Abstract
CYP2C8 is a member of Cytochrome P450 enzymes system. It plays an important role in metabolizing a wide range of exogenous and endogenous compounds. CYP2C8 is involved in the metabolism of more than 100 drugs, typical substrates include: anticancer agents, antidiabetic agents, antimalarial agents, lipid lowering drugs and many others that constitute 20% of clinically prescribed drugs. Genetic variations of CYP2C8 have been reported with different frequencies in different populations. These genetic polymorphisms can lead to differences in the efficacy and safety of different types of medications metabolized by CYP2C8. The aim of this study was to investigate the allele frequencies of CYP2C8*3 (rs10509681 and rs11572080) and CYP2C8*4 (rs1058930) polymorphisms in three populations living in Jordan; Circassians and Chechens and Jordanian-Arabs and compare those frequencies with other populations. A total of 200 healthy Jordanians, 93 Circassians and 88 Chechens were included in this study. Genotyping of CYP2C8*3 and CYP2C8*4 polymorphisms was done by using polymerase chain reaction (PCR) followed by Restriction Fragment Length Polymorphism (RFLP). Using the Chi-square test, we found that the prevalence of CYP2C8*3 and *4 among the three populations were significantly different. Moreover, the mutant allele CYP2C8*3 (416A) was only detected in the Jordanian-Arab population with an allele frequency of 0.082, while the mutant allele CYP2C8*4 (792G) was detected with frequencies of 0.065, 0.122, 0.017 in Jordanian-Arabs, Circassians and Chechens, respectively. As our results show, CYP2C8*3 was undetectable in our Circassians and Chechens samples, on the other hand, Circassians had the highest allele frequency of CYP2C8*4 compared to Chechens and Jordanian-Arabs. These genetic variations of the gene encoding the CYP2C8 drug metabolizing enzymes can lead to clinical differences in drug metabolism and ultimately variations in drug effectiveness and toxicities. This study provides evidence for the importance of personalized medicine in these populations and can be the foundation for future clinical studies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The CYP2C8 Enzymes
The term pharmacogenetics first appeared in the literature in 1961 by Evans and Clarke and it was used to refer to “genetically determined variations in animal species that are revealed by the effects of drugs”[1]. Pharmacogenetics now has a boarder definition, it can be defined as the study of the effects of genes on drug disposition: absorption, distribution, metabolism, and excretion. The interest in pharmacogenetics stems from the fact that it allows for tailoring the right drug and right dose for each individual patient based on their genetic makeup, resulting in higher efficacy and lower adverse drug reactions. The liver is considered the primary organ of detoxification. The majority of hepatically metabolized drugs are metabolized by cytochrome P450 (CYP), a heme-containing superfamily of enzymes. From a pharmacogenetics point of view, many changes in individual responses to drugs can be attributed to the genetic makeup of the CYP genes.
Drug metabolism reactions consist of two phases: phase I functionalization reactions and phase II conjugation reactions. Cytochrome P450-2C8 (CYP2C8), a member of the CYP family, is involved in the metabolism of at least 5% of drugs cleared by phase I metabolism, which accounts for about 20% of clinically prescribed drugs [2, 3]. According to Daily and Aquilante (2009) in their review article, CYP2C8 is involved in the metabolism of more than 100 drugs, and the typical substrates include: anticancer agents, antidiabetic agents, antimalarial agents, lipid lowering drugs and many others [4]. The major substrates of CYP2C8 -that account for more than 70% of their metabolism- are: amodiaquine [5], cerivastatin [6], daprodustat [7], enzalutamide [8], montelukast [9], pioglitazone [9] and repaglinide [9, 10].
The CYP2C8s are found primarily in the liver, but are also found in various other tissues, including the kidneys, brain, adrenal gland, mammary gland, ovaries and duodenum [11]. The variety of organs where the CYPs are expressed hints that these enzymes are not only important in the metabolism of drugs, but they also have biologically important functions. CYP2C8s are important in the epoxidation of arachidonic acid to biologically active epoxy eicosatrienoic acids [12, 13], the hydroxylation of the active metabolite of vitamin A (Retinol) [14], and the metabolism of the polyunsaturated essential fatty acid linoleic acid [15] as well as eicosapentaenoic acid [16]. Therefore, examining the pharmacogenetics of CYP2C8 and investigating CYP2C8 polymorphism could be a great resource for identifying ways to increase the efficacy and reduce the adverse drug events in patients using medications metabolized by the CYP2C8 enzymes.
CYP2C8 Polymorphisms
The CYP2C gene family spans nearly 400 kilo bases located on chromosome 10q24 and constitutes a cluster of polymorphic genes that share significant sequence homology. CYP2C8 is the smallest of the human CYP2C genes as it spans a region of 31-kb and contains 9 exons [11]. It shares 74% sequence homology with CYP2C9 [4], and although the main CYP2C enzymes (CYP2C8, 2C9, 2C18, and 2C19) share more than 80% amino acid sequence identity, the substrate specificity differs [17].
Several genetic polymorphisms of CYPs have been associated with altered metabolic activity, which could potentially play a crucial role in determining the efficacy and safety of a wide range of drugs metabolized by these enzymes [4]. To date nearly 100 nonsynonymous single nucleotide variations (SNV) have been identified. The majority are rare and occur at minor allele frequencies of 0.01 or less [4]. The wild type of the gene is denoted with CYP2C8*1 or CYP2C8*1A, and two SNPs have been reported in the 5-flanking region of the gene; CYP2C8*1B (C271A) and CYP2C8*1C (T370G) with no significant differences in the protein levels when compared with the wild type [11].
However, the major nonsynonymous variants of CYP2C8 are the three SNPs: CYP2C8*2, *3, and *4 [4]. The CYP2C8*3 polymorphism (rs10509681 and rs11572080) constitutes two nonsynonymous variants (G416A and A1196G) in exons 3 and 8 which are in an almost complete linkage disequilibrium, and leads to the amino acid substitutions Arg139Lys and Lys399Arg [4]3. While CYP2C8*4 polymorphism (rs1058930) constitute one change (C792G) in exon 5 and one amino acid change Ile264Met [3]. Greater AUC values and a decrease in drug clearance were observed in individuals with homozygous genotype (*3/*3) of CYP2C8*3, moreover, even in the presence of a drug-drug interaction like gemfibrozil (CYP2C8 inhibitor) and pioglitazone, the relative change in pioglitazone plasma exposure following gemfibrozil administration was still influenced by the CYP2C8 genotype [18]. Conversely, CYP2C8*3 and CYP2C8*4 genotypes were associated with slight decrease in paclitaxel clearance [19] and these genotypes were associated with increased risk of toxicities associated with paclitaxel treatment in patients with cancer [6]. However, one study found that AUC values of repaglinide were lower by approximately 40–50% in individuals with CYP2C8*1/*3 when compared to with individuals with the CYP2C8*1/*1 genotype, possibly, indicating a dose dependent effect [20]. Similarly, the CYP2C8*4 can have reduced clearance (paclitaxel 6α-hydroxylation and repaglinide), or increased clearance (cerivastatin), or can have no significant effects as shown in tanshinol borneol ester metabolism[21,22,23,24]. Thus, CYP2C8*3 and *4 influence on drug metabolism might be both substrate and dose dependent, suggesting that patients carrying the polymorphisms might be affected more or less depending on the medication and the dose they are using.
In this study, we focused on two polymorphisms (CY2C8*3 and CY2C8*4) for the following reasons; first, they are two of the most common nonsynonymous variables that are responsible for reduced enzyme activity, and second, their allele frequencies have been found to be variable between different populations.
The Circassian and Chechen Populations in Jordan
Many Circassians (also known as Adyghe or Čerkesy) and Chechens (Nokhchiy) migrated in the nineteenth century to Jordan. Due to their linguistic and ethnic differences along with the tendency of individuals to marry within their community, they have established independent communities that share a Northern Caucasus ancestry, [25,26,27,28,29]. Population genetic studies are used to better understand common genetic heritage among such populations. Such studies also have significant importance in medical applications; as some disease-causing alleles occur in clusters in specific populations living in particular geographic regions [30]. Recently, several studies have shown genetic variability between Jordanian-Arabs, Circassians and Chechens who live in Jordan with a certain focus on genetic variances in drug metabolism and pharmacogenes as shown in Table 1. These studies confirm the importance of studying the Circassian and Chechen population to help guide future applications of personalized medicine. As different populations have different frequencies of various alleles involved in drug metabolism, we aimed to determine the frequencies of CYP2C8*3 and CYP2C8*4 in our 3 populations of interest and then statistically compare them to each other and to other populations using previously reported allele frequencies, to report whether any of these populations have significant differences.
Studies identifying the genetic differences between ethnic minorities and the general population are crucial to understanding the interindividual differences in drug disposition that might be attributed to race or ethnicity which may change the risk–benefit use of certain medications. These studies will also set the stage for drug therapy that is tailored to each individual patient based on their genes. This is the foundation of personalized medicine or precision medicine that aims to maximize drug efficacy with minimum side effects.
Materials and Methods
Ethics
The IRB committee at the National Center for Diabetes Endocrinology and Genetics of Jordan reviewed and approved this study, and all participants were given a written informed consent before the collection of the blood samples.
Study Design and Samples Selection
The study recruited a total of 93 Circassians, 88 Chechens and 200 Jordanian-Arabs. A total of 9 ml of whole blood was drawn in EDTA tubes from the subjects by vacutainer system and the genomic DNA was isolated from whole blood using the phenol–chloroform protocol. Each Circassian and Chechen participant in the study completed an assessment that included lineage information, which included obtaining each participant's former three generations from both maternal and paternal sides. Our cross-sectional study included volunteers of random unrelated Circassians, Chechens and Jordanian-Arabs living in Jordan, Table 2 shows the demographics of the three groups.
DNA Quantification and Genotyping
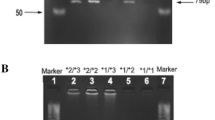
Absorbance was measured at 260 nm wavelength (A 260) using Ultraviolet (UV) Nano-Spectrophotometer to measure DNA concentration (ng/ml). Genotyping was done by polymerase chain reactions (PCRs) and by using Restriction Fragment Length Polymorphism (RFLP). The sequence of the primers used to study CYP2C8 gene was described previously by Nakajima et al. [31]. For CYP2C8*3, the forward primer sequence was (5'-AGG CAA TTC CCC AAT ATC TC-3') and the reverse primer was (5'-CAG GAT GCG CAA TGA AGA-3') with a produced size of 367BP. For CYP2C8*4 the forward primer sequence was (5'-AAA GTA AAA GAA CAC CAA GC-3') and the reverse primer was (5'-AAA CAT CCT TAG TTA ATT ACA-3') with a product size of 167BP. The thermal cycler conditions started with an initial denaturation at 94 °C for 3 min followed by denaturation, annealing and extension for 30 s at 94 °C, 55 °C and 72 °C, respectively. The number of cycles were 30 and a final extension for 5 min at 72℃. PCR reactions were run on 2% agarose gel electrophoresis for 45 min at 150 V and visualized using Red Safe Dye. PCR products with the desired length were digested by restriction enzymes; BseR I and Taq I for CYP2C8*3 and CYP2C8*4, respectively. Digestive products were mixed with a loading dye and run on 3.5% agarose gel electrophoresis and was observed by UV-trans illuminator. The digested fragments were read according to Table 3 which shows the genotype according to the bands’ length observed.
Statistical Analysis
For our main interest in this study, we tested our hypothesis that the allele frequencies between the three populations: Jordanian-Arabs, Circassians and Chechens are different (HA: PJ ≠ PC ≠ PCh, Ho: PJ = PC = PCh). We used the results of the Chi-square test and the p-value (with a significant level of α = 0.05) to determine if the evidence is suggestive of whether the allele frequencies are different between the three populations or in other words the possibility of rejecting the null hypothesis that the three populations have similar allele frequencies. We were also interested in comparing the allele frequencies from this study with other populations (HA: Ptested population in this study ≠ POther populations, Ho: Ptested population in this study = POther populations). We tested the main assumptions needed for the statistical analysis by determining if the expected frequencies are large enough (which represent expected counts if the H0 is true). We found that some expected frequencies were large enough (> 5) and in that case we reported the chi-square test (X2) result, when the expected frequencies were not large enough, we reported the result of the two-tailed Fisher-Exact test as an alternative. The JMP Pro 14 software was used for all statistical analyses.
Results
A total of 200 healthy Jordanians took part in this study and Table 2 shows their demographics. The low alleles frequencies of CYP2C8*3 and *4 found in this study is consistent with previous literature showing how rare these polymorphisms are, we were able to detect the CYP2C8*3 polymorphism only in Jordanian-Arabs with an allele frequency of 0.082, on the other hand, we were able to detect CYP2C8*4 in all three populations with allele frequencies of 0.065, 0.017 and 0.122 for Jordanian Arabs, Chechens and Circassians subpopulations, respectively (Table 4).
Since we were not able to detect the CYP2C8*3 polymorphism in the Circassians and Chechan populations, we concluded that Circassians and Chechens have significantly lower allele frequencies of CYP2C8*3 compared to the Jordanian-Arab population. While the CYP2C8*4 allele frequency of Circassians is higher compared to Chechens and Jordanian-Arabs. Upon comparing the three populations using the Fisher-Exact test, the results suggest that these three populations are statistically different at the level of CYP2C8*3 and *4 (p < 0.0001 and p < 0.05, respectively).
Discussion
Interindividual variability and genetic differences play a major role in determining the efficacy and toxicity of many prescribed medications. Genetic polymorphisms in genes coding drug transporters, drug metabolizing enzymes and drug target proteins are the best examples of these variations [32]. Polymorphisms of the CYP2C8 gene are present among several ethnic populations with different frequencies. Apart from the wild-type allele CYP2C8*1, three nonsynonymous variant alleles CYP2C8*2, CYP2C8*3 and CYP2C8*4 are the most frequently studied polymorphisms [33]. These polymorphisms have important clinical and physiological implications in individuals who are homozygous for the mutant alleles. For example, it has been shown that CYP2C8 encoded by CYP2C8*3 variant allele is defective in metabolizing two clinically important substrates: arachidonic acid and a chemotherapeutic drug, paclitaxel [34]. CYP2C8*3 allele is also present in an incomplete linkage disequilibrium with CYP2C9*2 allele [35]. Yasar et al. (2002) performed a study including 1468 subjects and concluded that about 96% of the subjects with CYP2C8*3 allele carried CYP2C9*2 allele and 85% of subjects who had CYP2C9*2 variant also carried CYP2C8*3 [5]. Moreover, Jiang and colleagues (2011) expressed and purified CYP2C8*4 polymorphic enzyme in E. coli and concluded that the enzymes expressed from this mutation have only 25% activity compared to the wild type [36].
The frequencies of CYP2C8*3 and *4 allele differ among different populations. They have been observed with higher frequencies primarily in Caucasians, and with lower frequencies in Africans and Asians as shown in Tables 5 and 6 for CYP2C8*3 and *4, respectively. Upon comparing the allele frequencies of CYP2C8*3 and *4 in Circassians and Chechens, we first noticed that they are statistically different from each other, indicating that they are two distinct populations. We found that both Circassians and Chechens were statistically different from Jordanian-Arabs as well. The genetic differences between these populations were confirmed in a study by AL-Eitan et al. [37]. This study reported using mitochondrial DNA (mtDNA) data that Circassian and Chechen have several common matrilineal ancestors, and that Circassian and Chechen are more closely related to each other than to the Jordanian population. Moreover, as a general trend for CYP2C8*3, we noticed that Circassians and Chechens are more similar to the Asian and African populations than they are to the European populations as shown in Table 5. On the other hand, CYP2C8*4, showed different patterns in Circassians and Chechens, Circassians were statistically different from the Asian and African populations and were only similar to one European population. Chechens had more similarities with the Asian and African populations. This supports the importance of studying these two populations separately and not grouped together as one subpopulation. Figure 1 shows a comparison between the frequencies of CYP2C8*3 and *4 within different populations. As for the clinical impact and the availability of our results, the effect of CYP2C8*3 on drug metabolism and pharmacokinetics has been recognized in many clinical studies [18, 20, 38] and we believe more clinical studies should focus on the use of CYP2C8 as a biomarker in subpopulations for ultimate efficacy and safety of drug use. However, as our results have shown, the CYP2C8*4 genotype is rare and has a lesser impact on the metabolism of CYP2C8 substrates, but the results are still valuable to add to the literature, especially for studies comparing SNP variations between different populations[38].
One of the struggles of pharmacotherapy is that the patient outcomes can be very variable, whether that is because of deviations of optimum efficacy or the occurrence of adverse drug reactions. However, one branch within the field of pharmacogenetics is using variations in genes to predict and tackle these variabilities. Therefore, determining the allele frequencies of polymorphisms in genes encoding drug metabolizing enzymes would enable not only the understanding of how the Circassian and Chechen populations might respond to certain medications differently than Jordanian-Arabs [2, 39], but also opens the door for future clinical studies. Table 1 shows similar studies investigating other drug metabolizing enzymes in these populations living in Jordan. These studies all highlight the genetic differences between these populations and indicate the importance of such pharmacogenetics studies when personalized medication is going to be implemented in the future. Application of pharmacogenetics in drug prescription well ensures that the right drug is prescribed for the right patient at the right dose at the right time.
Limitations
Some rare polymorphisms of CYP2C8 were not detected in this study which could be due to the relatively small sample size. Sample size was a limitation because the Circassian and Chechen populations living in Jordan are rather small, estimates range from 80,000 to 100,000 and 12,000–30,000, respectively. It was challenging to find unrelated individuals, who also do not have any non-Circassian or non-Chechen ancestry in their pedigree. It is also worth mentioning that the pedigree of the participants was collected for the previous three generations only. Circassians and Chechens immigrated to Jordan in the late nineteenth century and since then most of them remained endogamous. Circassian and Chechen populations are present in many countries other than Jordan, including, Turkey, Russia, Syria and Iraq, however, due to the founder effect, the findings in this study are descriptive only of the Circassian and Chechen communities in Jordan, and are not generalizable, though similar patterns with other Circassian and Chechen communities may exist.
Conclusion
Knowledge of the frequency of these polymorphisms in the Jordanian populations is a substantial asset for designing future pharmacokinetic and pharmacodynamics studies conducted with any of the CYP2C8 substrates. Furthermore, genotyping techniques may be useful tools to predict impaired metabolism when using drugs that are substrates of CYP2C8 and that have reduced efficacy or increased toxicities. In this study along with the results of previous research conducted on Circassians and Chechens we have been able to show how genetically different they are from Jordanian-Arabs and could potentially provide a baseline for future genome wide association studies on these populations and for the possibility of using them as biomarkers for personalized medicine applications.
References
Evans DA, Clarke CA. Pharmacogenetics. Br Med Bull. 1961;17:234–40.
Lee S-J, Lee S-S, Shin J-G. Chapter 6A. Pharmacogenetics of cytochrome P450. In: Bertino JS, DeVane CL, Fuhr U, Kashuba AD, Ma JD, editors. Pharmacogenomics Introd Clin Perspect. New York, NY: The McGraw-Hill Companies; 2013.
Totah RA, Rettie AE. Cytochrome P450 2C8: substrates, inhibitors, pharmacogenetics, and clinical relevance. Clin Pharmacol Ther. 2005;77:341–52.
Daily EB, Aquilante CL. Cytochrome P450 2C8 pharmacogenetics: a review of clinical studies. Pharmacogenomics. 2009;10:1489–510.
Yasar U, Lundgren S, Eliasson E, Bennet A, Wiman B, de Faire U, et al. Linkage between the CYP2C8 and CYP2C9 genetic polymorphisms. Biochem Biophys Res Commun. 2002;299:25–8.
Backman JT, Filppula AM, Niemi M, Neuvonen PJ. Role of cytochrome P450 2C8 in drug metabolism and interactions. Pharmacol Rev. 2016;68(1):168–241.
Johnson BM, Stier BA, Caltabiano S. Effect of food and gemfibrozil on the pharmacokinetics of the novel prolyl hydroxylase inhibitor GSK1278863. Clin Pharmacol Drug Dev. 2014;3:109–17.
Gibbons JA, de Vries M, Krauwinkel W, Ohtsu Y, Noukens J, van der Walt J-S, et al. Pharmacokinetic drug interaction studies with enzalutamide. Clin Pharmacokinet. 2015;54:1057–69.
Filppula AM, Laitila J, Neuvonen PJ, Backman JT. Reevaluation of the microsomal metabolism of montelukast: major contribution by CYP2C8 at clinically relevant concentrations. Drug Metab Dispos Biol Fate Chem. 2011;39:904–11.
Kajosaari LI, Laitila J, Neuvonen PJ, Backman JT. Metabolism of repaglinide by CYP2C8 and CYP3A4 in vitro: effect of fibrates and rifampicin. Basic Clin Pharmacol Toxicol. 2005;97:249–56.
Klose TS, Blaisdell JA, Goldstein JA. Gene structure of CYP2C8 and extrahepatic distribution of the human CYP2Cs. J Biochem Mol Toxicol. 1999;13:289–95.
Kaspera R, Totah RA. Epoxyeicosatrienoic acids: formation, metabolism and potential role in tissue physiology and pathophysiology. Expert Opin Drug Metab Toxicol. 2009;5:757–71.
Zeldin DC, Moomaw CR, Jesse N, Tomer KB, Beetham J, Hammock BD, et al. Biochemical characterization of the human liver cytochrome P450 arachidonic acid epoxygenase pathway. Arch Biochem Biophys. 1996;330:87–96.
Thatcher JE, Zelter A, Isoherranen N. The relative importance of CYP26A1 in hepatic clearance of all-trans retinoic acid. Biochem Pharmacol. 2010;80:903–12.
Bylund J, Kunz T, Valmsen K, Oliw EH. Cytochromes P450 with bisallylic hydroxylation activity on arachidonic and linoleic acids studied with human recombinant enzymes and with human and rat liver microsomes. J Pharmacol Exp Ther. 1998;284:51–60.
Barbosa-Sicard E, Markovic M, Honeck H, Christ B, Muller DN, Schunck W-H. Eicosapentaenoic acid metabolism by cytochrome P450 enzymes of the CYP2C subfamily. Biochem Biophys Res Commun. 2005;329:1275–81.
Ridderström M, Zamora I, Fjellström O, Andersson TB. Analysis of selective regions in the active sites of human cytochromes P450, 2C8, 2C9, 2C18, and 2C19 homology models using GRID/CPCA. J Med Chem. 2001;44:4072–81.
Aquilante CL, Kosmiski LA, Bourne DWA, Bushman LR, Daily EB, Hammond KP, et al. Impact of the CYP2C8 *3 polymorphism on the drug-drug interaction between gemfibrozil and pioglitazone. Br J Clin Pharmacol. 2013;75(1):217–26.
Bergmann TK, Brasch-Andersen C, Gréen H, Mirza M, Pedersen RS, Nielsen F, et al. Impact of CYP2C8*3 on paclitaxel clearance: A population pharmacokinetic and pharmacogenomic study in 93 patients with ovarian cancer. Pharmacogenomics J. 2011;11(2):113–20.
Niemi M, Backman JT, Kajosaari LI, Leathart JB, Neuvonen M, Daly AK, et al. Polymorphic organic anion transporting polypeptide 1B1 is a major determinant of repaglinide pharmacokinetics. Clin Pharmacol Ther. 2005;77:468–78.
Singh R, Ting JG, Pan Y, Teh LK, Ismail R, Ong CE. Functional role of Ile264 in CYP2C8: mutations affect haem incorporation and catalytic activity. Drug Metab Pharmacokinet. 2008;23:165–74.
Yu L, Shi D, Ma L, Zhou Q, Zeng S. Influence of CYP2C8 polymorphisms on the hydroxylation metabolism of paclitaxel, repaglinide and ibuprofen enantiomers in vitro. Biopharm Drug Dispos. 2013;34:278–87.
Kaspera R, Naraharisetti SB, Tamraz B, Sahele T, Cheesman MJ, Kwok P-Y, et al. Cerivastatin in vitro metabolism by CYP2C8 variants found in patients experiencing rhabdomyolysis. Pharmacogenet Genomics. 2010;20:619–29.
Liu D, Gao Y, Wang H, Zi J, Huang H, Ji J, et al. Evaluation of the effects of cytochrome P450 nonsynonymous single-nucleotide polymorphisms on tanshinol borneol ester metabolism and inhibition potential. Drug Metab Dispos. 2010;38:2259–65.
Shami S. Displacement, historical memory, and identity: the circassians in Jordan. Cent Migr Stud Spec Issues. 1994;11:189–201.
Zhemukhov S. Circassian World: Responses to the New Challenges. PONARS Eurasia. 2008;1.
Kailani W. Chechens in the Middle East: Between Original and Host Cultures. Casp Stud Program. 2002;
Dweik B. Linguistic and cultural maintenance among the Chechens of Jordan. Lang Cult Curric. 2000;13:184–95.
Richmond W. The Circassian Genocide. Press. U, editor. 2013.
Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, et al. Genetic structure of human populations. Science. 2002;298:2381–5.
Nakajima M, Fujiki Y, Noda K, Ohtsuka H, Ohkuni H, Kyo S, et al. Genetic polymorphisms of CYP2C8 in Japanese population. Drug Metab Dispos Biol Fate Chem. 2003;31:687–90.
Evans WE, Relling M V. Pharmacogenomics: translating functional genomics into rational therapeutics. science. American Association for the Advancement of Science 1999;286:487–91.
Pechandova K, Buzkova H, Matouskova O, Perlik F, Slanar O. Genetic polymorphisms of CYP2C8 in the Czech Republic. Genet Test Mol Biomark. 2012;16:812–6.
Dai D, Zeldin DC, Blaisdell JA, Chanas B, Coulter SJ, Ghanayem BI, et al. Polymorphisms in human CYP2C8 decrease metabolism of the anticancer drug paclitaxel and arachidonic acid. Pharmacogenetics. 2001;11:597–607.
Garcia-Martin E, Martinez C, Tabares B, Frias J, Agundez JAG. Interindividual variability in ibuprofen pharmacokinetics is related to interaction of cytochrome P450 2C8 and 2C9 amino acid polymorphisms. Clin Pharmacol Ther. 2004;76:119–27.
Jiang H, Zhong F, Sun L, Feng W, Huang Z-X, Tan X. Structural and functional insights into polymorphic enzymes of cytochrome P450 2C8. Amino Acids. 2011;40:1195–204.
AL-Eitan L, Saadeh H, Alnaamneh A, Darabseh S, AL-Sarhan N, Alzihlif M, et al. The genetic landscape of Arab Population, Chechens and Circassians subpopulations from Jordan through HV1 and HV2 regions of mtDNA. Gene 2020;729:144314
Tornio A, Backman JT. Cytochrome P450 in pharmacogenetics: An update. Adv Pharmacol. 2018;83:3–32.
Roden DM, Altman RB, Benowitz NL, Flockhart DA, Giacomini KM, Johnson JA, et al. Pharmacogenomics: challenges and opportunities. Ann Intern Med. 2006;145:749–57.
Abudahab S, Hakooz N, Jarrar Y, Al Shahhab M, Saleh A, Zihlif M, et al. Interethnic variations of UGT1A1 and UGT1A7 polymorphisms in the Jordanian population. Curr Drug Metab. 2019;20:399–410.
Al-Eitan LN, Mohammad NN, Al-Maqableh HW, Hakooz NM, Dajani RB. Genetic polymorphisms of pharmacogenomic VIP variants in the circassian subpopulation from Jordan. Curr Drug Metab. 2019;20:674–81.
Al-Eitan LN, Rababa’h DM, Hakooz NM, Alghamdi MA, Dajani RB. Genetic polymorphisms of pharmacogenes among the genetically isolated Circassian subpopulation from Jordan. J Pers Med 2020;10(1):2.
Minhas S, Setia N, Pandita S, Saxena R, Verma IC, Aggarwal S. Prevalence of CYP2C8 polymorphisms in a North Indian population. Genet Mol Res GMR. 2013;12:2260–6.
Arun Kumar AS, Chakradhara Rao US, Umamaheswaran G, Ramu P, Kesavan R, Shewade DG, et al. Haplotype structures of common variants of CYP2C8, CYP2C9, and ADRB1 genes in a South Indian population. Genet Test Mol Biomark. 2011;15:407–13.
Muthiah YD, Lee WL, Teh LK, Ong CE, Ismail R. Genetic polymorphism of CYP2C8 in three Malaysian ethnics: CYP2C8*2 and CYP2C8*3 are found in Malaysian Indians. J Clin Pharm Ther. 2005;30:487–90.
Kudzi W, Dodoo ANO, Mills JJ. Characterisation of CYP2C8, CYP2C9 and CYP2C19 polymorphisms in a Ghanaian population. BMC Med Genet. 2009;10:124.
Cavaco I, Stromberg-Norklit J, Kaneko A, Msellem MI, Dahoma M, Ribeiro VL, et al. CYP2C8 polymorphism frequencies among malaria patients in Zanzibar. Eur J Clin Pharmacol. 2005;61:15–8.
Bahadur N, Leathart JBS, Mutch E, Steimel-Crespi D, Dunn SA, Gilissen R, et al. CYP2C8 polymorphisms in Caucasians and their relationship with paclitaxel 6alpha-hydroxylase activity in human liver microsomes. Biochem Pharmacol. 2002;64:1579–89.
Yasar U, Bennet A, Eliasson E, Lundgren S, Wiman B, Faire U, et al. Allelic variants of cytochromes P450 2C modify the risk for acute myocardial infarction. Pharmacogenetics. 2004;13:715–20.
Halling J, Petersen MS, Damkier P, Nielsen F, Grandjean P, Weihe P, et al. Polymorphism of CYP2D6, CYP2C19, CYP2C9 and CYP2C8 in the Faroese population. Eur J Clin Pharmacol. 2005;61:491–7.
Cavaco I, Piedade R, Gil JP, Ribeiro V. CYP2C8 polymorphism among the Portuguese. Clin Chem Lab Med. 2006;44:168–70.
Acknowledgements
This study was supported by the University of Jordan in Amman and the Hashemite University in Zarqa, Jordan. Finally, the authors appreciative and acknowledge the support of Circassians and Chechens by their partnership on this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest, financial or otherwise.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Abudahab, S., Hakooz, N., Tobeh, N. et al. Variability of CYP2C8 Polymorphisms in Three Jordanian Populations: Circassians, Chechens and Jordanian-Arabs. J Immigrant Minority Health 24, 1167–1176 (2022). https://doi.org/10.1007/s10903-021-01264-x
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10903-021-01264-x