Abstract
Triple negative breast cancer (TNBC), an agressive subtype accounts nearly 15 % of all breast carcinomas. Conventional chemotherapy is the only treatment modality thus new, effective targeted therapy methods have been investigated. Epidermal growth factor receptor (EGFR) inhibitors give hope according to the recent studies results. Also therapeutic agents have been tried against aberrant p53 signal activity as TNBC show high p53 mutation rates. Our aim was to detect the incidence of mutations/amplifications identified in TNBC in our population. Here we used sequence analysis to detect HER2 (exon 18–23), p53 (exon 5–8) mutations; fluorescence in situ hybridization (FISH) method to analyse EGFR/chromosome 7 centromere gene status in 82 immunohistochemically TNBC. Basaloid phenotype was identified in 49 (59.8 %) patients. EGFR amplification was noted in 5 cases (6.1 %). All EGFR amplified cases showed EGFR overexpression by immunohistochemistry (IHC). p53 mutations were identified in 33 (40.2 %) cases. Almost 60 % of the basal like breast cancer cases showed p53 mutation. Only one case showed HER2 mutation (exon 20:g.36830_3). Our results showed that gene amplification is not the unique mechanism in EGFR overexpression. IHC might be used in the decision of anti-EGFR therapy in routine practice. p53 mutation rate was lower than the rates reported in the literature probably due to ethnic differences and low sensitivity of sanger sequences in general mutation screening. We also established the rarity of HER2 mutation in TNBC. In conclusion EGFR and p53 are the major targets in TNBC also for our population.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancers have been categorized into five groups according to the gene expression profiles: luminal A ( estrogen receptor (ER) (+) and/or progesterone receptor (PR) (+), human epidermal growth factor receptor 2 (HER2) (-), Ki-67 < 14 %); luminal B (ER + and/or PR+, HER2 -, Ki-67 ≥ 14 % or ER + and/or PR +, HER2 +); HER2-enriched (ER-, PR-, HER2+); basal cell-like and normal breast like types [1]. The luminal A subtype is associated with better prognosis, whereas the basal cell-like subtype is more aggressive [2].
Triple negative breast cancer (TNBC ) (ER-, PR-, HER2-) accounts nearly 15 % of all breast carcinomas. TNBC has aggressive biological behaviour [3]. Conventional chemotherapy is the only treatment modality currently; thus new, effective targeted therapy methods have been investigated. Most of the TNBCs show epidermal growth factor receptor (EGFR) overexpression and p53 mutation. EGFR inhibitors and poly (ADP-ribose) polymerase inhibitors gives hope according to the recent study reports [4, 5]. Also there are several preclinical and clinical trials targeting p53 pathway function [6, 7]. Our aim was to detect the incidence of p53 and HER2 mutations and EGFR amplification in TNBC cases in our population and shed light on targeted therapy decisions.
Materials and Methods
Eighty-two consecutive triple negative breast cancer cases diagnosed between 2008 and 2013 at Izmir Tepecik Training and Research Hospital were selected for the study. All specimens were formalin-fixed, paraffin embedded and processed according to the institute’s standardised protocols. The sections were prepared from archived paraffin blocks and were processed in parallel for immunohistochemistry (IHC), fluorescence in situ hybridization (FISH) and sanger sequence methods.
Triple negative status was confirmed by ER, PR, HER2 IHC and HER2 FISH methods. Cytokeratin 5/6 (CK5/6) and EGFR IHC analysis were performed to identify basaloid phenotype. The cases showing CK 5/6 positivity ( ≥ %5) and/or EGFR overexpression (2+, 3+) were considered as basaloid phenotype.
Immunohistochemistry
Immunohistochemical detection of EGFR (clone E30, 1/25 dilution, Dako), CK 5/6 (clone D5/16 B4, Dako), Ki67 (clone MIB1, Dako) was done on Autostainer Link 48 (Dako, Denmark).
EGFR positivity was considered when tumor cells showed weak or intense complete membranous staining (2+, 3+) with or without cytoplasmic reaction [8].
HER2 FISH Method
All of the cases were analysed in terms of HER2 and chromosome 17 centromere (CEP 17) gene status by instant quality (IQ) FISH (Dako, Denmark) method.
EGFR FISH Method
EGFR and chromosome 7 centromere copy number were investigated by EGFR FISH pharmDx Kit (Dako, Denmark). Sections were incubated at 58 °C for 30 min, deparaffinized in two series of xylol and rehydrated with ethanol series. Slides were pretreated with pretreatment solution in a water bath at 95 °C for 15 min. Enzymatic digestion was carried out with pepsin for 4 min at 37 °C on hybridizer. After dehydration, 10 μl of EGFR/cen7 probe mix was applied to each tissue section. The slides and probe were denatured at 82 °C for 5 min and hybridized at 45 °C for 12 h. After hybridization period the slides were washed with stringent wash buffer at 65 °C for 10 min in a water bath. Then the slides were dehydrated and 15 μl of fluorescence mounting medium containing 4’, 6-diamino-2-phenylindole (DAPI) was applied.
EGFR amplification was defined as high polysomy (≥4 copies in ≥40 % of cells) or gene amplification (EGFR gene clusters and ratio of ≥2) [9].
Sequence Analysis
Genomic DNA from paraffin embedded tissue was extracted using the Genomic DNA QIAamp DNA FFPE Tissue Extraction Kit according to the manufacturer’s instructions (Qiagen, Germany). PCR reaction was carried out in 50 μL of solution containing 100 ng of genomic DNA, 0.1 μmol/L of each primer, 100 μmol/L of each dNTP, 20 mmol/L of TrisHCl (pH 8.5), 50 mmol/L of KCl,3 mmol/L of MgCl2, and 1.0 U of Taq polymerase (Nanohelix, South Korea) using following M13 tagged pairs of TP53 exon 5, 6, 7, 8 and HER2 exon 18, 19, 20, 21 ,22, 23 primers (Table 1). The amplification was performed on thermocycler (Perkin Elmer 9700), with a predenaturing procedure for 10 min at 95 °C for 35 cycles (denaturing at 94 °C for 30 s, annealing at 60 °C for 1 min, and extension at 72 °C for 1 min), followed by an additional 7 min incubation at 72 °C. PCR products were purified by using Exosap-IT according to the manufacturer’s instructions (Affymetrix, USA) and subjected to automatic sequence analysis (Automated sequencer ABI 3130; Applied Biosystems, CA 94404, USA) by Big Dye terminator reaction according to the supplier’s instructions (ABI Prism Big Dye Terminator Cycle Sequencing Ready Reaction Kits Version 3.1; Applied Biosystems, CA 94404, USA) using M13 sequencing primer . The obtained sequences for exon 5 to 8 of TP53 gene and exon 18 to 23 of HER2 gene were analyzed by SeqScape version 2.6 software (ABI, USA).
Results
The mean age of the cases was 56.1 years (min: 24 years, max: 88 years) and the mean tumor diameter was 2.7 cm (min: 1 cm, max: 7 cm). The histological subtype was invasive ductal carcinoma in 76 (92.7 %), medullary carcinoma in 4 (4.9 %), metaplastic carcinoma in 2 (2.4 %) cases. Of the 82 cases 16 (19.5 %) were grade 2, 66 (80.5 %) were grade 3.
All cases were immunohistochemically negative for ER, PR and HER2. None of the cases showed HER2 amplification by HER2 IQ FISH method.
Cytokeratin 5/6 positivity was noted in 43 (52.4 %), EGFR overexpression in 38 (46.3 %) cases. Basal-like phenotype was identified in 49 (59.8 %) cases by using CK 5/6 and EGFR IHC. Mean Ki67 proliferation index was 49.1 % (min: 5 %, max:100 %), p53 positivity rate was 57.8 % (min: 5 %, max:100 %).
EGFR gene was amplified in 5 of 82 (6.1 %) cases by FISH method. Three of the amplified cases showed high level amplification with clusters, two of the FISH positive cases showed high polysomy. Of the five EGFR amplified cases, three showed (+++), 2 showed (++) immunoreactivity by IHC (Figs. 1 and 2). EGFR overexpression was statistically significantly correlated with EGFR amplification (p = 0.01).
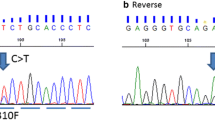
Sequence analysis revealed p53 mutations in 33 cases (40.2 %). The mutations were in exon 5 in 10 cases (30.3 %), exon 6 in 9 cases (27.3 %), exon 7 in 6 cases (18.2 %), exon 8 in 8 cases (24.2 %) (Fig. 3a and b). Of the 33 mutated cases, 19 (57.5 %) were missense, 5 (15.1 %) were nonsense, 1 (3.3 %) was frameshift insertion, 8 (24.1 %) were frameshift deletions. Almost 60 % (29/49) of the basal like breast cancer cases showed p53 mutation. p53 mutation was statistically significantly correlated with basaloid phenotype (p = 0.00). Though there was no significant relation between p53 mutation and EGFR overexpression, EGFR amplification, Ki67 proliferation index (p = 0.44, p = 0.38, p = 0.16 respectively).
Sequence analysis revealed HER2 gene mutation in only one case (mutation in exon 20:g.36830_3). This case was 60 years old. Histologic subtype was invasive ductal carcinoma (grade 3). HER2, EGFR and CK5/6 were negative by IHC. FISH and sequence analysis did not reveal EGFR, HER2 amplification and p53 mutation respectively.
Discussion
The most appropriate therapy for TNBC is an controversial issue. Conventional cytotoxic chemotherapy is the only treatment modality recommended by the National Comprehensive Cancer Network. But there are several preclinical and clinical studies on novel therapy modalities targeting EGFR and p53.
Nearly 75 % of TNBCs are basal like tumors. Basal like and triple negative tumors show different messenger RNA (mRNA) expression patterns in gene profiling studies. Basal like tumors tend to express c-kit, EGFR and mutant forms of p53, while TNBCs show more heterogeneous mRNA expression patterns [10, 11]. Park et al showed that combined alterations of EGFR, p53, PTEN together contribute to basal like breast cancer on human mammary epithelial cancer cell models [12]. In a study analysing the molecular portraits of human breast tumors, p53 mutation was reported in 12 % of luminal A, 32 % of luminal B, 75 % of HER2 and 84 % of basal-like breast cancers [13]. Triple negative/basal like tumors show high frequency of p53 mutations (60-88 %) [14, 15]. Grob et al found p53 mutations in 57.1 % of TNBC cases [16]. In our study almost 60 % of the basal like breast cancer cases showed p53 mutation. p53 mutation was statistically significantly correlated with basaloid phenotype (p = 0.00). There are so many ongoing preclinical and clinical studies on several agents targeting p53 pathway function in TNBC however optimal target has not been identified as p53 pathway function is not completely understood. So targeting mutant p53 or its downstream effectors is still challenging.
EGFR amplification is reported in 1–18 % of breast cancer cases [8, 17–19]. However TNBCs show high frequency of EGFR overexpression [20, 21]. EGFR is also overexpressed in up to 70 % of basal-like breast cancer [22]. Ryden et al compared EGFR overexpression and amplification rates between TNBC and non-TNBC. EGFR overexpression/amplification rates were 41 and 18 % in TNBC whereas 11 and 6 % in non-TNBC cases [19]. Gumuskaya et al reported EGFR gene amplification in one and high polysomy in 9 of 62 TNBCs [18]. Grob et al found true EGFR amplification in only one case (1/65) [16]. Shao et al reported EGFR amplification in 7 of 55 overexpressed cases [23]. In our study EGFR overexpression was determined in 38 (46.3 %) cases though EGFR gene was amplified in 5 (6.1 %) cases by FISH method. In our study we used IHC for determining EGFR protein expression and FISH for EGFR gene copy number. All of the EGFR amplified cases showed EGFR overexpression. But in the remaining 33 cases, protein overexpression was not due to the EGFR amplification. According to our knowledge there are so many mechanisms in protein overexpression out of gene amplification such as epigenetic factors, miRNAs and undefined mutations.
Because of high EGFR overexpression rates, novel approaches targeting growth-promoting proteins such as EGFR, poly ADP-ribose polymerase (PARP) are being investigated. Viale et al. recently found the EGFR protein expression score to be a prognostic factor among TNBC supporting EGFR-targeted therapy to be clinically relevant [24]. Tang et al reported that TNBC cases with EGFR overexpression have increased pathologic complete response rates to neoadjuvant chemotherapy when compared with non-TNBC cases [25]. In patients with metastatic lung and colorectal cancer, EGFR gene copy number is reported to be a superior predictive marker for EGFR-inhibiting treatment compared to EGFR protein expression [9, 26]. The response rates in EGFR overexpressed and amplified TNBC cases versus EGFR overexpressed but not amplified cases should be clarified in further clinical studies.
Studies on lung cancer showed that HER2 inhibitors might be effective in tumors with activating HER2 mutations. Li et al found HER2 exon 20 mutations in 2 of 107 lung adenocarcinoma cases of which one of them was insertion [27]. Grob et al found heterozygous missense mutation in exon 19 of the HER2 gene (p.L755S) in one of the 65 TNBC case [16]. We found mutation in exon 20:g.36830_3 in only one case.
Our results showed that gene amplification is not the only mechanism in EGFR overexpression. IHC can be used in the decision of anti-EGFR therapy in routine practice. P53 mutation rate was lower than the rates reported in the literature probably due to ethnic differences and low sensitivity of sanger sequences in general mutation screening. P53 mutation was correlated with basaloid phenotype. We also established the rarity of HER2 mutation in TNBC. In conclusion EGFR and p53 are the major targets in TNBC also in our population.
References
Sorlie T, Perou CM, Tibshirani R et al (2001) Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A 98(19):10869–10874
Carey LA, Perou CM, Livasy CA et al (2006) Race, breast cancer subtypes, and survival in the carolina breast cancer study. JAMA 295(21):2492–2502, PMID:16757721
Oakman C, Viale G, Di Leo A (2010) Management of triple negative breast cancer. Breast 19(5):312–321. doi:10.1016/j.breast.2010.03.026, PMID:20382530
Carey LA, Rugo HS, Marcom PK et al (2012) TBCRC 001: randomized phase II study of cetuximab in combination with carboplatin in stage IV triple-negative breast cancer. J Clin Oncol 30(21):2615–2623. doi:10.1200/JCO.2010.34.5579
O’Shaughnessy J, Osborne C, Pippen JE, Yoffe M, Patt D, Rocha C, Koo IC, Sherman BM, Bradley C (2011) Iniparib plus chemotherapy in metastatic triple-negative breast cancer. N Engl J Med 364(3):205–214. doi:10.1056/NEJMoa1011418
Langerod A, Zhao H, Borgan O, Nesland JM, Bukholm IR, Ikdahl T, Kåresen R, Borresen-Dale AL, Jeffrey SS (2007) TP53 mutation status and gene expression profiles are powerful prognostic markers of breast cancer. Breast Cancer Res 9(3):R30, PMID:17504517
Turner N, Moretti E, Siclari O, Migliaccio I, Santarpia L, D’Incalci M, Piccolo S, Veronesi A, Zambelli A, Del Sal G, Di Leo A (2013) Targeting triple negative breast cancer: is p53 the answer? Cancer Treat Rev 39(5):541–550. doi:10.1016/j.ctrv.2012.12.001
Bhargava R, Gerald WL, Li AR, Pan Q, Lal P, Ladanyi M, Chen B (2005) EGFR gene amplification in breast cancer: correlation with epidermal growth factor receptor mRNA and protein expression and HER-2 status and absence of EGFR-activating mutations. Mod Pathol 18(8):1027–1033, PMID:15920544
Hirsch FR, Varella-Garcia M, Bunn PA Jr, Di Maria MV, Veve R, Bremmes RM, Barón AE, Zeng C, Franklin WA (2003) Epidermal growth factor receptor in non-small-cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis. J Clin Oncol 21(20):3798–3807, PMID:12953099
Sorlie T, Tibshirani R, Parker J et al (2003) Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A 100(14):8418–8423, PMID:12829800
Bertucci F, Finetti P, Cervera N, Esterni B, Hermitte F, Viens P, Birnbaum D (2008) How basal are triple-negative breast cancers? Int J Cancer 123(1):236–240. doi:10.1002/ijc.23518, PMID:18398844
Pires MM, Hopkins BD, Saal LH, Parsons RE (2013) Alterations of EGFR, p53 and PTEN that mimic changes found in basal-like breast cancer promote transformation of human mammary epithelial cells. Cancer Biol Ther 14(3):246–253. doi:10.4161/cbt.23297
Network CGA (2012) Comprehensive molecular portraits of human breast tumours. Nature 490(7418):61–70. doi:10.1038/nature11412, PMID:23000897
Dumay A, Feugeas JP, Wittmer E et al (2013) Distinct tumor protein p53 mutants in breast cancer subgroups. Int J Cancer 132(5):1227–1231. doi:10.1002/ijc.27767, PMID:22886769
Shah SP, Roth A, Goya R et al (2012) The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature 486(7403):395–399. doi:10.1038/nature10933, PMID:22495314
Grob TJ, Heilenkötter U, Geist S, Paluchowski P, Wilke C, Jaenicke F, Quaas A, Wilczak W, Choschzick M, Sauter G, Lebeau A (2012) Rare oncogenic mutations of predictive markers for targeted therapy in triple-negative breast cancer. Breast Cancer Res Treat 134(2):561–567. doi:10.1007/s10549-012-2092-7, PMID:22610646
Burness ML, Grushko TA, Olopade OI (2010) Epidermal growth factor receptor in triple-negative and basal-like breast cancer: promising clinical target or only a marker? Cancer J 16(1):23–32. doi:10.1097/PPO.0b013e3181d24fc1, PMID:20164687
Gumuskaya B, Alper M, Hucumenoglu S, Altundag K, Uner A, Guler G (2010) EGFR expression and gene copy number in triple-negative breast carcinoma. Cancer Genet Cytogenet 203(2):222–229. doi:10.1016/j.cancergencyto.2010.07.118
Rydén L, Jirström K, Haglund M, Stål O, Fernö M (2010) Epidermal growth factor receptor and vascular endothelial growth factor receptor 2 are specific biomarkers in triple-negative breast cancer. Results from a controlled randomized trial with long-term follow-up. Breast Cancer Res Treat 120(2):491–498. doi:10.1007/s10549-010-0758-6, PMID:20135347
Corkery B, Crown J, Clynes M, O’Donovan N (2009) Epidermal growth factor receptor as a potential therapeutic target in triple-negative breast cancer. Ann Oncol 20(5):862–867. doi:10.1093/annonc/mdn710, PMID:19150933
Nogi H, Kobayashi T, Suzuki M, Tabei I, Kawase K, Toriumi Y, Fukushima H, Uchida K (2009) EGFR as paradoxical predictor of chemosensitivity and outcome among triple-negative breast cancer. Oncol Rep 21(2):413–417, PMID:19148516
Siziopikou KP, Cobleigh M (2007) The basal subtype of breast carcinomas may represent the group of breast tumors that could benefit from EGFR-targeted therapies. Breast 16(1):104–107, PMID:17097880
Shao MM, Zhang F, Meng G, Wang XX, Xu H, Yu XW, Chen LY, Tse GM (2011) Epidermal growth factor receptor gene amplification and protein overexpression in basal-like carcinoma of the breast. Histopathology 59(2):264–273. doi:10.1111/j.1365-2559.2011.03921.x, PMID:21884205
Viale G, Rotmensz N, Maisonneuve P et al (2009) Invasive ductal carcinoma of the breast with the “triple-negative” phenotype: prognostic implications of EGFR immunoreactivity. Breast Cancer Res Treat 116(2):317–328. doi:10.1007/s10549-008-0206-z, PMID:18839307
Tang Y, Zhu L, Li Y, Ji J, Li J, Yuan F, Wang D, Chen W, Huang O, Chen X, Wu J, Shen K, Loo WT, Chow LW (2012) Overexpression of epithelial growth factor receptor (EGFR) predicts better response to neo-adjuvant chemotherapy in patients with triple-negative breast cancer. J Transl Med 10(1):S4. doi:10.1186/1479-5876-10-S1-S4, PMID:23046633
Personeni N, Fieuws S, Piessevaux H et al (2008) Clinical usefulness of EGFR gene copy number as a predictive marker in colorectal cancer patients treated with cetuximab: a fluorescent in situ hybridization study. Clin Cancer Res 14(18):5869–5876. doi:10.1158/1078-0432.CCR-08-0449, PMID:18794099
Li C, Hao L, Li Y, Wang S, Chen H, Zhang L, Ke B, Yin Y, Suo H, Sun B, Zhang B, Wang C (2014) Prognostic value analysis of mutational and clinicopathological factors in non-small cell lung cancer. PLoS ONE 9(9):e107276. doi:10.1371/journal.pone.0107276, PMID:25198510
Acknowledgments
None of the parts of this research article entitled as ‘Determination of HER2 and p53 Mutations by Sequence Analysis Method and EGFR/Chromosome 7 Gene Status by Fluorescence in Situ Hybridization for the Predilection of Targeted Therapy Modalities in Immunohistochemically Triple Negative Breast Carcinomas: Results from Turkish Population’ has had prior or duplicate publication or submission elsewhere. The manuscript has been read and approved by all authors and the requirements for authorship have been met. Furthermore, each author believes that the manuscript represents honest work The authors has no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Pala, E.E., Bayol, U., Keskin, E.U. et al. Determination of HER2 and p53 Mutations by Sequence Analysis Method and EGFR/Chromosome 7 Gene Status by Fluorescence in Situ Hybridization for the Predilection of Targeted Therapy Modalities in Immunohistochemically Triple Negative Breast Carcinomas in Turkish Population. Pathol. Oncol. Res. 21, 1223–1227 (2015). https://doi.org/10.1007/s12253-015-9956-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12253-015-9956-1