Abstract
Application of the compound 1,4-dimethylnaphthalene (DMN) has been found to reduce premature sprouting in stored potato tubers. The mechanism of action for DMN has yet to be elucidated but transcriptional changes are known to occur following exposure. In this study non-dormant potato tubers (Solanum tuberosum L., cv. Russet Burbank) were treated with varying amounts of DMN resulting in an increasing residue on the tuber surface. RNA sequencing was used to measure transcriptome changes in excised meristems from tubers having increasing DMN exposure. Treatment of tubers with DMN that resulted in surface residue levels greater than 2 ppm was associated with a decrease in 45 transcripts that encoded for proteins linked with plastid development and function and an increase in the expression of 15 transcripts that encoded for WRKY-type transcription factors. qt-PCR analysis showed that repression of plastid transcripts appeared to recover 7 days after DMN exposure but induction of WRKY transcripts was maintained up to 35 days post treatment. The data suggests DMN may inhibit plastid development short term but also results in long-term changes in some regions of the transcriptome.
Resumen
Se ha encontrado que la aplicación del compuesto 1,4-dimetilnaftaleno (DMN) reduce la brotación prematura en tubérculos de papa almacenados. Aún tiene que elucidarse el mecanismo de acción del DMS, pero se sabe que ocurren cambios transcripcionales después de la exposición. En este estudio, a tubérculos de papa (Solanum tuberosum L. cv. Russet Burbank) no en reposo se les trató con diferentes cantidades de DMN, lo que resultó en un aumento de residuo en la superficie del tubérculo. Se usó secuenciación de ARN para medir los cambios en el transcriptoma en meristemos cortados de tubérculos que tuvieron exposición en aumento de DMN. El tratamiento de tubérculos con DMN que dio por resultado niveles de residuos en la superficie mayores a 2 ppm, se asoció con una disminución en 45 transcriptos que codificaban para proteínas ligadas con el desarrollo y función de plástidos, y en un aumento en la expresión de 15 transcriptos que codificaban para factores de transcripción del tipo WRKY. El análisis de qt-PCR mostró que la represión de transcriptos de plástidos parecía recuperarse siete días después de la exposición a DMN, pero la inducción de los transcriptos de WRKY se mantuvo hasta 35 días después del tratamiento. Los datos sugieren que DMN pudiera inhibir el desarrollo de plástidos por un período corto, pero también resulta en cambios a largo plazo en algunas regiones del transcriptoma.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Potato is a major agricultural crop that relies on the storage of a tuber, which is a modified stem. Upon harvest, potato tubers are in an endodormant state, which is a repressed state of growth that facilitates storage. The termination of endodormancy results in sprouting and loss of tuber quality for both processing and fresh market. Hormonal and metabolic processes control the sprouting of potato tubers (Sonnewald 2001) suggesting that there are multiple avenues for the regulation of sprouting in stored tubers. The phytohormones abscisic acid, auxin, and ethylene have been shown to have some ability to suppress sprout growth (Suttle 1995, 2003) but hormone applications are not often used to prevent premature sprouting on a commercial scale. A common practice to maintain tuber quality during storage and prevent sprouting is the application of growth suppressants. One of the most common compounds used to prevent premature sprouting in potato tubers during storage is chlorpropham (CIPC). CIPC functions through the disruption of mitotic spindles and the prevention of cell division (Vaughn and Lehnen 1991). There have been some concerns regarding the possible health effects of residual CIPC and the use of CIPC on tubers precludes their use as seed stock as resumption of normal growth processes can be significantly disrupted. Thus, there is interest in developing alternative compounds that can be used to control postharvest sprouting in potato tubers. The compound 1,4-dimethylnaphthalnene (DMN), originally isolated from potato tubers, has been shown to be useful as a sprout control agent with the ability to reversibly prevent sprouting in seed stock (Beveridge et al. 1981). How DMN functions as a sprout control agent is unknown. What is known is that CIPC and DMN do not function through a similar mechanism. CIPC arrests cell division in the mitotic phase of the cell cycle while DMN arrests cells in the S-phase prior to DNA replication (Campbell et al. 2010). Gene expression analysis using microarrays has shown that DMN alters gene expression in potato meristems and it may do so by increasing the expression of the cell cycle inhibitors KRP1 and KRP2 (Campbell et al. 2011).
In this study we expanded on the functional analysis of DMN as a sprout control agent by conducting detailed expression studies through the use of RNA-seq followed by qt-PCR assessment of specific transcripts. This approach enabled us to examine gene expression on a global scale, map the specific gene changes to the potato genome, and begin to build a transcriptional map outlining sprout control in potato. Elucidation of transcriptional changes associated with sprout suppression may open new avenues, and targets, for the genetic or physiological regulation of sprout control in potato.
Methods
Plant Material
Potato tubers were harvested in the fall of 2013 and stored at 10 C until dormancy release occurred. Release from dormancy was indicated by the presence of meristem peeping in a subset of tubers removed to 20 C. Tubers had exited dormancy by early February.
Ten to twelve tubers were placed in a single layer at the bottom of a 9.5 Liter BBL GasPak chamber (http://www.bd.com/ds/productCenter/ES-Gaspak.asp). Whatman filter paper treated with DMN was suspended in wire racks above the tubers and the chambers were sealed and incubated at 20 C in the dark for 2 days. DMN amounts ranged from 0, 1, 7.5, and 10 ul of DMN per liter of chamber air space. Following the 2-day incubation chambers were opened in a fume hood and tubers were removed to a wire basket and placed in a growth chamber overnight at 20 C. Two cm periderm plugs were then taken from each tuber immediately after the 2-day DMN treatment as well as after intervals of 7 and 35 days post-treatment. The 2 cm plugs were sent to Dichlor Analytical Laboratory (Meridian, ID) for DMN residue analysis. Two tuber plugs were placed in 10 ml of 70 % reagent alcohol, 30 % 2,2,4-trimethylpentane (TMP), and 10 ppm 2-ethylnaphthalene as an internal standard. Addition of 0.2 M NaCl was added and the resulting TMP layer was analyzed using GC and FID detection. The average ppm DMN residue was determined for each treatment. The experiment was replicated four times using two chambers for both control and DMN treatments.
A second set of tubers were harvested in the fall of 2014 and stored at 4 C until dormancy termination was detected. Tubers were treated with DMN as described above to yield a tuber residue of 2.9 ppm. Meristems were evaluated for transcript changes immediately after a 2-day exposure to DMN and then after 7 and 35 days incubation at 20 C.
RNA-Seq
Tubers meristems were excised using a 1 mm microcurette, quick frozen in liquid nitrogen, and stored at −80 C until RNA isolation. Total RNA was isolated by grinding meristems to a powder in a mortar and pestle followed by extraction using a Ribopure Kit (www.ambion.com). RNA was quantified using a BioSpec Nano and quality was measured using an Agilent 2100 Bioanalyzer (www.agilent.com). Samples having an RNA Integrity Number (RIN) of greater than 7.6 were used for analysis. Samples were shipped to the Nucleic Acid Core Facility at Penn State University Park for Illumina sequencing. Sequences were mapped to the double haploid potato genome (Solanum tuberosum phureja) using the Galaxy suite (https://usegalaxy.org) and the program Tophat. Following mapping gene expression changes between different DMN treatments were determined using CuffDiff v2.1.1 (Trapnell et al. 2012) and were based on differences in fragments per kilobase exon model per million mapped reads (FPKM).
For the 2013 harvest season RNA-seq was conducted in triplicate for samples treated with no DMN, 1 ul DMN/liter, 5 ul/liter, 7 ul/liter, and 10 ul/liter. Total reads were filtered for quality with 100 % of all reads meeting a minimum Phred score of 20.
qt-PCR
Potatoes were treated with DMN resulting in a residue of 2.5 ppm. This amount DMN was chosen based on the results of the RNA-seq analysis. RNA was isolated from potato meristems at 0, 7, and 35 days after DMN exposure. Total RNA samples with RIN scores of greater than 7.6 were used for synthesis of first strand cDNA. One ug of total RNA was converted to cDNA using oligo dT primers and a SuperScript First-Strand System (www.invitrogen.com). Gene changes between different DMN treatments were determined using a ΔΔCT method with the gene EF1-α as the internal control. EF1-α was chosen as the reference based on the RNA-seq expression data, which showed no statistical expression difference between different DMN treatments.
WRKY Gene Analysis
The Solanum tuberosum phureja (double haploid genome) http://solgenomics.net was searched using tBLASTx for WRKY-type transcriptions factors using known genes from the Arabidopsis thaliana genome. The putative peptide sequences were aligned using MAFFT (http://www.ebi.ac.uk/Tools/msa/mafft/) and a guide tree was constructed (Katoh and Standley 2013).
Results
DMN Residues
Treatment of potato tubers with varying amounts of DMN per liter of BBL GasPak chamber headspace resulted in changes in DMN residue on tuber surfaces that ranged from 0.15 (no DMN control), to 1.38, 2.15, and 4.2 ppm of DMN residue on tubers following 2 days of treatment. Following DMN exposure no visible differences were detected in association with increasing tuber residues. DMN exposure did not alter the ability to isolate RNA from potato meristems.
Transcriptome Changes Following DMN Treatment
A total of 1043 transcripts showed statistically significant change (q < 0.05) following treatments that resulted in DMN residues above the control (Supplemental Table 1). Comparisons between all residue levels revealed that 2142 transcripts changed indicating that some transcript expression was exclusive to low DMN residues while others were restricted to high levels of DMN exposure (Supplemental Table 2).
Changes in Transcripts Associated with Plastid Function and Development
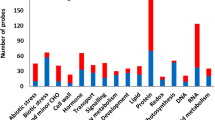
Exposure of potato tubers to DMN resulted in the decrease of multiple transcripts having a variety of functions in plastids (Table 1). DMN treatments that resulted in a residue level of greater than 2 ppm resulted in a decrease in plastid related proteins (Fig. 1). A decrease in transcripts encoding for proteins in photosystem II and I suggests a concomitant reduction in photosynthetic electron transport proteins following DMN exposure. Similarly a reduction in chlorophyll a/b binding protein should result in suppression of chlorophyll stabilization in plastids following DMN treatment. qt-PCR analysis of a subset of plastid transcript repressed by DMN exposure showed a return to control levels after 7 days of incubation at 20 C. This suggests that DMN repression of plastid development is probably short-lived (Fig. 7). The potato genome was searched using tBLASTx for homologs of the Arabidopsis GLK transcription factors. Nine genes having E-values of less than 1e−44 where found and five exhibited transcript expression in excised potato tuber meristems. However, no statistically significant difference based on CuffDiff analysis was found for any of the potato GLK homologs in meristems isolated from controls or those treated with DMN. The potato gene PGSC0003DMG400009378, a homolog of GLK1, did increase in a dose dependent manner after DMN treatment, which may suggest a mechanism of plastid suppression by DMN (Supplemental Table 2).
WRKY Transcript Changes Following DNA Treatment
A number of WRKY transcripts are altered by DMN exposure (Table 2). It is possible to group the WRKY transcripts by similar levels of expression in response to the amount of DMN residue. One group is composed of WRKY transcription factors that increase in expression at DMN residue levels of 1.375 ppm but are inhibited at 4.2 ppm (Fig. 2). A second group of WRKY transcription factors are induced by DMN residues of 1.375 and 2.125 ppm but return to control levels when residue levels reach 4.2 ppm (Fig. 3). A third class of WRKY transcription factor show induction at 1.375 ppm and a decrease to similar baseline levels when treatments result in DMN residues that are 2.125 ppm or above (Fig. 4). The final class of WRKY transcription factors reached a maximum level of induction when DMN residues are at 2.125 ppm but return to rates found in controls at 4.2 ppm (Fig. 5). The potato genome was searched for WRKY-type genes and a multiple alignment was created to show similarity between transcripts (Figs. 6 and 7). MAFFT alignment indicated that the WRKY transcripts that change in response to DMN exposure are diverse and represent multiple WRKY-type proteins.
MAFFT alignment of putative amino acid sequences for WRKY-type transcription factors found in the potato double haploid genome (Solanum tuberosum phureja). Accessions marked with * have been found to change in expression after exposure of potato tubers to DMN. The blue box represents a related group of WRKY transcripts that respond to DMN
qt-PCR and time-course analysis of select plastid genes following exposure to DMN resulting in a residue of 2.9 ppm. Samples collected immediately after DMN treatment were used as the baseline and transcript levels for tubers incubated a 20 C in the dark were compared using ef1-alpha as an internal control (ΔΔCT method). Zero time is two days post-DMN exposure
Discussion
Previous studies using microarrays have shown that transcripts encoding for Kip-Related Proteins (KRPs) are induced by DMN (Campbell et al. 2011). KRPs have been shown to inhibit cell division through the interaction with D-type cyclins (reviewed in Inzé and De Veylder 2006) and are implicated in maintaining suppression of meristem growth in potato tubers. Recent annotation of the potato genome (Consortium 2011) has resulted in a detailed catalog of putative transcripts. A search of the potato genome for mRNAs that encode for proteins having similarity to KRP proteins in Arabidopsis thaliana reveals that there are potentially 40 different genes for KRP in potato (data not shown). RNA-seq analysis revealed two transcripts encoding for KRP-like genes are inhibited by DMN (PGSC0003DMG400007708 and PGSC0003DMG400018989) suggesting that DMN inhibition of the cell cycle, if it does function through changes in KRP transcription, occurs by alteration of only a few of the KRP genes in potato.
Removal of tubers from storage did result in exposure of tubers to light, which may have initiated plastid development causing a shift from leucoplasts to chloroplasts. A decrease in transcripts associated with photosynthetic electron transport and chlorophyll a/b binding demonstrates that DMN inhibits chloroplast function and development. The decrease in transcripts associated with photosynthetic processes suggests that treatment with DMN yielding tuber residue levels of greater than 2 ppm should reduce plastid development and greening of tissues. RNA-seq data from tubers treated and then placed in storage indicates that the suppression of plastid development is maintained for at least 7 days post DMN exposure but qt-PCR analysis demonstrates recovery by 10 days. Thus, the temporal regulation of plastid transcripts by DMN is unclear. GLK transcription factors have been shown to regulate chloroplast development in plants (Waters et al. 2008; Yasumura et al. 2005) and a search of the potato genome did reveal genes encoding for GLK-like transcription factors but no significant change in expression of these genes could be associated with DMN exposure. There was some dose dependent shifts in expression for a transcript with similarity to GLK1 but it is unclear if the DMN repression of chloroplasts genes is linked to GLK activity. At this time it is unclear on the mode of DMN repression of plastid development.
WRKY transcription factors are represented by a large number of genes in plants that are involved with abiotic and biotic stress, seedling development, senescence, and seed dormancy reviewed in Rushton et al. (2010). Analysis of the tomato genome revealed 81 WRKY-type genes and these genes were expressed in response to abiotic and biotic stress (Huang et al. 2012). The putative function of WRKY-type transcriptional factors that shift in expression following DMN exposure are found in Table 3. In a number of plants the responses to abiotic stress involve a complex interaction of multiple WRKY transcription factors (Chen et al. 2012). Transcriptional changes occurring in potato tubers treated with DMN reveal WRKY-type transcripts changing as well as many genes associated with plastid and develop of photosystems. A link between WRKY transcription factors and plastid developed has been indicated by (Tang et al. 2013). The tomato mutant hp1, which is an over producer of chlorophyll and carotenoids, exhibits increased expression of the WRKY transcription factors 16 and 51. WRKY 16 (PGSC0003DMG401031196) shows a decrease in expression following DMN treatment, as does a jasmonic acid-induced WRKY protein (PGSC0003DMG400019824). WRKY 16 transcription factors have been linked to the hypersensitivity, or cell death response, in Arabidopsis (Gao et al. 2011), which may suggest a link between DMN and growth suppression as well as repression of plastid development. There is evidence that programmed cell death in tuber apical meristems has impact on sprouting in lateral meristems (Teper-Bamnolker et al. 2012).
The phytohormone jasmonic acid is associated with plant responses to biotic and abiotic stress (Delker et al. 2006). DMN induces the expression of transcription factors encoding for jasmonic acid 2 (PGSC0003DMG400015342). Jasmonic acid has been shown to regulate the expression of dehydration-induced WRKY transcription factors in tobacco BY-2 cell cultures (Rabara et al. 2013). A recent review of WRKY transcription factors and drought stress suggests significant cross-talk between abiotic signaling webs (Tripathi et al. 2014). One theory that has been proposed is that dormant non-sprouting meristems are symplastically isolated (Sonnewald and Sonnewald 2014). Abiotic stress does alter gene expression associated with plasmadesmata and cell-cell communication (Burch-Smith and Zambryski 2012), which suggests a linkage between DMN exposure and intercellular connection. This concept is supported by previous microarray-based experiments examining transcript changes in DMN potatoes that revealed an increase in the expression of a number of genes associated with response to water deprivation, desiccation, salt stress, and water channel activity (Campbell et al. 2011).
We conclude that DMN treatment results in a decrease in the expression of plastid and photosynthesis related transcripts. These results suggest that DMN may have application in the prevention of greening of tubers for fresh market. This level of DMN exposure is also linked to changes in a number of WRKY-type transcription factors that are associated with stress responses such as dehydration and wounding.
Abbreviations
- DMN:
-
Dimethylnaphthalene
- CIPC:
-
Chlorpropham
References
Beveridge, J., J. Dalziel, and H. Duncan. 1981. Dimethylnaphthalene as a sprout suppressent for seed and ware potatoes. Potato Research 24: 77–88.
Burch-Smith, T.M., and P.C. Zambryski. 2012. Plasmodesmata paradigm shift: Regulation from without versus within. Annual Review of Plant Biology 63: 239–260.
Campbell, M., A. Gleichsner, R. Alsbury, D. Horvath, and J. Suttle. 2010. The sprout inhibitors chlorpropham and 1,4-dimethylnaphthalene elicit different transcriptional profiles and do not suppress growth through a prolongation of the dormant state. Plant Molecular Biology 73: 181–189.
Campbell, M., A. Gleichsner, L. Hilldorfer, D. Horvath, and J. Suttle. 2011. The sprout inhibitor 1,4-dimethylnaphthalene induces the expression of the cell cycle inhibitors KRP1 and KRP2 in potatoes. Functional & Integrative Genomics 12: 533–541.
Chen, L., Y. Song, S. Li, L. Zhang, C. Zou, and D. Yu. 2012. The role of WRKY transcription factors in plant abiotic stresses. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms 1819: 120–128.
Consortium TPGS. 2011. Genome sequence and analysis of the tuber crop potato. Nature 475: 189–195.
Delker, C., I. Stenzel, B. Hause, O. Miersch, I. Feussner, and C. Wasternack. 2006. Jasmonate biosynthesis in arabidopsis thaliana - enzymes, products, regulation. Plant Biology 8: 297–306.
Gao, Q.-M., S. Venugopal, D. Navarre, and A. Kachroo. 2011. Low oleic acid-derived repression of jasmonic acid-inducible defense responses requires the WRKY50 and WRKY51 proteins. Plant Physiology 155: 464–476.
Huang, S., Y. Gao, J. Liu, X. Peng, X. Niu, Z. Fei, S. Cao, and Y. Liu. 2012. Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Molecular Genetics and Genomics 287: 495–513.
Inzé, D., and L. De Veylder. 2006. Cell cycle regulation in plant development1. Annual Review of Genetics 40: 77–105.
Katoh, K., and D.M. Standley. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780.
Rabara, R.C., P. Tripathi, J. Lin, and P.J. Rushton. 2013. Dehydration-induced WRKY genes from tobacco and soybean respond to jasmonic acid treatments in BY-2 cell culture. Biochemical and Biophysical Research Communications 431: 409–414.
Rushton, P.J., I.E. Somssich, P. Ringler, and Q.J. Shen. 2010. WRKY transcription factors. Trends in Plant Science 15: 247–258.
Sonnewald, U. 2001. Control of potato tuber sprouting. Trends in Plant Science 6: 333–335.
Sonnewald, S., and U. Sonnewald. 2014. Regulation of potato tuber sprouting. Planta 239: 27–38.
Suttle, J.C. 1995. Postharvest changes in endogenous ABA levels and ABA metabolism in relation to dormancy in potato tubers. Physiologia Plantarum 95: 233–240.
Suttle, J. 2003. Auxin-induced sprout growth inhibition: Role of endogenous ethylene. American Journal of Potato Research 80: 303–309.
Tang, X., Z. Tang, S. Huang, J. Liu, J. Liu, W. Shi, X. Tian, Y. Li, D. Zhang, J. Yang, Y. Gao, D. Zeng, P. Hou, X. Niu, Y. Cao, G. Li, X. Li, F. Xiao, and Y. Liu. 2013. Whole transcriptome sequencing reveals genes involved in plastid/chloroplast division and development are regulated by the HP1/DDB1 at an early stage of tomato fruit development. Planta 238: 923–936.
Teper-Bamnolker, P., Y. Buskila, Y. Lopesco, S. Ben-Dor, I. Saad, V. Holdengreber, E. Belausov, H. Zemach, N. Ori, A. Lers, and D. Eshel. 2012. Release of apical dominance in potato tuber is accompanied by programmed cell death in the apical bud meristem. Plant Physiology 158: 2053–2067.
Trapnell, C., A. Roberts, L. Goff, G. Pertea, D. Kim, D.R. Kelley, H. Pimentel, S.L. Salzberg, J.L. Rinn, and L. Pachter. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature Protocols 7: 562–578.
Tripathi, P., R. Rabara, and P. Rushton. 2014. A systems biology perspective on the role of WRKY transcription factors in drought responses in plants. Planta 239: 255–266.
Vaughn, K., and G. Lehnen. 1991. Mitotic disrupter herbicides. Weed Science 39: 450–457.
Waters, M.T., E.C. Moylan, and J.A. Langdale. 2008. GLK transcription factors regulate chloroplast development in a cell-autonomous manner. The Plant Journal 56: 432–444.
Yasumura, Y., E.C. Moylan, and J.A. Langdale. 2005. A conserved transcription factor mediates nuclear control of organelle biogenesis in anciently diverged land plants. The Plant Cell Online 17: 1894–1907.
Acknowledgments
DNA sequencing was provided by the Penn State Genomics Core Facility - University Park, PA. Funding was provided in part by the 1,4-Group, Meridian ID and the Penn State Erie undergraduate competitive grants program to O.D.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Table 1
(XLSX 174 kb)
Supplementary Table 2
(XLSX 18505 kb)
Rights and permissions
About this article
Cite this article
Campbell, M.A., D’Annibale, O. Exposure of Potato Tuber to Varying Concentrations of 1,4-Dimethylnaphthalene Decrease the Expression of Transcripts for Plastid Proteins. Am. J. Potato Res. 93, 278–287 (2016). https://doi.org/10.1007/s12230-016-9504-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12230-016-9504-x