Abstract
Cytochrome P450 (CYP) 2D6 is present in less than about 2% of all CYP enzymes in the liver, but it is involved in the metabolism of about 25% of currently used drugs. CYP2D6 is the most polymorphic among the CYP enzymes. We determined alleles and genotypes of CYP2D6 in 3417 Koreans, compared the frequencies of CYP2D6 alleles with other populations, and observed the differences in pharmacokinetics of metoprolol, a prototype CYP2D6 substrate, depending on CYP2D6 genotype. A total of 3417 unrelated healthy subjects were recruited for the genotyping of CYP2D6 gene. Among them, 42 subjects with different CYP2D6 genotypes were enrolled in the pharmacokinetic study of metoprolol. The functional allele *1 and *2 were present in frequencies of 34.6 and 11.8%, respectively. In decreased functional alleles, *10 was the most frequent with 46.2% and *41 allele was present in 1.4%. The nonfunctional alleles *5 and *14 were present at 4.5 and 0.5% frequency, respectively. The *X × N allele was present at a frequency of 1.0%. CYP2D6*1/*1, *1/*2 and *2/*2 genotypes with normal enzyme activity were present in 12.1%, 8.6% and 1.4% of the subjects, respectively. CYP2D6*5/*5, *5/*14, and *14/*14 genotypes classified as poor metabolizer were only present in 4, 2, and 1 subjects, respectively. Mutant genotypes with frequencies of more than 1% were CYP2D6*1/*10 (32.0%), *10/*10 (22.3%), *2/*10 (11.7%), *5/*10 (3.7%), *1/*5 (2.5%), and *10/*41 (1.2%). The relative clearance of metoprolol in CYP2D6*1/*10, *1/*5, *10/*10, *5/*10, and *5/*5 genotypes were 69%, 57%, 24%, 14% and 9% of CYP2D6*wt/*wt genotype, respectively. These results will be very useful in establishing a strategy for precision medicine related to the genetic polymorphism of CYP2D6.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cytochrome P450 (CYP) enzymes are classified into 10 classes according to the domain architecture and cellular location of CYP enzymes and redox proteins. According to amino acid sequence homology of CYP enzymes, there are 57 genes in CYP superfamily (Cook et al. 2016). The enzymatic activity of CYP enzymes can vary widely depending on the gene variation, and currently the variant alleles of 31 CYP enzymes are summarized in the Pharmacogene Variation Consortium (http://www.pharmvar.org/genes).
CYP2D6 is present in less than about 2% of all CYP450 enzymes in the liver, but it is involved in the metabolism of about 25% of currently used drugs (Ingelman-Sundberg 2005; Ingelman-Sundberg et al. 2007). To date, more than 113 different human CYP2D6 variant and subvariant alleles (CYP2D6*1B to *113) have been identified (http://www.pharmvar.org/gene/CYP2D6). However, at present only nine alleles constitute more than 95% of CYP2D6 diplotypes. Among them, *1 and *2 are fully functional alleles, *3, *4, *5, and *6 are nonfunctional alleles and *10, *17, and *41 are reduced functional alleles. These alleles show significant differences in distribution between race and ethnicity, and the CYP2D6*2, *5, and *10 alleles, along with the CYP2D6 gene duplication, are the most clinically important and widely distributed polymorphisms in East Asians. However, the distribution of these alleles in East Asians including Koreans, Chinese, and Japanese is somewhat different from one another (Roh et al. 2001; Hosono et al. 2009; Man et al. 2010; Park et al. 2011; Ota et al. 2015; Goh et al. 2017; Zhou et al. 2017), and the information on the enzymatic activity of each CYP2D6 genotype is insufficient. Accurate information on the distribution of alleles and genotypes of polymorphic drug metabolizing enzyme genes in a population and the enzymatic activity of each genotype is important to establish a strategy for precision medicine. Therefore, in this study, we determined alleles and genotypes of CYP2D6 in 3417 Koreans, compared the frequencies of CYP2D6 alleles with other populations, and observed the differences in pharmacokinetics of metoprolol, a prototype CYP2D6 substrate, depending on CYP2D6 genotype.
Materials and methods
Subjects
A total of 3417 unrelated healthy subjects were recruited for the genotyping of CYP2D6 gene. After genotyping, 42 of the recruited subjects with different CYP2D6 genotypes were enrolled in the pharmacokinetic study of metoprolol.
All study procedures were carried out in accordance with the recommendations of the Declaration of Helsinki on biomedical research involving human subjects, and the Institutional Review Board of Sungkyunkwan University, Suwon, Republic of Korea approved the research protocol. Written informed consent was obtained from all subjects.
Genotyping
Genomic DNA was isolated from peripheral blood leukocytes for genotyping of the CYP2D6 with a commercial blood kit (Wizard® Genomic DNA Purification Kit, Promega, Madison, WI, USA).
Analyses of the CYP2D6*2, *4, *5, *10, and *X × N alleles were performed using polymerase chain reaction restriction fragment length polymorphisms (PCR-RFLP) and long PCR analyses, as described previously (Byeon et al. 2015). The samples carrying the 2850C>T mutation were further genotyped for CYP2D6*14 and *41.
Genotyping of CYP2D6*14 was performed by PCR-RFLP method (Wang et al. 1999) with minor modifications. For the PCR amplification of the CYP2D6*14 allele the forward (5′-GTG GAT GGT GGG GCT AAT GCC TT-3′) and the reverse primer (5′-CAG AGA CTC CTC GGT CTC TCG CT-3′) were used. PCR cycling conditions were as follows: pre-denaturation at 94 °C for 5 min, 35 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s and extension at 72 °C for 30 s, followed by a final extension at 72 °C for 10 min. The PCR amplification was carried out using the T100 Thermal Cycler (Bio-Rad, Hercules, CA, USA). After amplification, the PCR product was digested with 0.25 µl of the specific restriction enzyme Msp I (New England Biolabs, Ipswich, MA, USA) and incubated at 37 °C for 1 h. Digested PCR products were analyzed by gel electrophoresis on 2% agarose gels and stained with ethidium bromide, then directly visualized under the UV light.
Genotyping of CYP2D6*41 was performed using direct sequencing. Therefore, the genomic DNA was amplified in T100 Thermal Cycler (Bio-Rad, Hercules, CA, USA) using the forward primer 5′-GTA CTT CGA TGT CAC GGG ATG-3′ and the reverse primer 5′-TGA CAG GTG CAG AAT TGG AG-3′. After initial denaturation at 94 °C for 5 min, denaturation at 94 °C for 1 min, annealing at 60 °C for 1 min and extension at 72 °C for 1 min were repeated 40 times, and then terminated with a final extension at 72 °C for 10 min. The amplicons were subsequently purified using LaboPass PCR Purification Kit (CosmoGenetech, Seoul, Korea) and afterwards sequenced on ABI 3730xl DNA Analyzer using BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA).
Pharmacokinetic study
Forty-two subjects with different CYP2D6 genotypes were enrolled. All subjects were in good health as determined by their medical histories, physical examinations, vital signs (blood pressure, pulse rate and body temperature) and routine laboratory tests (blood chemistry, hematology and urine analysis). For the study, subjects were not permitted to ingest any medication, alcohol or caffeine-containing beverages for 10 days prior to the study and during the study. All subjects were given identical meals and then fasted from 10 h before to 4 h after drug administration. Standard meals were served for lunch and dinner 4 and 10 h after drug administration, respectively. A single oral dose of 100 mg metoprolol tablet (Betaloc Tab., AstraZeneca Korea, Seoul, Korea) was administered with 240 ml of water to each subject. Blood samples (7 ml) were collected before and at 0.5, 1, 1.5, 2, 2.5, 3, 4, 6, 8, 10, 12 and 24 h after dosing, and plasma samples from all blood samples were immediately separated for the determination of the concentrations of metoprolol and stored at − 70 °C until the completion of the analysis.
Determination of metoprolol
Metoprolol in the plasma samples was determined by high-performance liquid chromatography (HPLC) system, which consisted of a Waters 515 HPLC pump, a Waters 717 plus autosampler and a Waters 474 scanning fluorescence detector (Ex = 280, Em = 300 nm) (Waters Corporation, Milford, MA, USA). The separation was performed on a Sunfire C18 column (5 μm, 4.6 i.d. × 250 mm, Waters Corporation, Milford, MA, USA) using 10 mM KH2PO4 (pH 3.0) containing 12% acetonitrile and 15% methanol at a flow rate of 1 ml/min. Fifty microliter of 5 μg/ml atenolol (as an internal standard) and 100 μl of 1 M NaOH were added to 0.5 ml of plasma sample. After brief vortex mixing, 6 ml of dichloromethane was added. The mixture was mixed for 30 s and centrifuged at 3500 rpm for 10 min. The organic layer was transferred to a 10 ml tube and evaporated to dryness under nitrogen stream in a 50 °C bath. The residue was dissolved in 300 μl of mobile phase and 70 μl aliquot was injected into the HPLC system.
Statistical analysis
Data were compiled according to the genotype and allele frequencies. The frequencies of each allele are reported with 95% confidence intervals. Hardy–Weinberg equilibrium was evaluated by comparing the genotype frequencies with the expected values using a contingency table χ2 test. Statistical significance was determined by the χ2 test. The pharmacokinetic data are expressed as mean ± SD Statistical analysis for AUCinf, Cmax, and CL/F ratio among genotype groups were performed using one-way analysis of variance (ANOVA) with Bonferroni post hoc test or Kruskal–Wallis one-way ANOVA with Dunn’s post hoc test after normality test and equal variance test. All statistical analyses were carried out using the statistical program SigmaPlot 12.5® (Systat Software Inc., San Jose, CA, USA). P value less than 0.05 was considered statistically significant.
Results
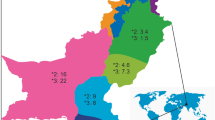
In this study, the frequency of CYP2D6 allele was measured in 3417 Korean subjects who were not related to each other. Then, these results were compared to the previous reports, and the frequency of major alleles of CYP2D6 measured in Koreans was compared with the results of previous studies that measured the frequencies of major CYP2D6 alleles in three East Asian populations (Korean, Japanese, and Chinese), Caucasians, and Africans.
CYP2D6 allele frequencies in Korean population
In 3417 Koreans, the functional allele *1 and *2 were present in frequencies of 34.6 and 11.8%, respectively. In the case of two decreased functional alleles, the CYP2D6*10 was the most frequent allele at 46.2% and the CYP2D6*41 allele was present in 1.4% of the subjects. The nonfunctional allele *5 and *14 were present at 4.5 and 0.5% frequency, respectively. The *X × N allele was present at a frequency of 1.0% (Table 1). The results of this study were similar to those of previous studies measured in Koreans (Lee et al. 2006, 2009; Man et al. 2010; Park et al. 2011, 2012).
Comparison of CYP2D6 allele frequencies among East Asian populations
Comparing the frequencies of major CYP2D6 alleles between Korean and Japanese populations, CYP2D6*1 frequency was lower in Koreans (34.6%) than in Japanese (43.6%) (P < 0.001), but CYP2D6*10 frequency was higher in Koreans (46.2%) than in Japanese (37.5%) (P < 0.001). CYP2D6*5 frequency was slightly lower in Koreans (4.5%) than in Japanese (5.5%) (P < 0.05). The frequencies of other alleles were not significantly different.
Comparing the frequencies of major CYP2D6 alleles between Korean and Chinese populations, CYP2D6*1 frequency was higher in Korean population (34.6% vs. 26.4%, P < 0.001), but the frequencies of CYP2D6*10 (46.2% vs. 52.5%, P < 0.001), *14 (0.5% vs. 1.2%, P < 0.001), and *41 (1.4% vs. 3.5%, P < 0.001) were lower in Korean population. The frequency of CYP2D6*X × N alleles was slightly lower in Koreans (1.0% vs. 2.3%, P < 0.001). The frequencies of other alleles were not significantly different.
The frequencies of CYP2D6*1 and *10 alleles showed remarkable differences among the three East Asian populations (Table 1).
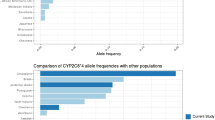
Comparison of CYP2D6 allele frequencies with Caucasian and African populations
In Caucasian population, the frequency of CYP2D6*1 allele was very similar to Japanese populations, but it was higher than Korean and Chinese populations (P < 0.001). The frequency of CYP2D6*2 allele was about twice higher in Caucasian population than in Koreans, Chinese and Japanese populations (P < 0.001). The frequency of CYP2D6*4 allele was very low in East Asian populations (0.3–0.5%), but in Caucasians, there was a very high frequency (19.1%, P < 0.001). The CYP2D6*5 allele was present in 4.5–5.5% of East Asian populations, but was present in 2.5% of Caucasian population (P < 0.001). CYP2D6*10 allele was present at a very high frequency of 37.5–52.5% in East Asian populations but very low (1.4%) in Caucasian population (P < 0.001). CYP2D6*14 and *17 existed at a very low frequency in East Asian and Caucasian populations. CYP2D6*41 was present in 1.4–3.5% of East Asian populations, but in 9.0% of Caucasian population (P < 0.001) (Table 1).
Compared to the African population, the frequency of CYP2D6*1 allele was higher in Japanese population, but lower in Korean and Chinese populations. The frequency of CYP2D6*2 allele was about twice higher in African population than in Koreans, Chinese and Japanese populations (P < 0.001). The frequency of CYP2D6*4 allele was higher in African population (5.1%) than in East Asian populations (0.3–0.5%, P < 0.001). The CYP2D6*5 allele frequency was similar between East Asian populations and African population. CYP2D6*10 allele was present at a very high frequency of 37.5–52.5% in East Asian populations but very low (4.2%) in African population (P < 0.001). CYP2D6*17 existed at a very low frequency in East Asian populations, but at higher frequency (20.5%) in African population (P < 0.001). CYP2D6*41 was present in 1.4–3.5% of East Asian populations, but in 6.0% of African population (P < 0.001) (Table 1).
Frequencies of CYP2D6 genotypes in Korean population
In 3417 Koreans, CYP2D6*1/*1, *1/*2 and *2/*2 genotypes with normal enzyme activity were present in 12.1% (CI 11.0–13.2%), 8.6% (CI 7.6–9.6%) and 1.4% (CI 1.0–1.8%), respectively. Number of subjects with CYP2D6*5/*5, *5/*14, and *14/*14 genotypes, who are classified as poor metabolizer (PM), were only 4, 2, and 1, respectively. Therefore, the frequency of CYP2D6 PM in Korean population was about 0.2%. Mutant genotypes with frequencies of more than 1% were CYP2D6*1/*10 (32.0%, CI 30.4–33.6%), *10/*10 (22.3%, CI 20.9–23.7%), *2/*10 (11.7%, CI 10.6–12.8%), *5/*10 (3.7%, CI 3.1–4.3%), *1/*5 (2.5%, CI 2.0–3.0%), and *10/*41 (1.2%, CI 0.8–1.6%). Any other genotypes were present in frequencies of less than 1% (Table 2).
Comparison of metabolic activity of CYP2D6 genotypes
The difference in the enzymatic activity of the CYP2D6 genotypes was observed by comparing the pharmacokinetic parameters of metoprolol, a prototype substrate drug of CYP2D6. Relative values of each genotype were calculated based on the average value of pharmacokinetic parameters of CYP2D6*wt/*wt (*wt = *1 or *2) genotype. The relative clearance of CYP2D6*1/*10, *1/*5, *10/*10, *5/*10, and *5/*5 genotypes were 69%, 57%, 24%, 14% and 9% of CYP2D6*wt/*wt genotype, respectively. The relative Cmax of CYP2D6*1/*10, *1/*5, *10/*10, *5/*10, and *5/*5 genotypes were 122%, 152%, 221%, 349% and 341% of CYP2D6*wt/*wt genotype, respectively. The relative AUCinf of CYP2D6*1/*10, *1/*5, *10/*10, *5/*10, and *5/*5 genotypes were 139%, 163%, 388%, 651% and 1062% of CYP2D6*wt/*wt genotype, respectively (Table 3, Fig. 1).
Individual clearance (a), AUCinf (b), and Cmax (c) ratio of metoprolol in subjects with different CYP2D6 genotypes. Individual ratio values were calculated as individual parameter value/mean of parameter values in CYP2D6*wt/*wt (*wt = *1 or *2) genotype. Each horizontal line indicates the mean of individual ratios
Discussion
When medication is administered, some patients do not respond to the drug, while some patients have serious adverse drug reactions. The proportion of these patients ranges from 40% to 70% depending on the drug (Eichelbaum et al. 2006; Lauschke and Ingelman-Sundberg 2016). 15–30% of these individual drug reactions are known to be caused by polymorphisms of genes encoding proteins that affect drug response such as drug metabolizing enzymes and drug transporters (Eichelbaum et al. 2006). In particular, the genetic polymorphisms of CYP enzymes have the greatest effect on individual differences in drug response, and among CYP enzymes, CYP2D6 is the most polymorphic enzyme (https://www.pharmvar.org/gene/CYP2D6).
This study reported the allele and genotype frequencies of CYP2D6 measured in the largest scale in Korean population. CYP2D6*10 was found to be the most frequent allele (47.3%), followed by CYP2D6*1 (34.2%), *2 (12.4%), *5 (3.8%), and *41 (1.1%). Allele frequencies were slightly different from the previous reports (Lee et al. 2006, 2009; Man et al. 2010; Park et al. 2011), but these differences were not meaningful.
Among East Asian populations (Koreans, Japanese and Chinese), the frequencies of CYP2D6*1 and *10 alleles were significantly different among ethnic groups. Because the enzyme activity of the CYP2D6*10 allele is greatly reduced, these differences can lead to differences in the drug action of CYP2D6 substrate drugs among the three East Asian populations.
Recently, an article provided a global distribution map of alleles with clinical importance by integrating whole-genome and exome sequencing data 56,945 unrelated individuals of five major populations (Zhou et al. 2017). In this article, the frequencies of CYP2D6*1 (13.6%), *2 (14%), *5 (6.5%), *10 (58.7%), *14 (1.6%), and *41 (3.0%) in East Asians were reported. However, the frequency of CYP2D6*1 seemed to be abnormally low, whereas the frequency of CYP2D6*10 seemed to be too high compared to the other studies including this study (Table 1).
Alleles with a significant frequency difference between East Asians and Caucasians were CYP2D6*2 (10.8–11.8% vs. 22.9%), *4 (0.3–0.5% vs. 19.1%), and *10 (37.5–52.5% vs. 1.4%). The functional alleles (CYP2D6*1 and *2) were in higher frequency in Caucasians than in East Asians. In addition, a nonfunctional allele CYP2D6*4 was also higher in frequency in Caucasians compared with East Asians. However, a decreased functional allele CYP2D6*10 was lower in frequency in Caucasians compared with East Asians. The frequency difference of the CYP2D6*10 allele between the two races was most characteristic.
The most distinctive difference between East Asians and Africans was the frequency difference of CYP2D6*10 (37.5–52.5% vs. 4.2%) and *17 alleles (0% vs. 20.5%). CYP2D6*4 allele also was in lower frequency in East Asians than in Africans (0.3–0.5% vs. 5.1%).
In Korean population, the frequency of CYP2D6 genotypes with normal enzyme activity was 22.1%, and only 7 of the 3417 subjects had CYP2D6 PM genotypes. Unlike Caucasians (8.45%) or Africans (2.57%) with a significant frequency of CYP2D6 PM (Gaedigk et al. 2017), CYP2D6 PM was very rare in Koreans, with frequency of about 0.2%. Most of the other subjects had genotypes with one nonfunctional allele, or one or two reduced functional alleles.
Generally, CYP2D6 enzymatic activity can be expressed in terms of four phenotypes: PM, intermediate metabolizer (IM), extensive metabolizer (EM), and ultra-rapid metabolizer. Genotype analysis has become the method of choice to predict a person’s metabolic status. However, there can be substantial differences in the number of genetic variants interrogated as well as differences in test interpretation. Furthermore, there is no standardized process of how a CYP2D6 genotype result is translated into a phenotype assignment (Hicks et al. 2014; LLerena et al. 2014; Fricke-Galindo et al. 2016).
The CYP2D6*10 allele is associated with reduced substrate affinity (Johansson et al. 1994) and many in vivo studies have shown that the metabolic capacity of homozygous CYP2D6*10 is between the capacities of EMs and PMs. Thus, the CYP2D6*10/*10 genotype is commonly categorized as an IM phenotype (Wang et al. 1993; Dahl et al. 1995; Owen et al. 2009). Variations in the activity of the CYP2D6 enzyme results from various combinations of functional alleles (activity = 1), nonfunctional alleles (activity = 0), and reduced functional alleles (activity = 0.5). Steimer et al. (2004) assigned a phenotypic classification based on pure allele activity, i.e., the semiquantitative gene dose. The classification of a semiquantitative gene dose is based on the sum of two allele activities, leading to six different phenotypes, i.e., semiquantitative gene doses of 0, 0.5, 1, 1.5, 2, and > 2. The clinical relevance of distinguishing allele activities (semiquantitative gene doses of 0.5, 1.0, and 1.5) remains open to question. The semiquantitative gene dose classification of 1.5 (1.0 + 0.5) for CYP2D6 is defined as EMs. Thus, subjects with the CYP2D6*1/*10 genotype are usually defined as EMs. However, Steimer et al. (2004) and ter Laak et al. (2010) suggested that this group should be defined as an IM in accordance with their clinical findings.
In this study, the CYP2D6*wt/*10 subjects demonstrated 1.22-fold higher Cmax, 1.39-fold higher AUCinf values and 31% lower CL/F than did CYP2D6*wt/*wt subjects. Although the differences are small, they support the hypothesis of ter Laak et al. (2010). Previous studies have also demonstrated the pharmacokinetic characteristics of diverse CYP2D6 substrates in heterozygous CYP2D6 EMs. In one study of healthy Korean volunteers, CYP2D6*1/*10 subjects showed a 1.9-fold higher Cmax and a 2.2-fold higher AUCinf of metoprolol compared to those of CYP2D6*1/*1 subjects after a single oral dose of 100 mg of metoprolol tartrate (Jin et al. 2008). Another study showed that, after administration of a single 100 mg oral dose of tramadol hydrochloride, healthy Chinese subjects with the CYP2D6*2/*10 genotype had significantly higher t1/2 and AUC values and lower oral clearance of tramadol and showed the opposite effect on the pharmacokinetics of its metabolite compared to the levels in subjects with the CYP2D6*1/*1 genotype (Li et al. 2010). Therefore, CYP2D6*wt/*10 was considered separately from CYP2D6*wt/*wt, depending on the substrate.
In this study, the CYP2D6*10/*10 subjects demonstrated 2.21-fold higher Cmax and 3.88-fold higher AUCinf values. The CL/F in the CYP2D6*10/*10 subjects was only about a quarter of that in CYP2D6*wt/*wt. This shows that the enzyme activity of CYP2D6*10/*10 genotype is much lower than the mean value in IM subjects. The frequency of CYP2D6*5/*5 genotype was very low in Korean population, so only two subjects with this genotype participated in the pharmacokinetic study. In the CYP2D6*5/*5 genotype, CL/F was only 9% of that in CYP2D6*wt/*wt genotype, and AUCinf was 10.62 times higher than that in CYP2D6*wt/*wt genotype.
In conclusion, in 3417 Korean population, the functional allele *1 and *2 were present in frequencies of 34.6 and 11.8%, respectively. In decreased functional alleles, *10 was the most frequent with 46.2% and *41 allele was present in 1.4%. The nonfunctional alleles *5 and *14 were present at 4.5% and 0.5% frequency, respectively. The *X × N allele was present at a frequency of 1.0%. CYP2D6*1/*1, *1/*2 and *2/*2 genotypes with normal enzyme activity were present in 12.1, 8.6 and 1.4% of the subjects, respectively. CYP2D6*5/*5, *5/*14, and *14/*14 genotypes classified as poor metabolizer were only present in 4, 2, and 1 subjects, respectively. Mutant genotypes with frequencies of more than 1% were CYP2D6*1/*10 (32.0%), *10/*10 (22.3%), *2/*10 (11.7%), *5/*10 (3.7%), *1/*5 (2.5%), and *10/*41 (1.2%). The relative clearance of metoprolol in CYP2D6*1/*10, *1/*5, *10/*10, *5/*10, and *5/*5 genotypes were 69%, 57%, 24%, 14% and 9% of CYP2D6*wt/*wt genotype, respectively. These results will be very useful in establishing a strategy for precision medicine related to the genetic polymorphism of CYP2D6.
References
Byeon JY, Kim YH, Na HS, Jang JH, Kim SH, Lee YJ, Bae JW, Kim IS, Jang CG, Chung MW, Lee SY (2015) Effects of the CYP2D6*10 allele on the pharmacokinetics of atomoxetine and its metabolites. Arch Pharm Res 38(11):2083–2091
Cai WM, Nikoloff DM, Pan RM, de Leon J, Fanti P, Fairchild M, Koch WH, Wedlund PJ (2006) CYP2D6 genetic variation in healthy adults and psychiatric African-American subjects: implications for clinical practice and genetic testing. Pharmacogenomics J 6(5):343–350
Cook DJ, Finnigan JD, Cook K, Black GW, Charnock SJ (2016) Cytochromes P450: history, classes, catalytic mechanism, and industrial application. Adv Protein Chem Struct Biol 105:105–126
Dahl ML, Yue QY, Roh HK, Johansson I, Säwe J, Sjöqvist F, Bertilsson L (1995) Genetic analysis of the CYP2D locus in relation to debrisoquine hydroxylation capacity in Korean, Japanese and Chinese subjects. Pharmacogenetics 5:159–164
Dandara C, Masimirembwa CM, Magimba A, Sayi J, Kaaya S, Sommers DK, Snyman JR, Hasler JA (2001) Genetic polymorphism of CYP2D6 and CYP2C19 in east- and southern African populations including psychiatric patients. Eur J Clin Pharmacol 57(1):11–17
de Leon J, Susce MT, Johnson M, Hardin M, Maw L, Shao A, Allen AC, Chiafari FA, Hillman G, Nikoloff DM (2009) DNA microarray technology in the clinical environment: the AmpliChip CYP450 test for CYP2D6 and CYP2C19 genotyping. CNS Spectr 14(1):19–34
Dong Y, Xiao H, Wang Q, Zhang C, Liu X, Yao N, Sheng H, Li H (2015) Analysis of genetic variations in CYP2C9, CYP2C19, CYP2D6 and CYP3A5 genes using oligonucleotide microarray. Int J Clin Exp Med 8(10):18917–18926
Eichelbaum M, Ingelman-Sundberg M, Evans WE (2006) Pharmacogenomics and individualized drug therapy. Annu Rev Med 57:119–137
Fricke-Galindo I, Céspedes-Garro C, Rodrigues-Soares F, Naranjo ME, Delgado Á, de Andrés F, López-López M, Peñas-Lledó E, LLerena A (2016) Interethnic variation of CYP2C19 alleles, ‘predicted’ phenotypes and ‘measured’ metabolic phenotypes across world populations. Pharmacogenomics J 16(2):113–123
Gaedigk A, Bradford LD, Marcucci KA, Leeder JS (2002) Unique CYP2D6 activity distribution and genotype–phenotype discordance in black Americans. Clin Pharmacol Ther 72(1):76–89
Gaedigk A, Simon SD, Pearce RE, Bradford LD, Kennedy MJ, Leeder JS (2008) The CYP2D6 activity score: translating genotype information into a qualitative measure of phenotype. Clin Pharmacol Ther 83(2):234–242
Gaedigk A, Sangkuhl K, Whirl-Carrillo M, Klein T, Leeder JS (2017) Prediction of CYP2D6 phenotype from genotype across world populations. Genet Med 19(1):69–76
Goh LL, Lim CW, Sim WC, Toh LX, Leong KP (2017) Analysis of genetic variation in CYP450 genes for clinical implementation. PLoS ONE 12(1):e0169233
Griese EU, Asante-Poku S, Ofori-Adjei D, Mikus G, Eichelbaum M (1999) Analysis of the CYP2D6 gene mutations and their consequences for enzyme function in a West African population. Pharmacogenetics 9(6):715–723
Hicks JK, Swen JJ, Gaedigk A (2014) Challenges in CYP2D6 phenotype assignment from genotype data: a critical assessment and call for standardization. Curr Drug Metab 15(2):218–232
Hosono N, Kato M, Kiyotani K, Mushiroda T, Takata S, Sato H, Amitani H, Tsuchiya Y, Yamazaki K, Tsunoda T, Zembutsu H, Nakamura Y, Kubo M (2009) CYP2D6 genotyping for functional-gene dosage analysis by allele copy number detection. Clin Chem 55(8):1546–1554
Ingelman-Sundberg M (2005) Genetic polymorphisms of cytochrome P450 2D6 (CYP2D6): clinical consequences, evolutionary aspects and functional diversity. Pharmacogenomics J 5(1):6–13
Ingelman-Sundberg M, Sim SC, Gomez A, Rodriguez-Antona C (2007) Influence of cytochrome P450 polymorphisms on drug therapies: pharmacogenetic, pharmacoepigenetic and clinical aspects. Pharmacol Ther 116(3):496–526
Ishiguro A, Kubota T, Sasaki H, Yamada Y, Iga T (2003) Common mutant alleles of CYP2D6 causing the defect of CYP2D6 enzyme activity in a Japanese population. Br J Clin Pharmacol 55(4):414–415
Jin SK, Chung HJ, Chung MW, Kim JI, Kang JH, Woo SW, Bang S, Lee SH, Lee HJ, Roh J (2008) Influence of CYP2D6*10 on the pharmacokinetics of metoprolol in healthy Korean volunteers. J Clin Pharm Ther 33(5):567–573
Johansson I, Oscarson M, Yue QY, Bertilsson L, Sjöqvist F, Ingelman-Sundberg M (1994) Genetic analysis of the Chinese cytochrome P4502D locus: characterization of variant CYP2D6 genes present in subjects with diminished capacity for debrisoquine hydroxylation. Mol Pharmacol 46:452–459
Kubota T, Yamaura Y, Ohkawa N, Hara H, Chiba K (2000) Frequencies of CYP2D6 mutant alleles in a normal Japanese population and metabolic activity of dextromethorphan O-demethylation in different CYP2D6 genotypes. Br J Clin Pharmacol 50(1):31–34
Lauschke VM, Ingelman-Sundberg M (2016) The importance of patients-specific factors for hepatic drug response and toxicity. Int J Mol Sci 17:E1714
Lee SY, Sohn KM, Ryu JY, Yoon YR, Shin JG, Kim JW (2006) Sequence-based CYP2D6 genotyping in the Korean population. Ther Drug Monit 28(3):382–387
Lee SJ, Lee SS, Jung HJ, Kim HS, Park SJ, Yeo CW, Shin JG (2009) Discovery of novel functional variants and extensive evaluation of CYP2D6 genetic polymorphisms in Koreans. Drug Metab Dispos 37(7):1464–1470
Li Q, Wang R, Guo Y, Wen S, Xu L, Wang S (2010) Relationship of CYP2D6 genetic polymorphisms and the pharmacokinetics of tramadol in Chinese volunteers. J Clin Pharm Ther 35(2):239–247
LLerena A, Naranjo ME, Rodrigues-Soares F, Penas-LLedó EM, Fariñas H, Tarazona-Santos E (2014) Interethnic variability of CYP2D6 alleles and of predicted and measured metabolic phenotypes across world populations. Expert Opin Drug Metab Toxicol 10(11):1569–1583
Man M, Farmen M, Dumaual C, Teng CH, Moser B, Irie S, Noh GJ, Njau R, Close S, Wise S, Hockett R (2010) Genetic variation in metabolizing enzyme and transporter genes: comprehensive assessment in 3 major East Asian subpopulations with comparison to Caucasians and Africans. J Clin Pharmacol 50(8):929–940
McGrane IR, Loveland JG (2016) Pharmacogenetics of Cytochrome P450 Enzymes in American Indian and Caucasian Children Admitted to a Psychiatric Hospital. J Child Adolesc Psychopharmacol 26(4):395–399
Nishida Y, Fukuda T, Yamamoto I, Azuma J (2000) CYP2D6 genotypes in a Japanese population: low frequencies of CYP2D6 gene duplication but high frequency of CYP2D6*10. Pharmacogenetics 10(6):567–570
Ota T, Kamada Y, Hayashida M, Iwao-Koizumi K, Murata S, Kinoshita K (2015) Combination analysis in genetic polymorphisms of drug-metabolizing enzymes CYP1A2, CYP2C9, CYP2C19, CYP2D6 and CYP3A5 in the Japanese population. Int J Med Sci 12(1):78–82
Owen RP, Sangkuhl K, Klein TE, Altman RB (2009) Cytochrome P450 2D6. Pharmacogenet Genomics 19:559–562
Park HS, Choi JY, Lee MJ, Park S, Yeo CW, Lee SS, Shin JG, Park BW (2011) Association between genetic polymorphisms of CYP2D6 and outcomes in breast cancer patients with tamoxifen treatment. J Korean Med Sci 26(8):1007–1013
Park IH, Ro J, Park S, Lim HS, Lee KS, Kang HS, Jung SY, Lee S (2012) Lack of any association between functionally significant CYP2D6 polymorphisms and clinical outcomes in early breast cancer patients receiving adjuvant tamoxifen treatment. Breast Cancer Res Treat 131(2):455–461
Pietarinen P, Tornio A, Niemi M (2016) High frequency of CYP2D6 ultrarapid metabolizer genotype in the Finnish population. Basic Clin Pharmacol Toxicol 119(3):291–296
Qin S, Shen L, Zhang A, Xie J, Shen W, Chen L, Tang J, Xiong Y, Yang L, Shi Y, Feng G, He L, Xing Q (2008) Systematic polymorphism analysis of the CYP2D6 gene in four different geographical Han populations in mainland China. Genomics 92(3):152–158
Rasmussen JO, Christensen M, Svendsen JM, Skausig O, Hansen EL, Nielsen KA (2009) CYP2D6 gene test in psychiatric patients and healthy volunteers. Scand J Clin Lab Invest 66:129–136
Roh HK, Chung JY, Oh DY, Park CS, Svensson JO, Dahl ML, Bertilsson L (2001) Plasma concentrations of haloperidol are related to CYP2D6 genotype at low, but not high doses of haloperidol in Korean schizophrenic patients. Br J Clin Pharmacol 52(3):265–271
Steimer W, Zöpf K, von Amelunxen S, Pfeiffer H, Bachofer J, Popp J, Messner B, Kissling W, Leucht S (2004) Allelespecific change of concentration and functional gene dose for the prediction of steady-state serum concentrations of amitriptyline and nortriptyline in CYP2C19 and CYP2D6 extensive and intermediate metabolizers. Clin Chem 50:1623–1633
Tateishi T, Chida M, Ariyoshi N, Mizorogi Y, Kamataki T, Kobayashi S (1999) Analysis of the CYP2D6 gene in relation to dextromethorphan O-demethylation capacity in a Japanese population. Clin Pharmacol Ther 65(5):570–575
ter Laak MA, Temmink AH, Koeken A, van’t Veer NE, van Hattum PR, Cobbaert CM (2010) Recognition of impaired atomoxetine metabolism because of low CYP2D6 activity. Pediatr Neurol 43:159–162
Wan YJ, Poland RE, Han G, Konishi T, Zheng YP, Berman N, Lin KM (2001) Analysis of the CYP2D6 gene polymorphism and enzyme activity in African-Americans in southern California. Pharmacogenetics 11(6):489–499
Wang SL, Huang JD, Lai MD, Liu BH, Lai ML (1993) Molecular basis of genetic variation in debrisoquin hydroxylation in Chinese subjects: polymorphism in RFLP and DNA sequence of CYP2D6. Clin Pharmacol Ther 53:410–418
Wang SL, Lai MD, Huang JD (1999) G169R mutation diminishes the metabolic activity of CYP2D6 in Chinese. Drug Metab Dispos 27(3):385–388
Wennerholm A, Johansson I, Hidestrand M, Bertilsson L, Gustafsson LL, Ingelman-Sundberg M (2001) Characterization of the CYP2D6*29 allele commonly present in a black Tanzanian population causing reduced catalytic activity. Pharmacogenetics 11(5):417–427
Yee MM, Josephson C, Hill CE, Harrington R, Castillejo MI, Ramjit R, Osunkwo I (2013) Cytochrome P450 2D6 polymorphisms and predicted opioid metabolism in African American children with sickle cell disease. J Pediatr Hematol Oncol 35(7):e301–e305
Zhou Q, Yu XM, Lin HB, Wang L, Yun QZ, Hu SN, Wang DM (2009) Genetic polymorphism, linkage disequilibrium, haplotype structure and novel allele analysis of CYP2C19 and CYP2D6 in Han Chinese. Pharmacogenomics J 9(6):380–394
Zhou Y, Ingelman-Sundberg M, Lauschke VM (2017) Worldwide distribution of cytochrome P450 alleles: a meta-analysis of population-scale sequencing projects. Clin Pharmacol Ther 102(4):688–700
Acknowledgements
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT, and Future Planning (NRF-2016R1A2B4007381).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no potential conflict of interest with respect to the authorship and/or publication of this article.
Rights and permissions
About this article
Cite this article
Byeon, JY., Kim, YH., Lee, CM. et al. CYP2D6 allele frequencies in Korean population, comparison with East Asian, Caucasian and African populations, and the comparison of metabolic activity of CYP2D6 genotypes. Arch. Pharm. Res. 41, 921–930 (2018). https://doi.org/10.1007/s12272-018-1075-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12272-018-1075-6