuji J, Morioka S (2014) TAK1 control of cell death. Cell Death Differ 21:1667–1676. https://doi.org/10.1038/cdd.2014.123
Nesseler N, Launey Y, Aninat C, Morel F, Malledant Y, Seguin P (2012) Clinical review: the liver in sepsis. Crit Care 16:235. https://doi.org/10.1186/cc11381
Nguyen T, Nioi P, Pickett CB (2009) The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J Biol Chem 284:13291–13295. https://doi.org/10.1074/jbc.R900010200
Qi GY, Mi YS, Fan R, Li RN, Wang YW, Li XY, Huang SX, Liu XB (2017) Tea polyphenols ameliorate hydrogen peroxide-and constant darkness-triggered oxidative stress via modulating the Keap1/Nrf2 transcriptional signaling pathway in HepG2 cells and mice liver. RSC Adv 7:32198–32208. https://doi.org/10.1039/c7ra05000c
Quan LH, Min JW, Sathiyamoorthy S, Yang DU, Kim YJ, Yang DC (2012) Biotransformation of ginsenosides Re and Rg1 into ginsenosides Rg2 and Rh1 by recombinant beta-glucosidase. Biotechnol Lett 34:913–917. https://doi.org/10.1007/s10529-012-0849-z
Ren Y, Wang JL, Zhang X, Wang H, Ye Y, Song L, Wang YJ, Tu MJ, Wang WW, Yang L, Jiang B (2017) Antidepressant-like effects of ginsenoside Rg2 in a chronic mild stress model of depression. Brain Res Bull 134:211–219. https://doi.org/10.1016/j.brainresbull.2017.08.009
Scholzen T, Gerdes J (2000) The Ki-67 protein: from the known and the unknown. J Cell Physiol 182:311–322. https://doi.org/10.1002/(sici)1097-4652(200003)182:3%3c311::Aid-jcp1%3e3.0.Co;2-9
Taguchi K, Kensler TW (2020) Nrf2 in liver toxicology. Arch Pharm Res 43:337–349. https://doi.org/10.1007/s12272-019-01192-3
Villarino AV, Kanno Y, Ferdinand JR, O’Shea JJ (2015) Mechanisms of Jak/STAT signaling in immunity and disease. J Immunol 194:21–27. https://doi.org/10.4049/jimmunol.1401867
Walker SR, Frank DA (2012) Screening approaches to generating STAT inhibitors: allowing the hits to identify the targets. JAKSTAT 1:292–299. https://doi.org/10.4161/jkst.22662
Yang YQ, Yan XT, Wang K, Tian RM, Lu ZY, Wu LL, Xu HT, Wu YS, Liu XS, Mao W, Xu P, Liu B (2018) Triptriolide alleviates lipopolysaccharide-induced liver injury by Nrf2 and NF-kappaB signaling pathways. Front Pharmacol 9:999. https://doi.org/10.3389/fphar.2018.00999
Yuan HD, Kim DY, Quan HY, Kim SJ, Jung MS, Chung SH (2012) Ginsenoside Rg2 induces orphan nuclear receptor SHP gene expression and inactivates GSK3beta via AMP-activated protein kinase to inhibit hepatic glucose production in HepG2 cells. Chem Biol Interact 195:35–42. https://doi.org/10.1016/j.cbi.2011.10.006
Zapelini PH, Rezin GT, Cardoso MR, Ritter C, Klamt F, Moreira JC, Streck EL, Dal-Pizzol F (2008) Antioxidant treatment reverses mitochondrial dysfunction in a sepsis animal model. Mitochondrion 8:211–218. https://doi.org/10.1016/j.mito.2008.03.002
Acknowledgements
This research was supported by National Research Foundation of Korea (2019R1C1C100733112).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All the authors declared that they have no conflict of interest to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Nguyen, T.L.L., Huynh, D.T.N., Jin, Y. et al. Protective effects of ginsenoside-Rg2 and -Rh1 on liver function through inhibiting TAK1 and STAT3-mediated inflammatory activity and Nrf2/ARE-mediated antioxidant signaling pathway. Arch. Pharm. Res. 44, 241–252 (2021). https://doi.org/10.1007/s12272-020-01304-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12272-020-01304-4