Abstract
Sprout suppression is a crucial aspect of maintaining postharvest Solanum tuberosum (potato) tuber quality. 1,4-dimethylnaphthalene (DMN) has demonstrated effective sprout suppression during long-term storage of potatoes. Its mode of action, however, remains unknown, and previous studies utilizing single cultivars preclude identification of a common response to treatment. Thus, the goal of this study was to identify common transcriptomic responses of multiple potato cultivars of varying dormancy lengths to DMN exposure during two dormancy stages. RNA-seq gene expression profiling supported differing sensitivity to DMN treatment dependent upon cultivar and dormancy stage. A limited number of genes with similar expression patterns were common to all cultivars. These were primarily identified in ecodormant tubers and were associated with cell cycle progression, hormone signaling, and biotic and abiotic stress response. DMN treatment resulted in significant upregulation of members of ANAC/NAC and WRKY transcription factor families. Investigation of affected protein-protein interaction networks revealed a small number of networks responsive to DMN in all cultivars. These results suggest that response to DMN is largely cultivar and dormancy stage-dependent, and the primary response is governed by a limited number of stress and growth-related genes and protein-protein interactions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Potatoes, which consistently rank among the top 5 crops in the world in terms of production, are stored, processed, and marketed in a dormant state. The quality of the crop is greatest when tubers are in an endodormant (EN) state, a period of arrested growth of the meristem regulated by endogenous signals that prevent growth even under ideal conditions (Lang et al. 1987). During EN, glucose and fructose (reducing sugars) are low, while starch content is high (Hu et al. 2023). As tubers enter ecodormancy (EC), a state regulated by exogenous signals (Lang et al. 1987), metabolic activity increases, reducing sugars accumulate, and meristematic sprouts develop. This activity contributes to decreased fresh weight, the “sweetening” of tubers and the potential loss of marketable products. The transition from EN to EC is governed by factors such as genotype and pre- and post-harvest conditions, although the exact mechanisms regulating the transition remain unknown.
Sprout inhibitors and suppressants are commonly utilized to prolong storage and maintain tuber quality. Globally, the most widely used inhibitor is chlorpropham (CIPC) due in part to its comparatively low cost and its demonstrated efficacy. Use of CIPC is associated with reduced starch degradation and reducing sugar accumulation (Khurana et al. 1985; Yang et al. 1999) and weight loss (Blenkinsop et al. 2002). Most importantly, CIPC maintains commercially acceptable chip and fry color quality standards even after several months of storage (Blenkinsop et al. 2002; Krause et al. 2023). However, its use has raised concerns regarding environmental and health effects, which led to the 2020 decision by the European Union to ban CIPC (Visse-Mansiaux et al. 2021a; Thoma et al. 2022).
1,4-dimethylnaphthalene (DMN), a compound naturally found in potatoes (Meigh et al. 1973), is another effective sprout suppressant (Beveridge et al. 1981) and promising alternative to CIPC (Yang et al. 1999; Nyankanga et al. 2018; Visse-Mansiaux et al. 2021b; Krause et al. 2023). While the mechanism by which DMN suppresses sprout growth remains unknown, recent studies have found that DMN halts cell cycle progression of dormant potatoes after the G1/S phase transition (Campbell et al. 2010, 2012), induces expression of WRKY transcription factors (Campbell et al. 2012; Campbell and D’Annibale 2016), and is associated with upregulation of multiple genes involved in biotic and abiotic stress responses (Campbell et al. 2020).
Although these studies have elucidated upon the response of dormant potatoes to DMN treatment, they were limited in their use of cultivars, each only utilizing a single long-dormancy cultivar. Thus, it remains unknown whether the changes induced are consistent among various cultivars of differing dormancy lengths. The goal of this study was to describe transcriptomic changes of multiple potato cultivars of varying dormancy lengths following DMN treatment in two stages of dormancy.
Materials and Methods
Plant Material
Field grown potato cultivars were gifted by commercial producer Troyer Farms of Waterford, Pennsylvania, USA, in the fall of 2015, 2021, and 2022. Following harvest, tubers were held at ambient temperature (approximately 15–18 °C) for 1 week to promote wound healing after which they were transferred to Penn State University for long term storage at 7°C with 90% relative humidity. Cultivars used in this study were short dormancy Colomba (2022), medium-long dormancy La Chipper (2015), and long dormancy Lamoka (2021). La Chipper and Lamoka tubers were grown for chipping purposes and Colomba was grown for seed. Tubers were not treated with sprout suppressants prior to or upon receipt.
Treatments
Tubers were treated at two cultivar-dependent stages of dormancy, EN and EC. Tubers were defined as EN upon initial storage, shortly after harvest. To determine the end of EN, and confirm EC status, several untreated tubers of a given variety were periodically removed from storage and incubated at 22°C for 1 week. Termination of dormancy was defined as peeping of tuber meristems following incubation (Campbell et al. 2010).
Each treatment involved approximately eight tubers, placed in a single layer at the bottom of a 9.5 Liter BBL GasPak chamber (MG Scientific, Pleasant Prairie, WI, USA), exposed to either water (control) or DMN at a rate of 67.5 μl per chamber, approximately 7.11 μl per liter of head space, with six replicates per treatment. Chambers were stored at 25°C in dark conditions for two days. Following treatment, tuber meristems were immediately harvested using a 1-mm micro curette, frozen in liquid nitrogen, and stored at −80°C (all cultivars).
RNA-Seq
Total RNA was extracted from frozen meristems using a Zymo Quick-RNA Plant Kit (Orange, California, USA) according to kit protocol. Extracted RNA was quantified on a Thermo Scientific NanoDrop One (Thermo Fisher Scientific, Waltham, MA, USA) and sent to the Penn State University Nucleic Acid Core Facility for quality assessment using an Agilent 2100 Bioanalyzer. Samples were sequenced using an Illumina HiSeq generating 150 bp single-end reads for the 2015 data, and Illumina NextSeq generating 100 bp single-end reads for all 2021 and 2022 data. The 2015 La Chipper analysis was completed using four biological replicates per treatment and dormancy state, while the 2021 Lamoka and 2022 Colomba analyses were completed for three biological replicates per treatment and dormancy state (Table S1).
Differential expression analysis was performed on the Galaxy (2022) platform, where sequences were assessed for quality using FastQC, then mapped to the double monoploid potato S. tuberosum genome assembly version 6.1 (Pham et al. 2020) using RNASTAR. Differential expression analysis was performed using DESeq2 and tested for significance with a q value of <0.05 and a log2 fold change absolute value >1. The double monoploid genome, genome annotation, and GO category annotations were downloaded from the Spud DB database (Hirsch et al. 2014).
Gene Set Enrichment Analysis (GSEA)
The Plant Gene Set Enrichment Analysis (GSEA) Toolkit (Yi et al. 2013) was used to identify enriched gene ontology (GO) biological process terms and transcription factor targets (TFT). Differentially expressed Solanum tuberosum genes (DEG) were translated to their associated Arabidopsis thaliana locus IDs as defined by the SpudDB working gene models for the current potato genome. These IDs were queried against the A. thaliana gene set using Fisher’s exact test with Benjamini-Hochberg correction for multiple testing at a value of q<0.01. Analysis was performed for each dormancy state for all cultivars, as well as for lists of genes observed with similar expression patterns in all cultivars in a given dormancy state.
Protein-Protein Interaction (PPI) Network Analysis
Network analysis of significant DEGs was performed to describe protein-protein interactions (PPI) using Cytoscape v3.10.1 (Shannon et al. 2003). DEGs were translated to their associated A. thaliana locus IDs, as described by SpudDB, and queried against the A. thaliana species set in the STRING database version 2.0.1 (Doncheva et al. 2019). Networks with a confidence score >0.90 were visualized.
Results
RNA-Seq Mapping and Gene Expression
Sequencing of the 2015 La Chipper samples produced 165,769,684 raw reads, while sequencing of the 2021 Lamoka and 2022 Colomba samples produced 585,053,964 and 460,312,027 raw reads, respectively. Sequences were assessed for quality prior to mapping, and it was determined that no sequences required trimming based upon FastQC analysis. Following mapping, 101,700,000 (approximately 61.4%) of La Chipper reads, 514,324,979 (approximately 88.2%) of Lamoka reads, and 419,421,856 (approximately 91.1%) of Colomba reads mapped uniquely. Of uniquely mapped reads, 89,400,000 (approximately 87.9%) from La Chipper, 458,110,000 (approximately 89.1%) from Lamoka, and 249,824,650 (approximately 59.6%) from Colomba were uniquely assigned to genomic features. The remaining reads were unassigned due to failure to map, ambiguities, no features associated with the reads or multi-mapping of the reads.
In total, 7642 DEGs were identified in DMN-treated tubers when compared to controls. Of these, 2310 and 1696 were downregulated and upregulated, respectively, in response to DMN during EN, while 3051 and 3935 were downregulated and upregulated, respectively, during EC. Seven hundred twenty-five genes were uniquely expressed in EN tubers and 3587 were uniquely expressed in EC tubers. Pronounced differences in gene expression patterns were observed between the EN and EC stages of La Chipper and Lamoka tubers. For these cultivars, significantly more genes showed a response to DMN during EC than during EN (Table 1).
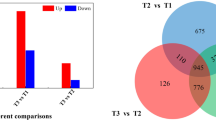
Small percentages of genes within each cultivar showed similar expression patterns in both dormancy states. Specifically, approximately 0.10% of La Chipper and 13.6% of Lamoka downregulated genes were common to both dormancy stages, while 0.30% of La Chipper and 11.6% of Lamoka upregulated genes were observed in both dormancy stages (Table S2). Colomba gene expression patterns did not exhibit the large differences between EN and EC that were observed for the other cultivars (Table 1; Table S2), such that 58.1% of genes were downregulated and 59.2% were upregulated in both stages. When considering all three cultivars, 656 genes exhibited the same expression pattern in each dormancy state. Approximately 0.5% (12 genes) of all downregulated genes (Fig. 1a) and 0.9% (16 genes) of all upregulated genes (Fig. 1b) showed similar expression patterns in all cultivars during EN. Of these, only three downregulated genes were observed exclusively during EN, while the rest were observed in both dormancy stages. In contrast, approximately 9.7% (296 genes) of downregulated genes (Fig. 1c) and 9.1% (357 genes) of upregulated genes (Fig. 1d) were common to all cultivars in the EC state, of which 287 and 341, respectively, were exclusive to EC.
Statistically significant changes to gene expression of three potato cultivars in response to DMN treatment during two stages of dormancy. Counts shown are downregulated (a) or upregulated (b) during EN, and downregulated (c) or upregulated (d) during EC at a significance q-value < 0.05 and a log2 fold change absolute value > 1
A limited number of genes were observed in the subset of genes shared by all EN, and as such, no gene function patterns were observed (Table S3). Numerous patterns were observed in the EC subset, however. Eighteen genes were associated with ethylene response and signaling, of which two were downregulated and 16 were upregulated (Table S3). Six potato homologs within the ethylene response factor (ERF) family (ERF1/AT3G23240; ERF-1/AT4G17500; ERF2/AT5G47220; ERF106/AT5G07580; ERF114/AT5G61890), three HCHIB (AT3G12500) homologs, and three pathogenesis-related 4 (PR4/AT3G04720) homologs were upregulated in DMN-treated tubers. Additionally, 12 potato genes related to cytokinin signaling, brassinosteroid biosynthesis, and cell cycle progression were downregulated following DMN treatment, while five genes related to negative regulation of cytokinin response were upregulated (Table S3). Of note, expression of cell cycle homologs of F-Box-Like protein 17 (FBL17/AT3G54650), Cell Division Cycle Protein 20.1 (CDC20.1/AT4G33270), and cyclin 3B (CYC3B/AT5G11300), CYCD3;2 (AT5G67260), and CYCB1;1 (AT4G37490) was suppressed by DMN, while expression of three homologs of KISS ME DEADLY 1 (KMD1/AT1G80440) and two of KMD2 (AT1G15670) was enhanced by DMN treatment. Fourteen genes related to auxin signaling and response genes were downregulated, and two were upregulated (Table S3). This included two downregulated homologs of PIN1 (AT1G73590), and one each of indole-3-acetic acid inducible 14 (IAA14/AT4G14550), YUCCA8 (YUC8/AT4G28720), and WUSCHEL-related homeobox 1 (WOX1/AT3G18010). Twenty potato genes were observed to exhibit similar expression patterns in both dormancy states (Table S3). Six genes were downregulated following DMN treatment, of which three were homologs of AT3G45770, a gene related to fatty acid biosynthesis. The remaining 14 genes were upregulated in both dormancy stages. These were primarily associated with flavonoid and lignin biosynthesis (GT72B1/AT4G01070, HCT/AT5G48930, MYB48/AT3G46130, and TT7/AT5G07990), and stress response (AT2G47710, AtKTI5/AT1G17860, DjC53/AT1G56300, GAD/AT5G17330, and GT72B1/AT4G01070). Three homologs of CYP76G1 (AT3G52970), a gene associated with terpenoid metabolism, were also upregulated following DMN treatment.
Functional Analysis of Differentially Expressed Genes
In EN tubers of individual cultivars, a total of two GO categories related to biological processes were identified as significantly downregulated and 26 as upregulated in response to DMN (Table S4). No significant GO categories were observed in EN La Chipper tubers. However, both downregulated and 24 upregulated terms were shared by the Lamoka and Colomba cultivars. Downregulated GO categories were associated with cell specification and commitment, while upregulated terms were associated with defense and immune responses, as well as response to heat, fungi, light intensity, and biotic, temperature, water, ethylene, jasmonic acid (JA), and abscisic acid (ABA) stimuli. When considering the EN subset, no GO categories were identified as either significantly upregulated or downregulated.
In EC tubers, 28 GO categories were downregulated in total, of which five were shared by all three cultivars, two by Lamoka and Colomba, nine by Lamoka and La Chipper, and 12 by La Chipper and Colomba (Table S4). A total of 162 GO categories were upregulated, of which 101 were observed for all three cultivars. The remaining upregulated GO categories were all shared between two cultivars, such that 40 were shared by Lamoka and Colomba, 17 by Lamoka and La Chipper, and four by La Chipper and Colomba (Table S4). Downregulated terms common to all cultivars were associated with cuticle development, very-long-chain fatty acid metabolic processes, response to UV-B, and response to sucrose and brassinosteroid stimuli. Upregulated GO categories observed in all cultivars were associated with immune, defense, biotic, and abiotic stress responses. Specifically, response to hypoxia, fungus, ethylene, salicylic acid (SA), and JA stimuli, inorganic substances, metal ions, chitin, nitrate, endoplasmic reticulum stress, and oxygen were upregulated. Metabolic processes of ethylene, SA, flavonoid, reactive oxygen species, hydrogen peroxide, and toxins were positively enriched, as were biosynthetic processes of lignin, alkene, ethylene, and SA. Additionally, GO categories associated with aging, senescence, cell death, and MAPK cascade, and signaling pathways of JA, SA, ABA, and ethylene were identified as significantly upregulated.
In the EC gene subset, 18 downregulated and 80 upregulated GO categories were identified in response to DMN (Table S4). Downregulated terms were associated with anatomical structure development and morphogenesis, flower, cuticle, and organ development, developmental processes, very long chain fatty acid and lipid metabolic processes, and multidimensional cell growth. Upregulated terms, on the other hand, were similar to those observed in the individual cultivars and were associated with cell death, response to ethylene, JA, SA and ABA stimuli, response to salinity, water, and wounding, and SA, phenylpropanoid, and ethylene biosynthesis and metabolism. Additional upregulated terms included defense and immune responses, systemic acquired resistance, and response to biotic stimulus, bacterium, and fungus.
Transcription Factor Target Enrichment Analysis
A total of 18 TFTs were downregulated and 29 upregulated in response to treatment with DMN at a significance cutoff value of q<0.01 during EN (Table S4). As with functional analysis of DEGs, no TFTs were significantly down- or upregulated in La Chipper tubers at this stage, and instead, all were shared by Lamoka and Colomba.
In EC tubers, a total of 17 TFTs were downregulated and 63 were upregulated (Table S4). Of the downregulated targets, five were observed in all cultivars, nine in Lamoka and Colomba, two in Lamoka and La Chipper, and one in La Chipper and Colomba cultivars. Of the upregulated targets, 36 were observed in all cultivars, 20 in Lamoka and Colomba, two in Lamoka and La Chipper, and five in La Chipper and Colomba.
AGAMOUS-LIKE15 (AGL15) and AGAMOUS-LIKE63 (AGL63), both members of the MIKC subfamily of MADS-box domain transcription factors, were significantly suppressed following DMN treatment in EN and EC, respectively. Numerous NAC domain protein family members were significantly induced in response to DMN, 10 in EN and 12 in EC, only two of which were observed in both dormancy states, ANAC092 and NAC2. Members of the WRKY gene family were the most highly represented of the upregulated TFTs for both dormancy states. In total, 24 WRKY TFTs were identified, of which 13 were significant during both EN and EC.
No significant TFTs were identified in the EN subset of genes. Seventy-one TFTs were downregulated in the EC subset (Table S4), of which MYB protein family members were the most highly represented. Additional downregulated targets of interest included APETALA 1 (AP1) and AP3 binding sites and AP2 targets, and phytochrome-interacting transcription factor 5 (PIF5) binding sites. One hundred eleven TFTs were upregulated, 30 of which were members of the ANAC/NAC protein family, and 23 of which were from the WRKY gene family. Other noteworthy TFTs included SOMBRERO (SMB), and cryptochrome 2 (CRY2), ethylene-insensitive3 (EIN3) and LEAFY binding sites.
Protein-Protein Interaction Network Analysis
DEGs were translated to their related A. thaliana proteins for PPI network analysis. It is important to note that many potato genes currently do not have associated A. thaliana genes or proteins, thus analysis was limited to those described to date. Only the largest subnetworks for each condition are reported herein.
The EN and EC gene subsets were used to identify significant PPI networks shared by all cultivars. No PPI networks were identified in down- (Fig. 2a) or upregulated genes (Fig. 2b) of EN tubers. However, a limited number of small networks were identified for down- (Fig. 2c) and upregulated genes (Fig. 2d) in EC tubers. The largest downregulated network was associated with brassinosteroid biosynthesis (Fig. 2c), while smaller two-protein networks were associated with flavonoid and anthocyanin, auxin, and wax biosynthesis, polyamine synthesis and conversion, and cell cycle progression (Fig. 2c). The largest upregulated and second largest networks were associated with pathogen and stress response, respectively (Fig. 2d).
PPI networks of individual cultivars were also considered. During EN, no networks were identified from downregulated genes of La Chipper (Fig. 3a). However, a network related to cell cycle progression was observed during EC for this cultivar (Fig. 3b). EN Lamoka tubers had a two-protein network related to flavonoid biosynthesis (Fig. 3c) and a larger network associated with photosynthesis in EC (Fig. 3d). Colomba had the largest downregulated network during EN, which was composed of clusters related to cell cycle progression, and sterol, flavonoid, and phenylpropanoid biosynthesis (Fig. 3e). Similarly, EC Colomba tubers had a network composed of clusters related to cell cycle progression and sterol, flavonoid and phenylpropanoid biosynthesis, as well as photosynthesis (Fig. 3f).
PPI subnetworks downregulated in response to DMN. The largest subnetworks are portrayed for endodormant (a) and ecodormant (b) La Chipper, endodormant (c) and ecodormant (d) Lamoka, and endodormant (e) and ecodormant (f) Colomba cultivars. All networks identified using Cytoscape with confidence scores >0.90
A simple two-protein network was identified from upregulated genes of EN La Chipper tubers, which was associated with anthocyanin production (Fig. 4a). Three networks of equal size were observed during EC for this cultivar. These networks were associated with starch production and abiotic stress response, water and temperature stress, and lignin biosynthesis (Fig. 4b). EN Lamoka tubers produced three small three-protein networks. These were associated with anthocyanin production, metal ion transport, and glycolysis (Fig. 4c). The largest subnetwork observed for this cultivar during EC was composed of proteins related to glycolysis and stress response (Fig. 4d). The EN subnetwork of Colomba tubers was primarily composed of proteins associated with stress response and glycolysis (Fig. 4e), while the EC subnetwork had clusters associated with JA biosynthesis, glycolysis, and stress response (Fig. 4f).
PPI subnetworks upregulated in response to DMN. The largest subnetworks are portrayed for endodormant (a) and ecodormant (b) La Chipper, endodormant (c) and ecodormant (d) Lamoka, and endodormant (e) and ecodormant (f) Colomba cultivars. All networks identified using Cytoscape with confidence scores >0.90
Discussion
Differentially Expressed Genes Vary by Cultivar and Dormancy State in Response to DMN
Gene expression changes in response to DMN were both cultivar and dormancy state-dependent, though cultivation year, the effect of which could not be directly assessed, may also have played a role. Both La Chipper (2015) and Lamoka (2021), which are medium-long to long dormancy varieties respectively, exhibited low response to DMN during EN, while Colomba (2022), a short dormancy variety, exhibited pronounced response to the treatment. In contrast, La Chipper and Lamoka showed increased response during EC, while the response of Colomba during this stage was similar to that observed during EN. These differences in gene expression levels and response suggest the short dormancy Colomba may have never fully entered an EN state following harvest. Additionally, although thousands of genes responded to DMN, particularly in the EC state, these were primarily cultivar specific and only a small fraction of genes exhibited similar expression patterns in all cultivars. These results agree with previous research which demonstrated differential response of tubers to DMN treatment dependent upon dormancy state (Campbell et al. 2020). Therefore, to elucidate the primary effect of DMN, the following discussion will be limited to responses which were common to all cultivars.
Ethylene Response Is Significantly Induced by DMN
Ethylene has been paradoxically shown to both suppress meristem growth and shorten tuber dormancy, with consistent exogenous exposure suppressing growth (Rylski et al. 1974; Prange et al. 1998) and cessation of exposure resulting in growth resuming almost immediately (Rylski et al. 1974; Foukaraki et al. 2016). In this study, DMN exposure during EC induced expression of numerous ethylene response (ERF1, ERF-1, ERF2, ERF106, PR4) and signaling (HCHIB, also known as PR3) genes but was not found to induce genes with confirmed roles in ethylene biosynthesis. While the lack of genes related to ethylene biosynthesis supports previous research which observed only a small transient increase in ethylene production following DMN treatment (Suttle 2003), it is important to note that ethylene concentration was not quantified in the present study and changes to its biosynthesis, therefore, cannot be confirmed. Future studies to assess ethylene concentrations following DMN exposure may help to elucidate the potential relationships between DMN and ethylene biosynthesis and response.
Although the mechanisms by which ethylene suppresses sprout growth remain unknown, exogenous exposure has been observed to induce expression of genes related to stress and defense (Yang et al. 2020; Tosetti et al. 2021). ERF (Thirugnanasambantham et al. 2015) and PR genes (Islam et al. 2023) have known roles in abiotic and biotic stress response, and expression of multiple genes from both families has been observed to increase following ethylene exposure (Yang et al. 2020). No previous studies of dormant tubers treated with DMN have noted changes to ERFs. However, DMN treatment has been observed to induce a stress response (Campbell et al. 2010, 2012, 2020; Campbell and D’Annibale 2016) and increase expression of PR4 and PR5 (Campbell et al. 2020). Together, this suggests that DMN and ethylene act on some of the same pathways to suppress sprout development.
DMN Affects Cell Cycle Signaling, Cell Division, and Expansion
Previous studies of dormant tubers exposed to DMN treatment have observed changes in expression of genes related to cell cycle regulation (Campbell et al. 2010, 2012, 2020; Campbell and D’Annibale 2016). Here, we observed downregulation of gene sets involved in anatomical structure development and morphogenesis, developmental processes, and multidimensional cell growth. This included several genes involved in cell cycle regulation, including CDC20.1, CYC3B, CYCB1;1, CYCD3;2, and FBL17, as well as two genes involved in cytokinin signaling, KMD1 and KMD2, and four involved in auxin biosynthesis, signaling and response, PIN1, IAA14, YUC8, and WOX1. Importantly, these genes exhibited similar expression patterns in all three evaluated EC cultivars, highlighting the potential importance of these genes in sprout growth suppression induced by DMN treatment.
Although previous research utilizing DMN suggested the sprout suppressant blocks cell division after the G1/S phase transition (Campbell et al. 2010), current results suggest that it suppresses cell cycle progression at multiple stages. CYCD3;2 (Swaminathan et al. 2000) and FBL17 (Noir et al. 2015), genes which regulate the G1/S phase transition, were downregulated following DMN exposure. CYCD3;2, an important rate-limiter of cytokinin response, plays a role in the regulation of cell number in developing lateral organs (Dewitte et al. 2007) and may be especially important for vascular development (Collins et al. 2015). Reduced expression of CYCD3;2 contributed to upregulation of KIP-Related Protein 1 (KRP1), KRP2, E2Fa, E2Fb, cyclin-dependent kinases (CDK) and CDK subunits in Dimocarpus longan embryos (Zhao et al. 2022), and loss of FBL17 function induced DNA-damage responses and contributed to KRP2 accumulation in A. thaliana (Gentric et al. 2020). Previous research has identified significant increases in expression of KRP1 and KRP2 following DMN exposure (Campbell et al. 2012), and although the current study did not observe significant changes to these genes across all cultivars, decreased expression of genes involved in their regulation was a common response. CDC20.1 (Kevei et al. 2011), CYC3B, and CYCB1;1 (Ito 2000), which play important roles in regulation of G2/M phase transition, were also downregulated in all cultivars following DMN treatment. CDC20, which interacts with anaphase promoting complex subunits, is co-expressed with its B-type cyclin targets, including CYCB1;1 (Yang et al. 2017). Reduced expression of CYCB1;1 was related to reduced growth and yield parameters in tomato (Liu et al. 2018) and delayed cellularization and reduced viability of rice (Guo et al. 2010).
Cytokinin levels and sensitivity are low in dormant potatoes and increase with dormancy release (Turnbull and Hanke 1985; Suttle 2004) to promote cell division. In the current study, two negative regulators of cytokinin signaling were significantly upregulated in response to DMN. KMD1 and KMD2 are thought to directly interact with and destabilize type-B Arabidopsis response regulators (ARR), specifically ARR1 and ARR12 leading to cytokinin insensitivity (Kim et al. 2013a, b). Increased expression of KMD1 and KMD2, as well as KMD4, has been documented to occur as floral development ceases and overexpression of KMD2, specifically, was found to accelerate meristem arrest in A. thaliana (Martínez-Fernández et al. 2020). These findings were consistent with previous research in which overexpression of KMD1 and KMD2 resulted in a dwarf phenotype and premature termination of primary root growth (Kim et al. 2013a). Taken together, expression of KMD1 and KMD2 plays an important role in regulation of meristem growth and development.
Like cytokinin, auxin concentration is crucial for cell division and expansion (Perrot-Rechenmann 2010), but its role in potato dormancy is not well understood. However, an increase in free and conjugated IAA concentration was seen in tuber meristems at dormancy release (Sorce et al. 2000), and genes related to auxin biosynthesis, transport, and signaling upregulation have been observed in dormant potatoes after treatment with GA3 (Hartmann et al. 2011). In EC tubers treated with DMN, we observed down regulation of YUC8, a key enzyme in auxin biosynthesis, as well as PIN1 and WOX1, regulators of auxin transport. WOX1, WOX3, and WOX5 have been implicated as having potential redundant roles in auxin biosynthesis necessary for leaf expansion through positive regulation of YUC genes (Zhang et al. 2020), and overexpression of WOX1 was correlated with significant upregulation of PIN1 and AUX1 expression at the shoot apical meristem of A. thaliana (Nakata et al. 2018). Localization of PIN1 is crucial for proper organogenesis and positioning in both the root and the shoot (Vernoux et al. 2000; Heisler et al. 2010; Krogan et al. 2016). The results of this study suggest DMN halts cell cycle progression and expansion through negative regulation of auxin and cytokinin signaling in potato meristems. Further studies quantifying auxin and cytokinin levels following DMN treatment are needed to elucidate whether the observed changes in hormone signaling are correlated with changes to endogenous concentrations.
DMN Elicits Water Stress Response in Dormant Tubers
Previous research has observed increases in transcripts and gene sets related to multiple biotic and abiotic stress responses following DMN exposure (Campbell et al. 2010, 2012, 2020). In the current study, DMN treatment induced numerous stress-related biological processes. Importantly, DMN treatment resulted in upregulation of gene sets related to cell death, and responses to salt and water. These results support previous work which identified transcripts associated with salt stress, water deprivation (Campbell et al. 2010, 2012), and osmotic stress (Campbell et al. 2012, 2020) in DMN-treated tubers. DMN treatment also induced biological processes related to hormonal response. Specifically, responses to SA, JA, ABA, and ethylene were upregulated following treatment. Similar results were observed when DMN elevated gene sets associated with SA and JA (Campbell et al. 2020). JA and SA are important regulators of plant defense signaling that experience considerable positive and negative crosstalk (Clarke et al. 2000; Tamaoki et al. 2013; Caarls et al. 2015). ABA and ethylene are known to be involved in dormancy induction and maintenance (Suttle and Hultstrand 1993; Li et al. 2021), as well as response to biotic and abiotic stresses (Cao et al. 2006; Yan et al. 2016; Gietler et al. 2020). All these hormones have also been found to play important roles in drought response (Manavella et al. 2006; Arraes et al. 2015; Sharma et al. 2017; Wang et al. 2021). Tubers lose weight during storage from transpiration and respiration activity, which is partially mitigated by sustaining low temperatures and high relative humidity to reduce the vapor pressure difference at the surface of the tubers. Tubers treated with DMN experience less weight loss in storage than untreated tubers (Krause et al. 2023), suggesting reduced water loss. Because DMN is a volatile organic compound that coats the surface of the tuber when applied, it is likely affecting transpiration and vapor pressure perception, signaling the occurrence of a potential water deficit and the need for water conservation.
Defense and Stress Response-Related Transcription Factor Targets Are Induced by DMN
In this study, 111 TFTs were commonly upregulated in all cultivars in response to DMN exposure, of which nearly half were members of the NAC and WRKY families. Both NAC and WRKY are major plant-specific transcription factor families with important roles in biotic and abiotic stress response as well as diverse physiological and developmental processes (Chen et al. 2017; Singh et al. 2021). To date, approximately 110 NAC (Singh et al. 2013) and 79 WRKY genes (Zhang et al. 2017) have been identified in Solanum tuberosum.
In a study identifying and describing potato NAC genes and their stress-related responses, salt and drought stresses induced the most genes, followed by heat (Singh et al. 2013). In the current study, 30 ANAC/NAC TFTs were upregulated in response to DMN, 11 of which have been previously found to be induced by drought, heat, salt, wound, and pathogen stresses and hormone or chemical exposure (Table 2; Singh et al. 2013). Of these, ANAC017 and VND6 (ANAC101) both increased expression when exposed to drought, heat, salt, or ABA stress (Singh et al. 2013). NST1 (ANAC043) increased when exposed to drought or salt, and ANAC096 increased when exposed to heat or salt stress (Singh et al. 2013).
Of 72 potato WRKY genes, 63%, 69%, and 74% were found to be upregulated in response to salt, heat, and drought stresses, respectively (Filiz and Kurt 2021). In the current study, 23 WRKY genes were upregulated in response to DMN exposure, 16 of which have documented upregulation in response to salt, heat, drought, SA, or a combination of these stresses in potato species (Table 2; Zhang et al. 2017; Villano et al. 2020; Feliz and Kurt 2021). Expression of WRKY8 increased in response to drought and heat stress (Zhang et al. 2017, Villano et al. 2020) and WRKY6 increased in response to drought stress (Zhang et al. 2017, Villano et al. 2020) in S. tuberosum and S. commersonii, while expression of WRKY22 increased in response to drought and salt in S. tuberosum (Zhang et al. 2017; Filiz and Kurt 2021). These results suggest that DMN significantly induces multiple biotic and abiotic stress responses, the most highly represented of which is response to drought stress.
DMN Response Involves Limited Protein-Protein Interaction
EN tubers treated with DMN did not exhibit changes to subnetworks. However, individual peptides did change in expression across cultivars following DMN treatment. Five peptides were downregulated by DMN in the EN subset (Table 1). Two of these peptides were associated with defense response, oxidative stress, wounding, and response to JA. JA has been linked to tuber initiation (Pelacho and Mingo-Castel 1991; Koda et al. 1991), but it is unclear if that developmental response is linked only to tuberization and not also to the onset of EN or tuber growth arrest. The downregulated genes associated with JA response may suggest a change in JA levels within tubers treated with DMN. DMN also resulted in the downregulation of a peptide with similarity to At3g45770, which has been assigned to the GO function of fatty acid biosynthesis and fatty acid metabolism. DMN is a highly hydrophobic molecule, and it is expected to interact with lipid structures, which may alter lipid metabolism. Twelve peptides were upregulated by DMN in the EN subset (Table 3) with no discernible subnetwork changes occurring. There was an increase in peptides with functions related to lipid metabolism (CYP76G1, At2g47710) and one involved with changes in membrane structure (FLOT1). Two of the peptides have GO biological process assignments involving flavonoid biosynthesis (HCT, TT7). The increase in expression of the transcription factor MYB48 by DMN has interesting implications. MYB transcription factors have been linked to a myriad of biological processes including response to biotic stress (Lee et al. 2001), flavonoid biosynthesis (Zhao et al. 2013), and axillary meristem initiation (Müller at al. 2006). More specifically, over expression of MYB48 from Zea mays in Arabidopsis resulted in increased drought tolerance (Wang et al. 2017). Drought is an environmental signal that induces dormancy and growth arrest in multiple perennial plants (Volaire et al. 2023).
In EC gene subsets, eight subnetworks were downregulated after DMN exposure (Table 3). The subnetwork SMO2-2:SMO1-1:STE1:DWF1 is involved with sterol and brassinosteroid (BR) biosynthesis. BRs function as growth promoting phytohormones and as regulators of abiotic stress in plants (Li et al. 2021). Thus, the suppression of BR synthesis pathways by DMN in meristems that have lost EN suggests a possible mechanism of growth inhibition in potato tubers. Previous work by Korableva et al. (2002) showed that application of the brassinosteroid 24-epibrassinolide (EB) to potato tubers increased endogenous ABA and ethylene levels and prolonged tuber dormancy. Thus, it appears that DMN suppression of growth and sterol biosynthesis occurs through an alternate mechanism than the brassinolide EB.
Downregulation of TT7:UF3GT by DMN suggests a suppression of the flavonoid biosynthetic pathway. Flavonoid biosynthesis has multiple roles in plant development including response to stress, protection from oxidative burst, regulation of cell cycle and growth (Kumar et al. 2018). It is unclear if the DMN suppression of proteins associated with the flavonoid pathway has a clear linkage to suppression of meristem activity and sprout growth in potato. The suppression of PAO5:SPDS2 subnetwork by DMN suggests an alteration of polyamine biosynthesis following DMN exposure. Downregulation of the KCS6:CER3 subnetwork by DMN should result in a decrease in long chain fatty acid production and cuticular waxes. An additional fatty acid associated subnetwork, CB5-B:FAH1, was also found to be downregulated by DMN. Suppression of cuticular, long-chain fatty acids has been shown to be associated with drought stress (Negin et al. 2023). A more direct link between DMN and cell division can be found in the suppression of the subnetwork CDC20.1:CYCB1.1. CDC20 is part of the anaphase-promoting complex (Willems and De Veylder 2022) and B-type cyclins are associated with the mitosis promoting factor.
Six PPI subnetworks are induced or upregulated by DMN across all EC potato cultivars (Table 4). The network OSM34:HCHIB:PRB1:PR4 involves protein interactions that center around phytohormones signaling and stress signaling. OSM34 encodes for an osmotin-like protein that is a positive regulator of ABA signaling (Park and Kim 2021). HCHIB encodes for a fungal pathogen induced endochitinase that also elevates genes controlling ethylene levels (Kappagantu et al. 2017). PRB1 and PR4 are pathogenesis related proteins associated with ethylene response pathways (Kaur et al. 2022). The remaining PPI subnetworks (NILR1:SIP1:AT4G01970, ERF1:ERF2, RPM1:RIPK, and AT1G70420:TyrA1) are associated with stress responses mitigated by ethylene and JA. The induction of PPI stress associated networks may be a function of ethylene response. Application of ethylene has been used to control sprouting in stored potato tubers (Prange et al. 1998), which may be part of the mechanism of action for DMN. A caveat to this concept is that the DMN treatment used in this study was for a single application whereas commercial use of ethylene in stores to control sprouting is through continuous application.
Conclusion
No previous study has evaluated the effect of DMN on multiple potato cultivars of differing dormancy profiles. Here, we have affirmed that DMN treatment elicits differing responses dependent upon cultivar and dormancy state with EC tubers being more responsive than EN tubers. DMN was observed to suppress numerous genes related to cell-cycle progression and plant development and induce genes and TFTs related to multiple biotic and abiotic stress responses. Specifically, response to water stress was strongly induced, suggesting DMN may alter water movement and perception. Treatment with DMN resulted in thousands of genes changing expression, but genes common to all three cultivars were limited, and of those, fewer were identified as acting in PPI networks. Thus, DMN results in the global response of tubers to a perceived stress, but a small number of genes and protein interactions govern the primary response.
Data Availability
Raw sequencing reads were submitted to the NCBI SRA database under the accession of PRJNA1064110.
Abbreviations
- BR:
-
Brassinosteroids
- CIPC:
-
Chlorpropham
- DMN:
-
1,4-dimethylnaphthalene
- EN:
-
Endodormant/endodormancy
- EC:
-
Ecodormant/ecodormancy
- TFT:
-
Transcription factor target
- PPI:
-
Protein-protein interactions
References
Arraes FBM, Beneventi MA, Lisei de Sa ME, Paixao FR, Albuquerque EVS, Marin SRR, Purgatto E, Nepomuceno AL, Grossi-de-Sa MF (2015) Implications of ethylene biosynthesis and signaling in soybean drought stress tolerance. BMC Plant Biol 15:213. https://doi.org/10.1186/s12870-015-0597-z
Beveridge JL, Dalziel J, Duncan HJ (1981) The assessment of some volatile organic compounds as sprout suppressants for ware and seed potatoes. Potato Res 24:61–76. https://doi.org/10.1007/BF02362017
Blenkinsop RW, Copp LJ, Marangoni Yada RY., AG, (2002) Effect of chlorpropham (CIPC) on carbohydrate metabolism of potato tubers during storage. Food Res Int 35(7):651–655. https://doi.org/10.1016/S0963-9969(01)00168-5
Caarls L, Pieterse CMJ, Van Wees SCM (2015) How salicylic acid takes transcriptional control over jasmonic acid signaling. Front Plant Sci 6:170. https://doi.org/10.3389/fpls.2015.00170
Campbell MA, D’Annibale O (2016) Exposure of potato tuber to varying concentrations of 1,4-dimethylnaphthalene decrease the expression of transcripts for plastid proteins. Am J Potato Res 93:278–287. https://doi.org/10.1007/s12230-016-9504-x
Campbell MA, Gleichsner A, Alsbury R, Horvath D, Suttle J (2010) The sprout inhibitors chlorpropham and 1,4-dimethylnaphthalene elicit different transcriptional profiles and do not suppress growth through a prolongation of the dormant state. Plant Mol Biol 73:181–189. https://doi.org/10.1007/s11103-010-9607-6
Campbell MA, Gleichsner A, Hilldorfer L, Horvath D, Suttle J (2012) The sprout inhibitor 1,4-dimethylnaphthalene induces the expression of the cell cycle inhibitors KRP1 and KRP2 in potatoes. Funct Integr Genom 12:533–541. https://doi.org/10.1007/s10142-011-0257-9
Campbell MA, Gwin C, Tai HH, Adams R (2020) Changes in gene expression in potato meristems treated with the sprout suppressor 1,4-dimethylnaphthalene are dependent on tuber age and dormancy status. PLoS ONE 15(7):e0235444. https://doi.org/10.1371/journal.pone.0235444
Cao Y, Song F, Goodman RM, Zheng Z (2006) Molecular characterization of four rice genes encoding ethylene-responsive transcriptional factors and their expressions in response to biotic and abiotic stress. J Plant Physiol 163(11):1167–1178. https://doi.org/10.1016/j.jplph.2005.11.004
Chen F, Hu Y, Vannozzi A, Wu K, Cai H, Qin Y, Mullis A, Lin Z, Zhang L (2017) The WRKY transcription factor family in model plants and crops. Critic Rev Plant Sci 36(5–6):311–335. https://doi.org/10.1080/07352689.2018.1441103
Clarke JD, Volko SM, Ledford H, Ausubel FM, Dong X (2000) Roles of salicylic acid, jasmonic acid, and ethylene in cpr-induced resistance in Arabidopsis. The Plant Cell 12(11):2175–2190. https://doi.org/10.1105/tpc.12.11.2175
Collins C, Maruthi NM, Jahn CE (2015) CYCD3 D-type cyclins regulate cambial cell proliferation and secondary growth in Arabidopsis. J Exp Bot 66(15):4595–4606. https://doi.org/10.1093/jxb/erv218
Dewitte W, Scofield S, Alcasabas AA, Maughan SC, Menges M, Braun N, Collins C, Nieuwland J, Prinsen E, Sundaresan V, Murray JAH (2007) Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Biol Sci 104(36):14537–14542. https://doi.org/10.1073/pnas.0704166104
Doncheva NT, Morris JH, Gorodkin J, Jensen LJ (2019) Cytoscape StringApp: network analysis and visualization of proteomics data. J Prot Res 18(2):623–632. https://doi.org/10.1021/acs.jproteome.8b00702
Filiz E, Kurt F (2021) Expression and co-expression analyses of WRKY, MYB, bHLH and bZIP transcription factor genes in potato (Solanum tuberosum) under abiotic stress conditions: RNA-seq data analysis. Potato Res 64:721–741. https://doi.org/10.1007/s11540-021-09502-3
Foukaraki SG, Cools K, Chope GC, Terry LA (2016) Impact of ethylene and 1-MCP on sprouting and sugar accumulation in stored potatoes. Postharvest Biol Technol 114:95–103. https://doi.org/10.1016/j.postharvbio.2015.11.013
Gentric N, Masoud K, Journot RP, Cognat V, Chabouté M-E, Noir S, Genschik P (2020) The F-box-like protein FBL17 is a regulator of DNA-damage response and colocalizes with RETINOBLASTOMA RELATED1 at DNA lesion sites. Plant Physiol 83(3):1295–1305. https://doi.org/10.1104/pp.20.00188
Gietler M, Fidler J, Labudda M, Nykiel M (2020) Abscisic Acid—Enemy or Savior in the Response of Cereals to Abiotic and Biotic Stresses? International Journal of Molecular Sciences 21:4607. https://doi.org/10.3390/ijms21134607
Guo J, Wang F, Song J, Sun W, Zhang XS (2010) The expression of Orysa;CycB1;1 is essential for endosperm formation and causes embryo enlargement in rice. Planta 231:293–303. https://doi.org/10.1007/s00425-009-1051-y
Hartmann A, Senning M, Hedden P, Sonnewald U, Sonnewald S (2011) Reactivation of meristem activity and sprout growth in potato tubers require both cytokinin and gibberellin. Plant Physiol 155(2):776–96. https://doi.org/10.1104/pp.110.168252
Heisler MG, Hamant O, Krupinski P, Uyttewaal M, Ohno C, Jönsson H, Traas J, Meyerowitz EM (2010) Alignment between PIN1 polarity and microtubule orientation in the shoot apical meristem reveals a tight coupling between morphogenesis and auxin transport. PLoS Biol 8(10):e1000516. https://doi.org/10.1371/journal.pbio.1000516
Hirsch CD, Hamilton JP, Childs KL, Cepela J, Crisovan E, Vaillancourt B, Hirsch CN, Habermann M, Neal B, Buell CR (2014) Spud DB: a resource for mining sequences, genotypes, and phenotypes to accelerate potato breeding. The Plant Genome 7(1):plantgenome2013.12.0042. https://doi.org/10.3835/plantgenome2013.12.0042
Islam MM, El-Sappah AH, Ali HM, Zandi P, Huang Q, Soaud SA, Alazizi EMY, Wafa HA, Hossain MA, Liang Y (2023) Pathogenesis-related proteins (PRs) countering environmental stress in plants: a review. South Afr J Botan 160:414–427. https://doi.org/10.1016/j.sajb.2023.07.003
Ito M (2000) Factors controlling cyclin B expression. Plant Mol Biol 43:677–90. https://doi.org/10.1023/a:1006336005587
Kappagantu M, Bullock JM, Nelson ME, Eastwell KC (2017) Hop stunt viroid: effect on host (Humulus lupulus) transcriptome and its interactions with hop powdery mildew (Podospheara macularis). Mol Plant-Microbe Interact 30(10):842–851. https://doi.org/10.1094/MPMI-03-17-0071-R
Kaur S, Samota MK, Choudhary M, Choudhary M, Pandey AK, Sharma A, Thakur J (2022) How do plants defend themselves against pathogens-Biochemical mechanisms and genetic interventions. Physiol Mol Biol Plants 28:485–504. https://doi.org/10.1007/s12298-022-01146-y
Kevei Z, Baloban M, Da Ines O, Tiricz H, Kroll A, Regulski K, Mergaert P, Kondorosi E (2011) Conserved CDC20 cell cycle functions are carried out by two of the five isoforms in Arabidopsis thaliana. PLoS ONE 6(6):e20618. https://doi.org/10.1371/journal.pone.0020618
Khurana DS, Randhawa KS, Bajaj KL (1985) Carbohydrate content of potato (Solanum tuberosum L.) tubers treated with Isopropyl-N (3-chlorophenyl) carbamate under different storage conditions. J Sci Food Agric 36(10):959–962. https://doi.org/10.1002/jsfa.2740361009
Kim HJ, Chiang Y-H, Kieber JJ, Schaller GE (2013a) SCFKMD controls cytokinin signaling by regulating the degradation of type-B response regulators. PNAS 110(24):10028–10033. https://doi.org/10.1073/pnas.1300403110
Kim HJ, Kieber JJ, Schaller GE (2013b) The rice F-box protein KISS ME DEADLY2 functions as a negative regulator of cytokinin signalling. Plant Signal Behav 8:12. https://doi.org/10.4161/psb.26434
Koda Y, Kikuta Y, Tazaki H, Tsujino Y, Sakamura S, Yoshihara T (1991) Potato tuber-inducing activities of jasmonic acid and related compounds. Phytochemistry 30(5):1435–1438. https://doi.org/10.1016/0031-9422(91)84180-Z
Korableva N, Platonova T, Dogonadze M, Evsunina AS (2002) Brassinolide effect on growth of apical meristems, ethylene production, and abscisic acid content in potato tubers. Biol Plant 45:39–43. https://doi.org/10.1023/A:1015167616960
Krause MR, de Araujo FF, Moreira KF, de Araújo NO, de Jesus Tello JP, de Sousa Santos MN, Finger FL (2023) Carbohydrate metabolism dynamic in chlorpropham- and 1,4-dimethylnaphthalene-treated potatoes and its effect on the browning of French fries. Food Chemistry 429:136718. https://doi.org/10.1016/j.foodchem.2023.136718
Krogan NT, Marcos D, Weiner AI, Berleth T (2016) The auxin response factor MONOPTEROS controls meristem function and organogenesis in both the shoot and root through the direct regulation of PIN genes. New Phytol 212(1):42–50. https://doi.org/10.1111/nph.14107
Kumar V, Suman U, Rubal, Yadav SK (2018) Flavonoid secondary metabolite: biosynthesis and role in growth and development in plants. In: Yadav, S., Kumar, V., Singh, S. (eds). Recent Trends and Techniques in Plant Metabolic Engineering, Springer, Singapore. https://doi-org.ezaccess.libraries.psu.edu/https://doi.org/10.1007/978-981-13-2251-8_2
Lang GA, Early JD, Martin GC, Darnell RL (1987) EN, Para-, and EC: physiological terminology and classification for dormancy research. HortScience 22(5):371-377. https://doi.org/10.21273/HORTSCI.22.5.701b
Lee M-W, Qi M, Yang Y (2001) A novel jasmonic acid-inducible rice myb gene associates with fungal infection and host cell death. Mol Plant-Microbe Interact 14:527–535. https://doi.org/10.1094/MPMI.2001.14.4.527
Li P, Zheng T, Zhuo X, Zhang M, Yong X, Li L, Wang J, Cheng T, Zhang Q (2021) Photoperiod- and temperature-mediated control of the ethylene response and winter dormancy induction in Prunus mume. Horticultural Plant J 7(4):232–242. https://doi.org/10.1016/j.hpj.2021.03.005
Liu C-C, Ahammed GJ, Wang G-T, Xu C-J, Chen K-S, Zhou Y-H, Yu J-Q (2018) Tomato CRY1a plays a critical role in the regulation of phytohormone homeostasis, plant development, and carotenoid metabolism in fruits. Plant, Cell Environ 41(2):354–366. https://doi.org/10.1111/pce.13092
Manavella PA, Arce AL, Dezar CA, Bitton F, Renou J-P, Crespi M, Chan RL (2006) Cross-talk between ethylene and drought signalling pathways is mediated by the sunflower Hahb-4 transcription factor. Plant J 48(1):125–137. https://doi.org/10.1111/j.1365-313X.2006.02865.x
Martínez-Fernández I, de Moura SM, Alves-Ferreira M, Ferrándiz C, Balanzà V (2020) Identification of players controlling meristem arrest downstream of the FRUITFULL-APETALA2 pathway. Plant Physiol 184(2):945–959. https://doi.org/10.1104/pp.20.00800
Meigh DF, Filmer AAE, Self R (1973) Growth-inhibitory volatile aromatic compounds produced by Solanum tuberosum tubers. Phytochemistry 12(5):987–993. https://doi.org/10.1016/0031-9422(73)85004-6
Müller D, Schmitz G, Theres K (2006) Blind homologous R2R3 Myb genes control the pattern of lateral meristem initiation in Arabidopsis. Plant Cell 18(3):586–597. https://doi.org/10.1105/tpc.105.038745
Nakata MT, Tameshige T, Takahara M, Mitsuda N, Okada K (2018) The functional balance between the WUSCHEL-RELATED HOMEOBOX1 gene and the phytohormone auxin is a key factor for cell proliferation in Arabidopsis seedlings. Plant Biotechnol (Tokyo) 35(2):141–154. https://doi.org/10.5511/plantbiotechnology.18.0427a
Negin B, Hen-Avivi S, Almekias-Siegl E, Shachar L, Jander G, Aharoni A (2023) Tree tobacco (Nicotiana glauca) cuticular wax composition is essential for leaf retention during drought, facilitating a speedy recovery following rewatering. New Phytol 237(5):1574–1589. https://doi.org/10.1111/nph.18615
Noir S, Marrocco K, Masoud K, Thomann A, Gusti A, Bitrian M, Schnittger A, Genschik P (2015) The control of Arabidopsis thaliana growth by cell proliferation and endoreplication requires the F-box protein FBL17. Plant Cell 27(5):1461–1476. https://doi.org/10.1105/tpc.114.135301
Nyankanga RO, Murigi WW, Shibairo SI, Olanya OM, Larkin RP (2018) Effects of foliar and tuber sprout suppressants on storage of ware potatoes under tropical conditions. Am J Potato Res 95:539–548. https://doi.org/10.1007/s12230-018-9662-0
Park E-J, Kim T-H (2021) Arabidopsis OSMOTIN 34 functions in the ABA signaling pathway and is regulated by proteolysis. Int J Mol Sci 22(15):7915. https://doi.org/10.3390/ijms22157915
Pelacho AM, Mingo-Castel AM (1991) Jasmonic acid induces tuberization of potato stolons cultured in vitro. Plant Physiol 97(3):1253–1255. https://doi.org/10.1104/pp.97.3.1253
Perrot-Rechenmann C (2010) Cellular responses to auxin: division versus expansion. Cold Spring Harbor Perspect Biol 2(5):a001446. https://doi.org/10.1101/cshperspect.a001446
Pham GM, Hamilton JP, Wood JC, Burke JT, Zhao H, Vaillancourt B, Ou S, Jiang J, Buell CR (2020) Construction of a chromosome-scale long-read reference genome assembly for potato. GigaScience 9(9):giaa100. https://doi.org/10.1093/gigascience/giaa100
Prange PK, Kalt W, Danials-Lake BJ, Liew CL, Page RT, Walsh JR, Dean P, Coffin R (1998) Using ethylene as a sprout control agent in stored “Russet Burbank” potatoes. J Am Soc Hortic Sci 123(3):463–469. https://doi.org/10.21273/JASHS.123.3.463
Rylski I, Rappaport L, Pratt HK (1974) Dual effects of ethylene on potato dormancy and sprout growth. Plant Physiol 53(4):658–662. https://doi.org/10.1104/pp.53.4.658
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. https://doi.org/10.1101/gr.1239303
Sharma M, Gupta SK, Majumder B, Maurya VK, Deeba F, Alam A, Pandey V (2017) Salicylic acid mediated growth, physiological and proteomic responses in two wheat varieties under drought stress. J Proteom 163:28–51. https://doi.org/10.1016/j.jprot.2017.05.011
Singh AK, Sharma A, Pal AK, Acharya V, Ahuja PS (2013) Genome-wide organization and expression profiling of the NAC transcription factor family in potato (Solanum tuberosum L.). DNA Research 20(4):403–423. https://doi.org/10.1093/dnares/dst019
Singh S, Koyama H, Bhati KK, Alok A (2021) The biotechnological importance of the plant-specific NAC transcription factor family in crop improvement. J Plant Res 134:475–495. https://doi.org/10.1007/s10265-021-01270-y
Sorce C, Lorenzi R, Ceccarelli N, Ranalli P (2000) Changes in free and conjugated IAA during dormancy and sprouting of potato tubers. Funct Plant Biol 27(4):371–377. https://doi.org/10.1071/PP99150
Suttle JC (2003) Role of ethylene in naphthalene-mediated sprout growth inhibition in potato. Acta Horticulturae 619:383–388. https://doi.org/10.17660/ActaHortic.2003.619.45
Suttle JC (2004) Physiological regulation of potato tuber dormancy. Am J Potato Res 81:253–262. https://doi.org/10.1007/BF02871767
Suttle JC, Hultstrand JF (1993) Involvement of abscisic acid in ethylene-induced cotyledon abscission in cotton seedlings. Plant Physiol 101(2):641–646. https://doi.org/10.1104/pp.101.2.641
Swaminathan K, Yang Y, Grotz N, Campisi L, Jack T (2000) An enhancer trap line associated with a D-class cyclin gene in Arabidopsis. Plant Physiol 124(4):1658–1667. https://doi.org/10.1104/pp.124.4.1658
Tamaoki D, Seo S, Yamada S, Kano A, Miyamoto A, Shishido H, Miyoshi S, Taniguchi S, Akimitsu T, Gomi K (2013) Jasmonic acid and salicylic acid activate a common defense system in rice. Plant Signal Behav 8:e24260. https://doi.org/10.4161/psb.24260
The Galaxy Community (2022) The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2022 update. Nucl Acids Res 50(W1):W345–W351. https://doi.org/10.1093/nar/gkac247
Thirugnanasambantham K, Durairaj S, Saravanan S, Karikalan K, Muralidaran S, Islam VIH (2015) Role of ethylene response transcription factor (ERF) and its regulation in response to stress encountered by plants. Plant Mol Biol Rep 33:347–357. https://doi.org/10.1007/s11105-014-0799-9
Thoma JL, Cantrell CL, Zheljazkov VD (2022) Evaluation of essential oils as sprout suppressants for potato (Solanum tuberosum) at room temperature storage. Plants 11(22):3055. https://doi.org/10.3390/plants11223055
Tosetti R, Waters A, Chope GA, Cools K, Alamar MC, McWilliam S, Thompson AJ, Terry LA (2021) New insights into the effects of ethylene on ABA catabolism, sweetening and dormancy in stored potato tubers. Postharvest Biol Technol 173:111420. https://doi.org/10.1016/j.postharvbio.2020.111420
Turnbull CGN, Hanke DE (1985) The control of bud dormancy in potato tubers. Measurement of the seasonal pattern of changing concentrations of zeatin-cytokinins. Planta 165:366–376. https://doi.org/10.1007/BF00392234
Vernoux T, Kronenberger J, Grandjean O, Laufs P, Traas J (2000) PIN-FORMED 1 regulates cell fate at the periphery of the shoot apical meristem. Development 127(23):5157–5165. https://doi.org/10.1242/dev.127.23.5157
Villano C, Esposito S, D’Amelia V, Garramone R, Alioto D, Zoina A, Aversano R, Carputo D (2020) WRKY genes family study reveals tissue-specific and stress-responsive TFs in wild potato species. Sci Rep 10:7196. https://doi.org/10.1038/s41598-020-63823-w
Visse-Mansiaux M, Tallant M, Curty F, Schwärzel R, Brostaux Y, Dupuis B (2021a) Storage of processing potato varieties: the post-CIPC era. Agrarforschung Schweiz 12:175–186. https://doi.org/10.34776/afs11-175e
Visse-Mansiaux M, Tallant M, Brostaux Y, Delaplace P, Vanderschuren H, Dupuis B (2021b) Assessment of pre- and post-harvest anti-sprouting treatments to replace CIPC for potato storage. Postharvest Biol Technol 178:111540. https://doi.org/10.1016/j.postharvbio.2021.111540
Volaire F, Barkaoui K, Grémillet D, Charrier G, Dangles O, Lamarque LJ, Martin-StPaul N, Chuine I (2023) Is a seasonally reduced growth potential a convergent strategy to survive drought and frost in plants? Annals Botany 131(2):245–254. https://doi.org/10.1093/aob/mcac153
Wang Y, Wang Q, Liu M, Bo C, Wang X, Ma Q, Cheng B, Cai R (2017) Overexpression of a maize MYB48 gene confers drought tolerance in transgenic arabidopsis plants. J Plant Biol 60:612–621. https://doi.org/10.1007/s12374-017-0273-y
Wang X, Li Q, Xie J, Huang M, Cai J, Zhou Q, Dai T, Jian D (2021) Abscisic acid and jasmonic acid are involved in drought priming-induced tolerance to drought in wheat. Crop J 9(1):120–132. https://doi.org/10.1016/j.cj.2020.06.002
Willems A, De Veylder LD (2022) The plant anaphase-promoting complex/cyclosome. Ann Rev Cell Dev Biol 38:25–48. https://doi.org/10.1146/annurev-cellbio-120420-092421
Yan Q, Cui X, Lin S, Gan S, Xing H, Dou D (2016) GmCYP82A3, a soybean cytochrome P450 family gene involved in the jasmonic acid and ethylene signaling pathway, enhances plant resistance to biotic and abiotic stresses. PLoS ONE 11(9):e0162253. https://doi.org/10.1371/journal.pone.0162253
Yang W, Wightman R, Meyerowitz EM (2017) Cell cycle control by nuclear sequestration of CDC20 and CDH1 mRNA in plant stem cells. Mol Cell 68(6):1108–1119. https://doi.org/10.1016/j.molcel.2017.11.008
Yang X, Chen L, Yang Y, Guo X, Chen G, Xiong X, Dong D, Li G (2020) Transcriptome analysis reveals that exogenous ethylene activates immune and defense responses in a high late blight resistant potato genotype. Sci Rep 10:21294. https://doi.org/10.1038/s41598-020-78027-5
Yang J, Powers JR, Boylston TD, Weller KM (1999) Sugars and free amino acids in stored Russet Burbank potatoes treated with CIPC and alternative sprout inhibitors. J Food Sci 64(4):592-596. https://doi.org.ezaccess.libraries.psu.edu/https://doi.org/10.1111/j.1365-2621.1999.tb15091.x
Yi X, Du Z, Su Z (2013) PlantGSEA: a gene set enrichment analysis toolkit for plant community. Nucl Acids Res 41(W1):W98–W103. https://doi.org/10.1093/nar/gkt281
Zhang C, Wang D, Yang C, Kong N, Shi Z, Zhao P, Nan Y, Nie T, Wang R, Ma H, Chen Q (2017) Genome-wide identification of the potato WRKY transcription factor family. PLoS One 12(17):e0181573. https://doi.org/10.1371/journal.pone.0181573
Zhang Z, Runions A, Mentink RA, Kierzkowski D, Karady M, Hashemi B, Huijser P, Strauss S, Gan X, Ljung K, Tsiantis M (2020) A WOX/auxin biosynthesis module controls growth to shape leaf form. Curr Biol 30(24):4857-4868.e6. https://doi.org/10.1016/j.cub.2020.09.037
Zhao L, Gao L, Wang H, Chen X, Wang Y, Yang H, Wei C, Wan X, Xia T (2013) The R2R3-MYB, bHLH, WD40, and related transcription factors in flavonoid biosynthesis. Funct Integr Genom 13:75–98. https://doi.org/10.1007/s10142-012-0301-4
Zhao P, Zhang C, Song Y, Xu X, Wang J, Wang J, Zheng T, Lin Y, Lai Z (2022) Genome-wide identification, expression and functional analysis of the core cell cycle gene family during the early somatic embryogenesis of Dimocarpus longan Lour. Gene 821:146286. https://doi.org/10.1016/j.gene.2022.146286
Acknowledgements
We would like to thank Craig Praul of the Penn State Genomic Core Facility for his help and advice throughout this project. We would also like to thank Mark Troyer for providing the tubers used in this study, Jessie Johnson for her assistance in harvesting tubers, and Keegan Rupp and Ian Costello for technical assistance.
Funding
This work was supported by a grant from the 1,4Group of Meridian, ID and by the Lake Erie Regional Grape Research and Extension Center.
Author information
Authors and Affiliations
Contributions
EPD and MAC performed experiments and composed the manuscript. EPD performed all relevant data analyses and compiled data for publication.
Corresponding author
Ethics declarations
Conflict of Interest
Part of this work was supported by the 1,4Group who manufacture and sell the sprout inhibitor DMN. The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Dobry, E.P., Campbell, M.A. The Sprout Inhibitor 1,4-Dimethylnaphthalene Results in Common Gene Expression Changes in Potato Cultivars with Varying Dormancy Profiles. Potato Res. (2024). https://doi.org/10.1007/s11540-024-09772-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11540-024-09772-7