Abstract
The past 20 years has witnessed the development of technologies designed to measure changes in the expressed human genome, including the levels of RNA transcripts, proteins, and metabolites. Gene expression profiling, or the measurement of RNA transcripts, allows investigators to obtain a snapshot of a subject’s current physiological state and may be used to assess disease likelihood. In this review, we provide an overview of recent work using peripheral blood gene expression to assess coronary artery disease (CAD) and discuss the best approaches for developing and validating tests utilizing such gene expression signatures.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Over the past 20 years, tremendous gains have been made in understanding the structure and function of the human genome [1]. Advances in technology have allowed investigators to interrogate genes, transcripts, proteins, and metabolites at the genome-wide level, with innovation progressing at a rapid pace [2]. Due to the prominent genetic nature of cancer, oncology has led the implementation of genomic-based medicine and is the initial focus of the precision medicine initiative recently announced by the President of the USA [3, 4]. The near-term goals of this initiative are to utilize molecular signatures to develop targeted tumor therapies and to better understand mechanisms underlying drug resistance, including the development of new tumor cell models to aid in these tasks. A longer-term goal is to collect biological, clinical, and environmental data from one million volunteer subjects using electronic medical records and mobile health devices, allowing investigators to look beyond cancer and apply precision medicine to other diseases. Coronary artery disease (CAD) is one such disease that should benefit greatly from this initiative, as a firm foundation has already been established in the development of molecular- and genomic-based tools for the diagnosis and treatment of CAD [5].
Although it has been long known that familial history is a good predictor of CAD risk, and population-based studies such as the CARDIoGRAM consortium have independently replicated a moderate number of CAD-associated loci, these single-nucleotide polymorphisms (SNPs) only account for ~10 % of CAD heritability and have relatively low effect sizes [6]. Using systems biology and network-based approaches to incorporate existing SNPs and biomarkers with novel variants and other genomic markers that may be discovered in large studies such as those outlined in the precision health initiative may provide novel insights into complex disorders such as CAD [7, 8]. Gene expression profiling, which measures changes in gene transcript levels in response to alterations in biological state and can provide a dynamic measurement of a patient’s current physiological condition, holds great promise as platform that can be utilized in precision health. In this article, we review what has already been accomplished using peripheral blood gene expression profiling to assess CAD and discuss the best approaches for developing and validating such signatures.
Peripheral Blood Gene Expression Studies in CAD
It has long been known that the cellular and molecular basis of atherosclerotic plaque development has a strong systemic inflammatory component involving cells of both the innate and adaptive immune systems [9, 10]. The deposition of oxidized lipids into the vascular bed initiates the process with subsequent responses by endothelial, vascular smooth muscle cells, and circulating immune cells. A large body of evidence supports the role of monocytes/macrophages in the development and progression of CAD [10], and it has been recently demonstrated that neutrophils also play a key role in CAD progression and plaque instability [11, 12]. Work in recent years has also highlighted both an athero-protective and atherogenic role for the adaptive immune system in the development of atherosclerosis [13]. In sum, strong evidence supports a role for multiple immune cell types in the development and progression of CAD and suggests that gene expression profiling in peripheral blood is a viable approach for monitoring the presence and progression of CAD.
To date, a number of studies have been published examining whole blood gene expression profiling as a method to identify subjects at risk for CAD (Table 1) [14–18]. Comparison of significant genes has shown limited overlap (Fig. 1) [14, 16]; there are a number of reasons for why this might be. Lack of concordance in clinical phenotype can be a major contributor to intra-study differences. In two studies, control populations did not have angiographic evidence supporting disease absence, which could result in decreased power to detect CAD associations [15, 16]. Disease definitions varied between studies, as did patient populations. Different exclusions were applied to patients with a previous history of MI or CAD, as well as other conditions whose presence might confound CAD signals; diabetes and rheumatoid arthritis have each been shown to influence the expression of genes associated with CAD [18, 19] as has the use of steroids or immunosuppressive drugs [20–23]. A variety of gene expression profiling technologies exist (summarized in Table 2) including multiple microarray platforms, which differed between the above studies. In addition, independent technical confirmation and validation of candidate genes in separate cohorts were not consistently implemented. Lastly, poor agreement between genome association studies often indicates that true associations are weak and that individual studies are not well powered to detect associations; i.e., results of individual studies may be qualitatively correct, but marker effect size estimates are overstated [24]. In such underpowered studies, sets of genes that are associated with a given outcome can be unstable due to their correlative nature, a factor that is compounded in peripheral gene expression studies where gene expression patterns can be highly correlated [25, 26].
Venn diagram showing the overlap of genes in the CAD cohorts. Due to the use of different array platforms and annotations in the five coronary artery disease studies described, the diagram likely has missed shared genes and represents a slight underrepresentation of the true overlap. In addition, the list of the 365 genes described in Taurino et al. was not available and thus was not included in the analysis
Pathways for Clinical Test Development
In the previous section, we summarized the results from recent CAD gene expression studies and highlighted the lack of concordance between studies. Developing a gene expression-based diagnostic is complex with many critical factors to be considered. With this in mind, the remaining sections outline approaches for developing diagnostic tests. Using CAD as an example, we describe general stages of diagnostic test development and offer recommendations that focus on core considerations and common pitfalls.
Developing a diagnostic test requires clarity, particularly with respect to the clinical outcome, methods for clinical phenotyping, existing clinical and molecular prediction models and confounders, and intended use population. Test development is a multistage process starting with initial gene discovery and proceeding through test validation and beyond to post-validation studies. The stage definitions and boundaries vary in practice but generally can be defined as follows: (1) gene discovery, (2) prediction model building, (3) prediction model testing, (4) test development, and (5) test validation. Each stage is described below and may include multiple studies (Fig. 2).
Overview of the pathway for new diagnostic research, development, and commercialization, with key studies (a–f). Research is characterized by marker discovery, prediction model building, and testing (a) which may be performed in multiple independent studies (1–3) or in a single step (4). Development begins with transfer of assays from research platforms to commercial using technical replication studies (b) and includes analytical validation and clinical validation (c) which may continue post-development, to broaden support for diagnostic validity. Commercialization includes utility studies (e) or commercial registries, directed at answering questions related to decision impact and patient health outcomes. Additional studies may be performed to demonstrate economic benefits of testing (f)
Gene Discovery
Important technical considerations must be addressed prior to undertaking gene discovery, such as the type of RNA that will be assessed, how samples will be collected, and which technology will be used to assess gene expression levels; examples of these options are summarized in Tables 2, 3, and 4. Gene discovery itself involves the identification and selection of informative gene expression markers from a larger candidate pool; this pool can represent the content on a real-time quantitative polymerase chain reaction (RT-qPCR) panel or microarray or the total sum of detectable transcripts in a biological sample. Gene discovery studies require a variety of design considerations such as the disease phenotype and subphenotypes, study population(s), the disease prevalence in study population, the state of prior evidence supporting candidate markers, and the desired power of the study to detect modest or weak associations. Various study designs have been described in the literature including single step and sequential, and various designs have been implemented for CAD [32–34]. Examples for CAD marker discovery range from single-center matched and unmatched case-control studies [14–16] to multistaged multicentered prospectively recruited cohorts [18]. Arguably, the majority of markers thus far identified as associated with obstructive CAD risk have shown only modest to weak effect sizes. Consequently, two statistical pitfalls of marker discovery are important to note—multiple testing artifacts and winner’s curse.
Multiple Testing
Data sets used for gene discovery are commonly characterized by large initial gene sets and smaller sample sizes and are commonly examined using various statistical models (including and excluding covariates, normalizing and un-normalizing marker expression levels, defining and redefining endpoints, population subsets, etc.). When any large number of tests is performed, it is inappropriate to use traditional single model test thresholds (e.g., p ≤ 0.05) for significance testing, as many apparently significant observations occur by chance. Statistical methods exist for multiple testing corrections [35, 36]; however, these methods are accurate only when the full scope of multiple testing is defined and when the techniques are applied rigorously. Often, the iterative nature of research makes this challenging; in such cases, it may be simpler for researchers to adopt split-sample designs, i.e., to use one part of a cohort for unfettered discovery and hypothesis generation and a separate part for testing a strictly limited number of candidate hypotheses.
Winner’s Curse
It is common for large discovery studies to identify candidate markers whose statistical significance clusters near statistical rejection thresholds and for the initial findings to replicate poorly in subsequent studies. This phenomenon is expected when the disease is only modestly or weakly associated with many markers and where the studies are not reliably powered to detect such associations. In such cases, a subset of true disease associations may achieve statistical significance but do so by being overestimated, by chance. This statistical problem of biased effect size estimates, conditional on statistical significance, is referred to in the literature as the “winner’s curse” or “Beavis effect” and is pronounced when the power of the marker discovery study is low [37, 38]. Consequent failure to confirm an initial finding may thus be due to a true positive finding being overestimated.
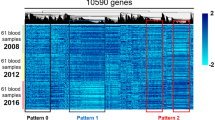
In addition to the above pitfalls, it should be recognized that known clinical risk factors describe a significant portion of CAD. For example, increased risk for CAD is associated with age, sex, smoking, and a variety of patient symptoms [39]; ignoring clinical predictors, or other clinical variables such as medicine usage, during gene discovery may yield markers correlated with different patient strata (Fig. 3). For example, any biological marker that changes value with patient age will therefore be associated with all other age-associated diseases, including CAD. It is important to note that it is possible to identify gene expression changes associated with clinical co-factors associated with CAD; identifying gene expression surrogates for clinical risk factors can be advantageous when clinical data is incomplete and to measure physiological responses to either environmental or biological phenomena, thus adding information beyond clinical data alone. For example, gene expression signatures have been identified for age [40–42], smoking [43–45], hyperlipidemia [46], and hypertension [47], all of which are associated with CAD risk. Finally, whole blood cell populations may change in response to disease states. For instance, the ratio of neutrophils to lymphocytes (N/L ratio) has been shown to be prognostic for MI [48], and a gene surrogate measurement of the N/L ratio has been correlated with the presence of obstructive CAD [18].
Prediction Model Building
The goal of prediction model building is to identify candidate predictors using a set of candidate genes and a family of prediction functions. Ideally, predictors should be chosen from families with the best general performance characteristics. In our experience, however, many families yield similar accuracy, and prediction functions are often chosen from families that are straightforward to interpret such as those where disease probability is modeled by a linear combination of independent variables.
Prediction model building need not be separate from gene discovery, and many statistical methods (e.g., stepwise regression, Random Forest, LASSO) can be used to simultaneously reduce candidate genes and combine them in a predictive model. However, it is useful to treat gene discovery and model building as separate steps, as they may proceed sequentially (e.g., when markers are identified from the literature or by univariate associations). Even when this is not the case, there are specific considerations to model development above and beyond those of gene discovery.
Decisions for prediction model building include the following: (1) model family selection, (2) gene selection, (3) determining model constraints, (4) use of clinical covariates in the model, (5) consideration of population heterogeneity, (6) the data set used for final model selection, and (7) criteria for model acceptance. Of these, the most critical are those that influence model evaluation and acceptance. It is technically valid to reuse gene discovery data for model building; however, use of such data can result in models with overstated performance, as naive reuse of gene discovery data leads to model overfitting and biased model performance [49]. When performing gene discovery and model building on the same data set, statistically valid methods such as cross-validation and bootstrapping can provide relatively unbiased overall performance estimates [50]. However, these methods only work when all steps of discovery are nested within the cross-validation loop [51]. For complex research workflows, it may be simpler to rely instead on an independent test set to evaluate candidate model performance.
The importance of evaluating clinical covariates continues through prediction model building. Clinical covariates or biomarker surrogates may be included in the model or distinct models developed for different covariate strata (e.g., separate models for separate sexes) to ensure a well-calibrated model. As such, matched case-control designs may be of limited value, though unmatched case-control designs may still be necessary for low-prevalence disease applications.
Prediction Model Testing
Disease prediction models are tested to assess diagnostic performance. Testing may occur on the gene discovery cohort (e.g., using cross-validation) or as a distinct step on a reserved sample set. In either case, prediction model testing does not invariably constitute clinical validation.
In many cases, prediction model testing is performed on a discovery cohort or using a split population design. In such cases, model performance cannot be known in advance and the performance testing set cannot be formally powered for confirmatory testing. In such cases, the focus during prediction model testing is to estimate performance. Split sample set designs offer the advantage of allowing both for estimation of performance (e.g., AUC) as well as precision (e.g., confidence intervals) in the reserved test set. Cross-validation approaches offer advantages of increased power for marker discovery and model building at the costs of some bias and difficulty in characterizing precision of performance estimates.
Many methods used to build prediction models do not require that individual model terms achieve statistical significance. Resulting models may perform well overall, though their use introduces an element of uncertainty in explaining or attributing performance. For instance, it may be difficult to know which terms are positively contributing to the model accuracy, which are not, and which (if any) are detrimental to model accuracy. In such cases, it is important to prespecify the baseline for overall model comparison (e.g., a competing clinical model or the base clinical terms of the full-disease prediction model).
Test Development
Diagnostic test development entails the translation of the test as performed in a research setting into a form and process amenable to clinical laboratory. Test development encompasses the complete system required to run the assay, including laboratory instrumentation, reagents, and software implementing the disease prediction model. Development is entered after final product requirements are accepted, with clearly defined design inputs and outputs and should be managed under a quality system. Technical replication, necessary when assay platforms are changed such as from microarray to RT-qPCR, can be performed using a subset of samples from the discovery studies, as the objective is to prove accuracy and precision of measurements across the assay’s dynamic range and not diagnostic validity of the test. The principal sample requirements for test development are that the samples represent the analytic and clinical ranges observed during discovery and that sufficient quantity of sample materials remains for repeat testing.
Test Validation
Test validation is composed of two components: analytical validation and clinical validation.
Analytical Validation
Analytical validation demonstrates that markers (e.g., analytes or RNA transcripts in the case of gene expression tests) are correctly measured by the new test. Analytical validation is performed through a series of studies designed to confirm test accuracy, precision, limits of detection or quantitation, robustness against likely interfering substances, stability of reagents and samples against their defined limitations of use, and, potentially, the robustness of the process to variance in assay conditions [52, 53].
Clinical Validation
Clinical validation studies demonstrate that the diagnostic performance of the test meets predefined acceptance criteria. Design considerations for such studies, including sample size calculations for categorical (e.g., diagnostic sensitivity and specificity) and continuous endpoints (e.g., AUC), are complex but well defined in the statistical literature [54]. As with analytical validation, clinical validation may appear to recapitulate findings of previous studies. However, clinical validation serves as a strong claim of diagnostic performance. Clinical validation studies have the following characteristics: (1) validation is performed in the intended use population; (2) the study cohort is independent of discovery cohorts; (3) diagnostic testing is performed using the final version of the diagnostic assay(s), with qualified materials and equipment, executed in the clinical laboratory setting; (4) clinical endpoints and performance claims are prespecified, with clear null and alternative hypotheses; and (5) the study is powered to meet the primary objective (or co-primary objectives, when claims of diagnostic sensitivity and specificity are desired) (for a more detailed description of diagnostic accuracy study reporting and evaluation considerations, see Bossuyt et al. and Whiting et al. [55, 56]). Clinical validation studies are not intrinsically single studies, as it may be necessary to validate multiple claims (e.g., in different intended use populations) or to strengthen initial claims (e.g., by raising the stringency of the null hypotheses). Indeed, in the case of any new diagnostic test, it may be difficult to perform the first validation study in the desired intended use population due to costs and risks of gold standard testing on all study participants, such as invasive coronary angiography. In such cases, initial validation studies may be focused on higher-risk populations [57, 58].
Beyond Gene Expression
The development and clinical validation of a peripheral blood gene expression diagnostic test can be challenging although achievable; the choice of the appropriate platform and adherence to rigorous experimental, clinical, and statistical approaches is paramount to success. As mentioned previously, although gene expression profiling in whole blood is a powerful approach, it has inherent limitations especially when applied to a disease such as CAD where the measurement of the disease process may be indirect.
To remedy this, the incorporation of other types of biomarkers, whether they are gene expression based such as the measurement of circulating RNAs or other types of markers (genetic, proteomic, metabolite, etc.), should be considered. A number of studies have examined the interactions between genetics and gene expression (for a general review, please see Cookson et al. [59]), and it has been suggested that examining genetic-gene expression interactions in CAD may be a powerful approach to further understanding coronary disease [8]. One example of this approach is illustrated in a study using the same subject set described in Joehanes et al. [60] (Table 1). In this study, the investigators identified co-expression modules that were differentially represented in either CHD cases or age- and sex-matched controls and demonstrated that these differential modules were enriched in CHD risk expression SNPs (eSNPs), loci known to be associated with increased CHD risk and also to alter gene expression. This approach led to the identification genes involved in B cell activation, immune response, and ion transport, as well as higher-level regulatory drivers. The same group of researchers also successfully employed a similar approach to investigate miRNA-mRNA-SNP interactions in the same set of subjects [61]. In addition to genetics, the inclusion of other biomarkers in a systems biology approach may strengthen the performance of a diagnostic test for CAD by incorporating orthogonal signals that may reflect different biological aspects of the disease [62].
As the field of genomics continues to develop and mature, the incorporation of precision medicine into clinical practice will continue to progress and holds great promise for altering the diagnosis and treatment of cardiovascular disease.
Abbreviations
- CAD:
-
Coronary artery disease
- MI:
-
Myocardial Infarction
- SNP:
-
Single-nucleotide polymorphism
- RT-qPCR:
-
Real-time quantitative polymerase chain reaction
- PBMC:
-
peripheral blood mononuclear cell
- RNAseq:
-
RNA sequencing
References
Lander, E. S. (2011). Initial impact of the sequencing of the human genome. Nature, 470(7333), 187–197.
Hayden, E. C. (2014). Technology: the $1,000 genome. Nature, 507(7492), 294–295.
Collins, F. S., & Varmus, H. (2015). A new initiative on precision medicine. The New England Journal of Medicine, 372(9), 793–795.
Fox, J. L. (2015). Obama catapults patient-empowered precision medicine. Nature Biotechnology, 33(4), 325.
Eagle, K. A., Ginsburg, G. S., Musunuru, K., Aird, W. C., Balaban, R. S., Bennett, S. K., et al. (2010). Identifying patients at high risk of a cardiovascular event in the near future: current status and future directions: report of a national heart, lung, and blood institute working group. Circulation, 121(12), 1447–1454.
Consortium, C. A. D, Deloukas, P., Kanoni, S., Willenborg, C., Farrall, M., Assimes, T. L., et al. (2013). Large-scale association analysis identifies new risk loci for coronary artery disease. Nature Genetics, 45(1), 25–33.
Bjorkegren, J. L., Kovacic, J. C., Dudley, J. T., & Schadt, E. E. (2015). Genome-wide significant loci: how important are they?: systems genetics to understand heritability of coronary artery disease and other common complex disorders. Journal of the American College of Cardiology, 65(8), 830–845.
Foroughi Asl, H., Talukdar, H. A., Kindt, A. S., Jain, R. K., Ermel, R., Ruusalepp, A., et al. (2015). Expression quantitative trait loci acting across multiple tissues are enriched in inherited risk for coronary artery disease. Circulation. Cardiovascular Genetics, 8, 305–315.
Hansson, G. K., Libby, P., Schonbeck, U., & Yan, Z. Q. (2002). Innate and adaptive immunity in the pathogenesis of atherosclerosis. Circulation Research, 91(4), 281–291.
Libby, P. (2002). Inflammation in atherosclerosis. Nature, 420(6917), 868–874.
Della Bona, R., Cardillo, M. T., Leo, M., Biasillo, G., Gustapane, M., Trotta, F., et al. (2013). Polymorphonuclear neutrophils and instability of the atherosclerotic plaque: a causative role? Inflammation Research, 62(6), 537–550.
Soehnlein, O. (2012). Multiple roles for neutrophils in atherosclerosis. Circulation Research, 110(6), 875–888.
Ammirati, E., Moroni, F., Magnoni, M., & Camici, P. G. (2015). The role of T and B cells in human atherosclerosis and atherothrombosis. Clinical and Experimental Immunology, 179(2), 173–187.
Sinnaeve, P. R., Donahue, M. P., Grass, P., Seo, D., Vonderscher, J., Chibout, S. D., et al. (2009). Gene expression patterns in peripheral blood correlate with the extent of coronary artery disease. PloS One, 4(9), e7037.
Taurino, C., Miller, W. H., McBride, M. W., McClure, J. D., Khanin, R., Moreno, M. U., et al. (2010). Gene expression profiling in whole blood of patients with coronary artery disease. Clinical Science (London), 119(8), 335–343.
Joehanes, R., Ying, S., Huan, T., Johnson, A. D., Raghavachari, N., Wang, R., et al. (2013). Gene expression signatures of coronary heart disease. Arteriosclerosis, Thrombosis, and Vascular Biology, 33(6), 1418–1426.
Wingrove, J. A., Daniels, S. E., Sehnert, A. J., Tingley, W., Elashoff, M. R., Rosenberg, S., et al. (2008). Correlation of peripheral-blood gene expression with the extent of coronary artery stenosis. Circulation. Cardiovascular Genetics, 1(1), 31–38.
Elashoff, M. R., Wingrove, J. A., Beineke, P., Daniels, S. E., Tingley, W. G., Rosenberg, S., et al. (2011). Development of a blood-based gene expression algorithm for assessment of obstructive coronary artery disease in non-diabetic patients. BMC Medical Genomics, 4(1), 26.
Niu, X., Lu, C., Xiao, C., Zhang, Z., Jiang, M., He, D., et al. (2014). The shared crosstalk of multiple pathways involved in the inflammation between rheumatoid arthritis and coronary artery disease based on a digital gene expression profile. PloS One, 9(12), e113659.
Blits, M., Jansen, G., Assaraf, Y. G., van de Wiel, M. A., Lems, W. F., Nurmohamed, M. T., et al. (2013). Methotrexate normalizes up-regulated folate pathway genes in rheumatoid arthritis. Arthritis and Rheumatism, 65(11), 2791–2802.
Czock, D., Keller, F., Rasche, F. M., & Haussler, U. (2005). Pharmacokinetics and pharmacodynamics of systemically administered glucocorticoids. Clinical Pharmacokinetics, 44(1), 61–98.
Julia, A., Barcelo, M., Erra, A., Palacio, C., & Marsal, S. (2009). Identification of candidate genes for rituximab response in rheumatoid arthritis patients by microarray expression profiling in blood cells. Pharmacogenomics, 10(10), 1697–1708.
van Baarsen, L. G., Wijbrandts, C. A., Gerlag, D. M., Rustenburg, F., van der Pouw Kraan, T. C., Dijkmans, B. A., et al. (2010). Pharmacogenomics of infliximab treatment using peripheral blood cells of patients with rheumatoid arthritis. Genes and Immunity, 11(8), 622–629.
Young, N. S., Ioannidis, J. P., & Al-Ubaydli, O. (2008). Why current publication practices may distort science. PLoS Medicine, 5(10), e201.
Preininger, M., Arafat, D., Kim, J., Nath, A. P., Idaghdour, Y., Brigham, K. L., et al. (2013). Blood-informative transcripts define nine common axes of peripheral blood gene expression. PLoS Genetics, 9(3), e1003362.
Simon, R. (2005). Roadmap for developing and validating therapeutically relevant genomic classifiers. Journal of Clinical Oncology, 23(29), 7332–7341.
Chen, F., Zhao, X., Peng, J., Bo, L., Fan, B., & Ma, D. (2014). Integrated microRNA-mRNA analysis of coronary artery disease. Molecular Biology Reports, 41(8), 5505–5511.
De Rosa, S., Curcio, A., & Indolfi, C. (2014). Emerging role of microRNAs in cardiovascular diseases. Circulation Journal, 78(3), 567–575.
Fichtlscherer, S., De Rosa, S., Fox, H., Schwietz, T., Fischer, A., Liebetrau, C., et al. (2010). Circulating microRNAs in patients with coronary artery disease. Circulation Research, 107(5), 677–684.
Jarinova, O., Stewart, A. F., Roberts, R., Wells, G., Lau, P., Naing, T., et al. (2009). Functional analysis of the chromosome 9p21.3 coronary artery disease risk locus. Arteriosclerosis, Thrombosis, and Vascular Biology, 29(10), 1671–1677.
Liu, Y., Sanoff, H. K., Cho, H., Burd, C. E., Torrice, C., Mohlke, K. L., et al. (2009). INK4/ARF transcript expression is associated with chromosome 9p21 variants linked to atherosclerosis. PloS One, 4(4), e5027.
Dudbridge, F., Gusnanto, A., & Koeleman, B. P. (2006). Detecting multiple associations in genome-wide studies. Human Genomics, 2(5), 310–317.
Skates, S. J., Gillette, M. A., LaBaer, J., Carr, S. A., Anderson, L., Liebler, D. C., et al. (2013). Statistical design for biospecimen cohort size in proteomics-based biomarker discovery and verification studies. Journal of Proteome Research, 12(12), 5383–5394.
Wallstrom, G., Anderson, K. S., & LaBaer, J. (2013). Biomarker discovery for heterogeneous diseases. Cancer Epidemiology, Biomarkers and Prevention, 22(5), 747–755.
Dudoit, S., & van der Laan, M. (2007). Multiple testing procedures with applications to genomics (Springer Series in Statistics). Springer.
Holm, S. (1979). A simple sequentially rejective multiple test procedure. Scandinavian Journal of Statistics, 6(2), 65–70.
Ioannidis, J. P. (2008). Why most discovered true associations are inflated. Epidemiology, 19(5), 640–648.
Xu, S. (2003). Theoretical basis of the Beavis effect. Genetics, 165(4), 2259–2268.
Chaitman, B. R., Bourassa, M. G., Davis, K., Rogers, W. J., Tyras, D. H., Berger, R., et al. (1981). Angiographic prevalence of high-risk coronary artery disease in patient subsets (CASS). Circulation, 64(2), 360–367.
Harries, L. W., Hernandez, D., Henley, W., Wood, A. R., Holly, A. C., Bradley-Smith, R. M., et al. (2011). Human aging is characterized by focused changes in gene expression and deregulation of alternative splicing. Aging Cell, 10(5), 868–878.
Hong, M. G., Myers, A. J., Magnusson, P. K., & Prince, J. A. (2008). Transcriptome-wide assessment of human brain and lymphocyte senescence. PloS One, 3(8), e3024.
Marttila, S., Jylhava, J., Nevalainen, T., Nykter, M., Jylha, M., Hervonen, A., et al. (2013). Transcriptional analysis reveals gender-specific changes in the aging of the human immune system. PloS One, 8(6), e66229.
Beineke, P., Fitch, K., Tao, H., Elashoff, M. R., Rosenberg, S., Kraus, W. E., et al. (2012). A whole blood gene expression-based signature for smoking status. BMC Medical Genomics, 5, 58.
Charlesworth, J. C., Curran, J. E., Johnson, M. P., Goring, H. H., Dyer, T. D., Diego, V. P., et al. (2010). Transcriptomic epidemiology of smoking: the effect of smoking on gene expression in lymphocytes. BMC Medical Genomics, 3, 29.
Lampe, J. W., Stepaniants, S. B., Mao, M., Radich, J. P., Dai, H., Linsley, P. S., et al. (2004). Signatures of environmental exposures using peripheral leukocyte gene expression: tobacco smoke. Cancer Epidemiology, Biomarkers and Prevention, 13(3), 445–453.
Inouye, M., Silander, K., Hamalainen, E., Salomaa, V., Harald, K., Jousilahti, P., et al. (2010). An immune response network associated with blood lipid levels. PLoS Genetics, 6(9), e1001113.
Huan, T., Esko, T., Peters, M. J., Pilling, L. C., Schramm, K., Schurmann, C., et al. (2015). A meta-analysis of gene expression signatures of blood pressure and hypertension. PLoS Genetics, 11(3), e1005035.
Horne, B. D., Anderson, J. L., John, J. M., Weaver, A., Bair, T. L., Jensen, K. R., et al. (2005). Which white blood cell subtypes predict increased cardiovascular risk? Journal of the American College of Cardiology, 45(10), 1638–1643.
Harrell, F. E., Jr., Lee, K. L., & Mark, D. B. (1996). Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Statistics in Medicine, 15(4), 361–387.
Efron, B., & Tibshirani, R. J. (1994). An introduction to the bootstrap (Chapman & Hall/CRC Monographs on Statistics & Applied Probability). Chapman and Hall/CRC.
Varma, S., & Simon, R. (2006). Bias in error estimation when using cross-validation for model selection. BMC Bioinformatics, 7, 91.
Jennings, L., Van Deerlin, V. M., Gulley, M. L., & College of American Pathologists Molecular Pathology Resource, C. (2009). Recommended principles and practices for validating clinical molecular pathology tests. Archives of Pathology and Laboratory Medicine, 133(5), 743–755.
Elashoff, M. R., Nuttall, R., Beineke, P., Doctolero, M. H., Dickson, M., Johnson, A. M., et al. (2012). Identification of factors contributing to variability in a blood-based gene expression test. PloS One, 7(7), e40068.
Qiu, P. (2005). The statistical evaluation of medical tests for classification and prediction. Journal of the American Statistical Association, 100(470), 705.
Bossuyt, P. M., Reitsma, J. B., Bruns, D. E., Gatsonis, C. A., Glasziou, P. P., Irwig, L. M., et al. (2003). The STARD statement for reporting studies of diagnostic accuracy: explanation and elaboration. The Standards for Reporting of Diagnostic Accuracy Group. Croatian Medical Journal, 44(5), 639–650.
Whiting, P. F., Rutjes, A. W., Westwood, M. E., Mallett, S., Deeks, J. J., Reitsma, J. B., et al. (2011). QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Annals of Internal Medicine, 155(8), 529–536.
Rosenberg, S., Elashoff, M. R., Beineke, P., Daniels, S. E., Wingrove, J. A., Tingley, W. G., et al. (2010). Multicenter validation of the diagnostic accuracy of a blood-based gene expression test for assessing obstructive coronary artery disease in nondiabetic patients. Annals of Internal Medicine, 153(7), 425–434.
Thomas, G. S., Voros, S., McPherson, J. A., Lansky, A. J., Winn, M. E., Bateman, T. M., et al. (2013). A blood-based gene expression test for obstructive coronary artery disease tested in symptomatic nondiabetic patients referred for myocardial perfusion imaging the COMPASS study. Circulation. Cardiovascular Genetics, 6(2), 154–162.
Cookson, W., Liang, L., Abecasis, G., Moffatt, M., & Lathrop, M. (2009). Mapping complex disease traits with global gene expression. Nature Reviews Genetics, 10(3), 184–194.
Huan, T., Zhang, B., Wang, Z., Joehanes, R., Zhu, J., Johnson, A. D., et al. (2013). A systems biology framework identifies molecular underpinnings of coronary heart disease. Arteriosclerosis, Thrombosis, and Vascular Biology, 33(6), 1427–1434.
Huan, T., Rong, J., Tanriverdi, K., Meng, Q., Bhattacharya, A., McManus, D. D., et al. (2015). Dissecting the roles of microRNAs in coronary heart disease via integrative genomic analyses. Arteriosclerosis, Thrombosis, and Vascular Biology, 35(4), 1011–1021.
Abraham, G., Bhalala, O. G., de Bakker, P. I., Ripatti, S., & Inouye, M. (2014). Towards a molecular systems model of coronary artery disease. Current Cardiology Reports, 16(6), 488.
Acknowledgments
The authors would like to thank Siw Daniels, Andrea Johnson, and Phil Beineke for their valuable comments in reviewing the manuscript.
Conflict of Interest
BR and JAW are employees and have equity interests or stock options in CardioDx.
Human/Animal Subjects
No human or animal studies were carried out by the authors for this article.
Funding
This study was funded by CardioDx.
Author information
Authors and Affiliations
Corresponding author
Additional information
Editor-in-Chief Jennifer L. Hall oversaw the review of this article
Rights and permissions
About this article
Cite this article
Rhees, B., Wingrove, J.A. Developing Peripheral Blood Gene Expression-Based Diagnostic Tests for Coronary Artery Disease: a Review. J. of Cardiovasc. Trans. Res. 8, 372–380 (2015). https://doi.org/10.1007/s12265-015-9641-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12265-015-9641-5