Abstract
The suppression of sprout growth is critical for the long-term storage of potato tubers. 1,4-Dimethylenapthlene (DMN) is a new class of sprout control agent but the metabolic mode of action for this compound has yet to be elucidated. Changes in transcriptional profiles of meristems isolated from potato tubers treated with the DMN were investigated using an Agilent 44 K 60-mer-oligo microarray. RNA was isolated from nondormant Russet Burbank meristems isolated from tubers treated with DMN for 3 days or activated charcoal as a control. RNA was used to develop probes that were hybridized against a microarray developed by the Potato Oligo Chip Initiative. Analysis of the array data was conducted in two stages: total array data was examined using a linear model and the software Limma and pathway analysis was conducted by linking the potato sequences to the Arabidopsis thaliana. DMN elicited a change in a number of transcripts associated with cold responses, water regulation, salt stress, and osmotic adjustment. DMN also resulted in a repression of cyclin or cyclin-like transcripts. DMN also resulted in a 50% decrease in thymidine incorporation suggesting a repression of the S phase of the cell cycle. Quantitative real-time polymerase chain reaction analysis demonstrated that DMN increased transcripts for the cell cycle inhibitors KRP1 and KRP2. We conclude the DMN results in alteration of genes associated with the maintenance of a G1/S phase block possibly through the induction of the cell cycle inhibitors KRP1 and KRP2.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Potato is the third largest agricultural commodity on the world market (http://faostat.fao.org) and a sizable portion of the yearly harvest is placed into long-term storage. Extended storage of harvested potato tubers is predicated on the ability to prevent sprouting of tuber meristems. Sprouting is accompanied by conversion of stored starch to sugars resulting in plant material unsuitable for either the fresh market or for processing. Thus, suppression of sprouting is key to the maintenance of potato postharvest quality. At harvest, potato tubers will not sprout and are in a state of endodormancy (Lang et al. 1987). Endodormancy acquisition, duration, and release are quantitative traits controlled by a suite of genes. Differences in these genes and their regulation results in variability in the length of dormancy between potato cultivars and, at times, between different harvests of the same cultivar (Simko et al. 1997; Hill et al. 2004). The onset and duration of endodormancy in potato is linked to abscisic acid (ABA) and ethylene levels in tuber tissue (reviewed in Suttle 2007). Genetic analysis has shown that genes linked to increased ABA levels prolong the endodormant state (Simko et al. 1997) and changes in expression of genes controlling ABA metabolism are altered as dormancy terminates (Destefano-Beltran et al. 2006). In a number of perennial plants, regulation of FLOWERING LOCUS T through circadian responses and cold has been implicated in establishment and maintenance of endodormancy (Böhlenius et al. 2006; Horvath 2010; Hsu et al. 2011).
In potato, the application of a growth inhibitor such as chlorpropham (CIPC) is often used to prevent premature sprouting following the termination of endodormancy during storage. CIPC is known to disrupt mitotic spindle formation in dividing cells and it has a long history of use as a sprout control agent on stored potatoes (Vaughn and Lehnen 1991). In 1996, the Environmental Protection Agency reregistered CIPC and reduced the allowable amounts of CIPC residue on potato tubers from 50 to 30 ppm (www.epa.gov/oppsrrd1/REDs/0271red.pdf). CIPC is not used on seed potatoes as its effects are irreversible and will prevent the sprouting of seed stock. Thus, there is interest in finding new methods or compounds that can be used to control premature sprouting in stored potato tubers. The naturally occurring compound 1,4-dimethylnaphthalene (DMN) was isolated from potato tubers and was shown to prevent premature sprouting (Beveridge et al. 1981). DMN has demonstrated the ability to reversibly prevent sprouting, making it attractive as a sprout inhibitor on seed tubers (Pinhero et al. 2009). The mode of action of DMN is unknown but recent experiments have determined that CIPC and DMN do not function through a similar mechanism of action and neither sprout inhibitor functions by prolongation of innate dormancy (Campbell et al. 2010).
Transcriptional profiling, using oligo arrays developed for potato, has been used to examine changes in gene expression during tuber initiation (Kloosterman et al. 2008) and in reactivated meristems following treatment with the phytohormones cytokinin and gibberellin (Hartmann et al. 2011). cDNA microarrays have been used to demonstrate transcriptional changes in potato tubers meristems in response to dormancy status (Campbell et al. 2008). In this study, potato tubers were treated with DMN and transcriptional profiling, via oligo array analysis, was used to identify genes that exhibit altered expression in response to DMN.
Experimental methods
Plant material
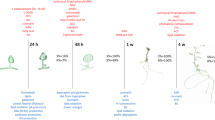
Field-grown certified Russet Burbank seed tubers were obtained from a commercial grower shortly after harvest. The tubers were allowed to wound-heal for 2 weeks at room temperature in the dark. The tubers were transferred to cold (3–4°C) storage. The tubers were periodically evaluated for dormancy status. Only nondormant tubers (100% sprouting after 2 weeks at 20°C) were used in these studies. Thus, the dormancy status of these meristems can be considered to be ecodormant and held in the growth-arrested state due to low temperatures. A diagram of the experimental design is found in Fig. 1.
DMN treatment
On the day of treatment, tubers were transferred from 3°C to 20°C and were immediately washed in running tap water. After drying at room temperature in the dark, the tubers were placed into 4 L airtight containers. A beaker containing ca. 5 g of activated charcoal was placed in the chamber containing the control tubers prior to sealing (control). DMN (0.15 mL liquid) was placed on cotton wool in a beaker and sealed in the treatment chamber (DMN3days). The chambers were incubated in the dark at 20°C for 3 days. At this time tubers were removed and the lateral meristems were isolated using a curette with the aid of a dissecting microscope and either used fresh (thymidine incorporation) or immediately frozen in liquid nitrogen and stored at −80°C as previously described (Campbell et al. 1996). One group of DMN-treated tubers was incubated for a further 2 days in air (dark, 20°C) prior to meristem isolation (DMN3days + 2). Samples were also collected from untreated nondormant buds at day 0 immediately after removal from 3°C storage; henceforth, referred to as ecodormant (Lang et al. 1987)
Thymidine incorporation
Freshly excised meristems were washed with deionized water and groups of 20 buds were incubated on 1 mL of buffer (10 mM Mes/KOH; pH 5.7) containing 0.9 μCi of 3H-thymidine (60 μCi/μmole; American Radiolabeled Chemicals Inc., St. Louis, MO, USA). The meristems were incubated on an oscillating shaker (100 rpm) at room temperature in the dark. After 3 h, meristems were removed from the incubation media, washed extensively with deionized water followed by two washes with incubation buffer containing 5 mM unlabeled thymidine, blotted dry, frozen in liquid nitrogen, and stored at −80°C. To determine incorporation, the frozen meristems were homogenized in 1 mL ice-cold 10% (w/v) TCA. After standing on ice for 3–4 h, the samples were vigorously mixed. An aliquot was removed and placed in a scintillation vial to determine total uptake. A second equal aliquot was removed and placed on a microfiber filter (GF/C; Whatman). The filter was sequentially washed under vacuum with 2 × 20 ml 5% (w/v) TCA and 20 ml 95% (v/v) ethanol (both at 4°C). The washed filter was then placed in a scintillation vial to determine total incorporation into TCA-precipitable material (DNA).
RNA isolation
RNA was isolated from frozen meristems by grinding to a fine powder in a mortar and pestle followed by isolation and purification using a Ribopure Kit (www.ambio.com). RNA was quantified using a BioSpec nano and quality was assessed by gel electrophoresis and visualization of ribosomal RNA. Total RNA (500 ng) was denatured in the presence of a T7 promoter primer and cDNA was synthesized using reverse transcriptase. The cDNA was used as a template for in vitro transcription by synthesis with T7RNA polymerase, which amplified target material and incorporated cyanine 3-labeled CTP. The labeled cRNA was purified using spin columns and quantified using a spectrophotometer. A sample of 1.65 μg of cyanine three-labeled linearly amplified cRNA was hybridized to an Agilent 44 K 60-mer-oligo microarray that was developed by the Potato Oligo Chip Initiative (POCI; Kloosterman et al. 2008). Probe generation and hybridization to the array were conducted through a contract to Gene Logic (www.genelogic.com). Twelve samples were used to probe against the POCI array; three samples from ecodormant meristems (harvested directly from 3°C storage), three samples from meristems isolated from tubers treated with DMN for 3 days in a closed container, three samples treated with water for 3 days in a closed container, three samples treated with DMN for 3 days and then exposed to air for 2 days. The three samples for each treatment were biological replicates.
Microarray analysis
Analysis of the microarray was conducted in two stages; total array data was examined using a linear model and the software Limma and pathway analysis was conducted by linking the potato sequences to the Arabidopsis thaliana. Analysis of the entire microarray data set was accomplished using R and the Limma package (Smyth 2005), or GeneMaths XT (Applied Maths, Inc.). The data was corrected for background and normalized between arrays using a loess method. Comparisons between treatments were established using an empirical Bayes method (Smyth 2004). The use of Limma for analysis of oligo arrays was accomplished by analyzing data from a single emission channel.
Pathway analysis was accomplished by using tBLASTx (Altschul et al. 1990) to search the A. thaliana genome for homologs. Transcripts that exhibited an e-value of less than 1.0e-5 were assigned a function according to the described function for the A. thaliana homolog. The array data was transformed to log base 2 and normalized between arrays using default parameters in GeneMathsXT software package (www.applied-maths.com). Probes had to have hybridization intensities of at least 2 SD greater than background in one or more treatments to be included in the dataset. Pathway Studio 8.0 (www.ariadnegenomics.com) was utilized to determine metabolic changes that were associated with DMN treated for 3 days compared to controls.
QT-PCR
A set of transcripts was selected for further analysis to confirm the microarray data and to determine the possible cell cycle position of tubers treated with DMN. Gene names and primers sequences used for quantitative real-time polymerase chain reaction (QT-PCR) are found in Table 1. The oligo nucleotide sequences spotted on the array were too short for adequate primer design. Therefore, primers were determined by isolating a longer DNA sequence from the DFCI-Potato Gene Index corresponding to the microarray oligonucleotide and searching that sequence for primers using the Applied Biosystems Primer Express 3.0 software (www.appliedbiosystems.com). cDNA template for QT-PCR was prepared from mRNA isolated from potato meristems treated with DMN for 3 days as described above. PCR reactions were run on a StepOneTM Real Time System and analyzed using Step One Software v2.0 (www.appliedbiosystems.com) to determine ΔΔCT values on a log10 scale. Three biological replicates and three technical replicates were analyzed for each sample. Primers amplifying ACTIN2 (TC133139) homologs were used as internal controls and to normalize across reactions. ACTIN2 was chosen as controls because expression was relatively constant across all arrays.
Results
3H-thymidine incorporation was used to determine the effects of DMN treatment on cell division. In meristems isolated from tubers incubated at 20°C for 3 days, between 12.5% and 14.3% of the labeled thymidine taken up was incorporated into DNA while in meristems isolated from DMN-treated tubers, thymidine incorporation was reduced to 6.5–6.7% of the label taken up (Table 1). The level of thymidine incorporation after DMN exposure (under 10%) was similar to the levels previously reported for endodormant potato meristems (Campbell et al. 1996).
Microarray analysis: 34,444 probes were considered expressed in at least one treatment; of these 13,125 were differentially expressed in at least one treatment (Benjamini Hochberg modified p value <0.005; Supplemental Table 1). These large numbers of differentially expressed genes suggest major modifications to the transcriptome for all treatments.
Comparison of gene expression between each treatment and the untreated ecodormant control was visualized using a Venn diagram (Fig. 2). Placing potato tubers in a closed container with or without DMN did result in a small change in meristem transcripts (1897). A total of 1,469 oligonucleotides exhibited expression unique to the tissues treated with DMN for 3 days and the number increased to 3,271 oligonucleotides being unique to the DMN-treated tissues for 3 days and then vented to the air for 2 days. This rise in transcripts changing in expression following DMN treatment and then exposure to air suggests an increasing shift in transcriptional profiles as tissues begin to respond to DMN exposure. Placing tubers in closed containers did result in a unique set of transcript changes independent from DMN-induced gene expression.
Venn diagram showing transcript changes between meristems isolated from tubers prior to treatment (cold control) and those exposed to no DMN for 3 days (3d warm control), DMN treated for 3 days (3d DMN3 treated), and DMN treated for 3 days and then vented to the air for 2 days (3d DMN treated + 2d recovery)
Pathway analysis highlights the similarities between meristems from ecodormant and DMN-treated tubers in comparison to meristems incubated at 20°C for 3 days. Of the ontologies, 294 were associated with genes that are preferentially expressed following incubation at 20°C compared to ecodormant tissues, and 251 ontologies were associated with genes that are preferentially expressed following incubation at 20°C compared to DMN-treated tubers. More than half of the ontologies were common between the comparisons (Supplemental Tables 2, 3). These common ontologies highlight significant changes in the transcriptome associated with cell cycle progression that were generally upregulated in meristems from tubers allowed to incubate at room temperatures in the enclosed container for 3 days relative to the ecodormant tissues (cold control). This is consistent with the increase in 3H-thymidine incorporation during the same time. Interestingly, these same meristems show similar increases in cell cycle progression when compared to meristems from tubers treated with DMN under the same conditions.
Confirmation of differential gene regulation by QT-PCR
QT-PCR was used to both confirm microarray data and to examine additional transcripts that were either not present on the POCI array or were eliminated from analysis after normalization and removal of systematic errors. The transcripts for CDKB1, CYCD3, PCNA, and UTPase, all of which are associated with G1/S phase or early S phase, were decreased following DMN treatment. Transcripts for the cell cycle inhibitor KRP1 and KRP2, which were not present in the microarray data but were implicated as having a role through subnetwork analysis, were elevated in tissues treated with DMN, as were transcripts for MYC2, WRKY, and CYCA3 (Fig. 3).
QT-PCR analysis of potato meristems transcripts isolated from treated with DMN for three 3 days followed by 2-day recovery. All transcripts were a result of data pooled from three biological replicates and three technical replicates. Genes marked by asterisks are derived from two biological replicates. Data represents ΔΔCT values on a log10 scale using ACTIN2 (TC133139) as the internal control. Samples were normalized against cDNA synthesized from RNA isolated from 3-day control tubers
Discussion
Although the growth-inhibiting properties as well as the commercial utility of DMN have been known and exploited for a number of years, the mechanisms through which DMN exerts its biological effects are currently unknown. Previous research has established that DMN does not exert its sprout-inhibiting activities by extending the natural period of tuber endodormancy (Campbell et al. 2010). However, the growth-inhibiting properties of DMN suggest that there is a disruption of cell cycle progression that results in the inhibition of cell division.
In both DMN-treated and ecodormant meristems, a large number of transcripts associated with water regulation, salt stress, and osmotic adjustment were preferentially upregulated relative to what was observed in 20°C untreated tuber meristems (Table 3). Some of these transcripts have also been associated with stress and/or ABA induction in other plant species (Okamoto et al. 2010). However, there were relatively fewer differences among genes associated with these ontologies in meristems from DMN-treated tubers compared to the ecodormant tubers than when ecodormant tubers were compared to tubers incubated at 20°C without DMN. However, there were still some ontologies associated with cold/drought/osmotic stress that were downregulated when tubers were moved to 20°C in the presence of DMN (Tables 1, 2, 3 and Supplemental table 5). This is consistent with previous research on potato indicating that DMN treatment did not result in an increase in ABA levels over control tissues (Campbell et al. 2010), and suggests that DMN treatment is at least partially capable of maintaining transcription profiles associated with cold/drought/osmotic stress in the absence of these stresses.
Cell cycle regulation by DMN
The ca. 50% reduction in thymidine incorporation into DNA in nondormant tuber meristems following DMN treatment (Table 1) and decreased expression of a large number of transcripts that encode cyclin or cyclin-like proteins (Table 2) strongly implicate a role of DMN in cell cycle progression. It has been shown that tubers exiting endodormancy are arrested in the G1 phase of the cell cycle (Campbell et al. 1996). The suppression by DMN of a diversity of cyclin transcripts, while tubers are in a G1-induced dormancy block, should prevent entry into the S phase of the cell cycle. Specifically, progression of the cell cycle through a G1/S phase block has been linked to the expression of the D and E-type cyclins (De Veylder et al. 2003; Doonan and Kitsios 2009), and initiation of meristem growth would require initial expression of the D and E-cyclins for meristem activation. Thus, we hypothesize that DMN maintains cell cycle arrest in the G1/S phase by directly or indirectly inhibiting transcription of D and E-type cyclins. DMN suppression of transcription for the potato homologs of the cyclins CYCD1, CYCD2, CYCD3 would limit protein components for the CDK/cyclin partners that will drive initiation of cell division in potato meristems that result in sprouting (Van Leene et al. 2010).
Cell cycle progression is also controlled by cyclin-dependent kinase inhibitors (CKIs; Inze and De Veylder 2006). In the early G1 phase of the cell cycle, CDKA forms a complex with CYCD3. The activity of this complex is inhibited by the CKIs KRP1 and KRP2, thereby repressing cell cycle advancement (Menges et al. 2005). KRP1 and KRP2 were found to have increased expression in tissues treated with DMN (Fig. 3). This upregulation corresponds with the downregulation of CYCD3 suggesting that DMN prevents sprouting by inhibiting the formation of the CDKA/CYCD3 complex thus resulting in a G1 cell cycle block. It has been suggested that CDKB1 indirectly increases the activity of the CDKA/CYCD3 by phosphorylation and inhibition of KRP2 (Verkest et al. 2005). QT-PCR analysis indicates that DMN decreases the transcription of CDKB1 suggesting that the G1 block induced by DMN is also a result of the maintenance of KRP2 inhibition of the CDK/CYCD3 complex. Deoxyuridine triphosphate (dUTPase) has been shown to be an early marker of dormancy termination prior to entry into the S phase of the cell cycle (Senning et al. 2010). Exposure to DMN resulted in a decrease of UTPase, which again suggests DMN is maintaining a cell cycle block prior to S phase. Figure 4 outlines the interaction of some of the proteins associated with cell cycle regulation. The increased expression of KRP1 and KRP2 suggests two possible positions for cell cycle repression, one in G1 and the other in S phase. The decreased expression of dUTPase suggests that a DMN-induced block via KRP expression is more likely occurring during G1 because the expression of dUTPase occurs prior to S phase entry. The low levels of thymidine incorporation following DMN exposure also support this hypothesis.
Nucleoside diphosphate kinase (NDPK) is downregulated by exposure to DMN (Table 2). The function of the NDPK protein is to regulate the cellular levels of nucleotides and it is linked to organogenesis in animals (Lakso et al. 1992), cell proliferation (Keim et al. 1992), and response to salt stress in plants (Kawasaki et al. 2001). There is evidence that NDPK expression is directly associated with response to stress and reactive oxygen and the alteration of the mitogen-activated protein kinases MPK3 and MPK6 (Moon et al. 2003).
In Saccharomyces cerevisiae, activation of HOG1 by salt stress or high osmoticum results in cell cycle arrest in the G1/S phase by directly affecting the SIC1 gene product (Escote et al. 2004). In potato, DMN treatment results in the increase of MPK4, 6 (Table 1) and KRP1, 2, (Fig. 3) homologues of HOG1 and SIC1, respectively. This suggests that in potato there is a HOG1-like cascade interacting with the G1/S phase regulators CIP/KRP and this pathway may be a target for sprout control by DMN. Transcripts assigned to water regulation and responses to salt stress were increased following DMN treatments (Table 2). It has been reported that a phenotypic response of potato tubers to DMN exposure is an increase in turgidity and maintenance of tuber fresh weight (J. Zalewsky, personal communication). Interestingly the KRP genes are known to decrease cell number and increase cell size when overexpressed (De Veylder et al. 2001).
DMN exposure also increases the transcription of the MPK genes. In yeast, the gene HOG1 is classified as a member of the mitogen-activated protein kinase (MAPK) family. In yeast and other fungi, HOG1 functions as MAPK that interacts within the signal transduction cascade linking growth signals to cell division regulation and it also functions in the sensing system for the regulation of osmotic potential (Brewster et al. 1993; Gustin et al. 1998). HOG1 expression results in a shift in transcriptional profiles, which results in a large change in gene expression in response to changes in osmolarity (O’Rourke and Herskowitz 2004). Exposure of potato tubers to DMN results in an increase both in a HOG1 homolog. The HOG1 cascade in yeast results in growth suppression by downregulation of the G1/S phase cyclins Cln1 and Cln2 (Clotet and Posas 2007). A tBLASTx analysis of the A. thaliana genome using the Cln1 gene from S. cerevisiae finds that the gene CYCA1 is the closest homolog. DMN treatment, which results in elevated expression of the HOG1 homologs MPK4 and MPK6, also results in a decrease in the Cln1 homolog CYCA1. This suggests that DMN growth arrest may result growth arrest due to decrease in the G1/S phase cyclins through the increase activity of MPK4/PK6.
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic Local Alignment Search Tool. J Mol Biol 215:403–410
Beveridge J, Dalziel J, Duncan HJ (1981) Dimethylnaphthalene as a sprout suppressent for seed and ware potatoes. Potato Res 24:77–88
Böhlenius H, Huang T, Charbonnel-Campaa L, Brunner AM, Jansson S, Strauss SH, Nilsson O (2006) CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 312:1040–1043
Brewster JL, Valoir TD, Dwyer ND, Winter E, Gustin MC (1993) An osmosensing signal transduction pathway in yeast. Science 259(5102):1760–1763
Campbell MA, Suttle J, Sell TW (1996) Changes in cell cycle status and expression of p34cdc2 kinase during potato tuber meristem dormancy. Physiol Plant 98:743–752
Campbell MA, Segear E, Beers L, Knauber D, Suttle J (2008) Dormancy in potato tuber meristems: chemically induced cessation in dormancy matches the natural process based on transcript profiles. Funct Integr Genom 8:317–328
Campbell MA, Gleichsner A, Alsbury R, Horvath D, Suttle J (2010) The sprout inhibitors chlorpropham and 1,4-dimethylnaphthalene elicit different trasncriptional profiles and do not supress growth through a prolongation of the dormant state. Plant Mol Biol 73:181–189
Clotet J, Posas F (2007) Control of cell cycle in response to osmostress: lessons from yeast. Methods Enzymol 428:63–76
De Veylder L, Beeckman T, Beemster GTS, Krols L, Terras F, Landrieu I, Van Der Schueren E, Maes S, Naudts M, Inzé D (2001) Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. The Plant Cell Online 13(7):1653–1668
De Veylder L, Joubes J, Inzé D (2003) Plant cell cycle transitions. Curr Opin Plant Biol 6:536–543
Destefano-Beltran L, Knauber D, Huckle L, Suttle JC (2006) Effects of postharvest storage and dormancy status on ABA content, metabolism, and expression of genes involved in ABA biosynthesis and metabolism in potato tuber tissues. Plant Mol Biol 61(4–5):687–697
Doonan J, Kitsios G (2009) Functional evolution of cyclin-dependent kinases. Mol Biotechnol 42(1):14–29
Escote X, Zapater M, Clotet J, Posas F (2004) Hog1 mediates cell-cycle arrest in G1 phase by the dual targeting of Sic1. Nat Cell Biol 6(10):997–1002
Gustin MC, Albertyn J, Alexander M, Davenport K (1998) MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev 62(4):1264–1300
Hartmann A, Senning M, Hedden P, Sonnewald U, Sonnewald S (2011) Reactivation of meristem activity and sprout growth in potato tubers require both cytokinin and gibberellin. Plant Physiol 155(2):776–796
Hill AE, Shachar-Hill B, Shachar-Hill Y (2004) What are aquaporins for? J Membr Biol 197(1):1–32
Horvath D (2010) Bud dormancy and growth. Plant Dev Biol-Biotechnol Perspect 2:53–70
Hsu CY, Adams JP, Kim H, No K, Ma C, Strauss SH, Drnevich J, Vandervelde L, Ellis JD, Rice BM, Wickett N, Gunter LE, Tuskan GA, Brunner AM, Page GP, Barakat A, Carlson JE, dePamphilis CW, Luthe DS, Yuceer C (2011) FLOWERING LOCUS T duplication coordinates reproductive and vegetative growth in perennial poplar. PNAS 108(26):10756–10761
Inze D, De Veylder L (2006) Cell cycle regulation in plant development. Annu Rev Genet 40:77–105
Kawasaki S, Borchert C, Deyholos M, Wang H, Brazille S, Kawai K, Galbraith D, Bohnert HJ (2001) Gene expression profiles during the initial phase of salt stress in rice. The Plant Cell Online 13(4):889–906
Keim D, Hailat N, Melhem R, Zhu XX, Lascu I, Veron M, Strahler J, Hanash SM (1992) Proliferation-related expression of p19/nm23 nucleoside diphosphate kinase. J Clin Investig 89(3):919–924
Kloosterman B, De Koeyer D, Griffiths R, Flinn B, Steuernagel B, Scholz U, Sonnewald S, Sonnewald U, Bryan GJ, Prat S, Bánfalvi Z, Hammond JP, Geigenberger P, Nielsen KL, Visser RG, Bachem CW (2008) Genes driving potato tuber initiation and growth: identification based on transcriptional changes using the POCI array. Funct Integr Genom 8(4):329–340
Lakso M, Steeq P, Westphal H (1992) Embryonic expression of nm23 during mouse organogenesis. Cell Growth Differ 3(12):873–879
Lang GA, Early JD, Martin GC, Darnell RL (1987) Endodormancy, paradormancy, and ecodormancy—physiological terminology and classification for dormancy research. HortSci 22(3):371–377
Menges M, de Jager S, Gruissem W, Murray JA (2005) Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J 41:546–566
Moon H, Lee B, Choi G, Shin D, Prasad DT, Lee O, Kwak SS, Kim DH, Nam J, Bahk J, Hong JC, Lee SY, Cho MJ, Lim CO, Yun DJ (2003) NDP kinase 2 interacts with two oxidative stress-activated MAPKs to regulate cellular redox state and enhances multiple stress tolerance in transgenic plants. Proc Natl Acad Sci 100(1):358–363
Okamoto M, Tatematsu K, Matsui A, Morosawa T, Ishida J, Tanaka M, Endo TA, Mochizuki Y, Toyoda T, Kamiya Y, Shinozaki K, Nambara E, Seki M (2010) Genome-wide analysis of endogenous abscisic acid-mediated transcription in dry and imbibed seeds of Arabidopsis using tiling arrays. Plant J 62(1):39–51
O’Rourke SM, Herskowitz I (2004) Unique and redundant roles for HOG MAPK pathway components as revealed by whole-genome expression analysis. Mol Biol Cell 15(2):532–542
Pinhero RG, Coffin R, Yada RY (2009) Post-harvest storage of potatoes. In: Singh J, Kaur L (eds) Advances in potato chemistry and technology. Academic, New York, pp 339–370
Senning M, Sonnewald U, Sonnewald S (2010) Deoxyuridine triphosphatase expression defines the transition from dormant to sprouting potato tuber buds. Mol Breed 26(3):525–531
Simko I, McMurray S, Yang HM, Manschot A, Davies PJ, Ewing EE (1997) Evidence from polygene mapping for a causal relationship between potato tuber dormancy and abscisic acid content. Plant Physiol 115:1453–1459
Smyth G (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3. doi:10.2202/1544-6115.1027
Smyth GK (2005) Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W (eds) Bioinformatics and computational biology solutions using R and bioconductor. Springer, New York, pp 397–420
Suttle JC (2007) Dormancy and sprouting. In: Vreugdenhill D, Bradshaw J, Gebhart C et al. (eds) Potato biology and biotechnology: advances and perspectives. Amsterdam, Elsevier
Van Leene J, Hollunder J, Eeckhout D, Persiau G, Van De Slijke E, Stals H, Van Isterdael G, Verkest A, Neirynck S, Buffel Y, De Bodt S, Maere S, Laukens K, Pharazyn A, Ferreira PC, Eloy N, Renne C, Meyer C, Faure JD, Steinbrenner J, Beynon J, Larkin JC, Van de Peer Y, Hilson P, Kuiper M, De Veylder L, Van Onckelen H, Inzé D, Witters E, De Jaeger G (2010) Targeted interactomics reveals a complex core cell cycle machinery in Arabidopsis thaliana. Mol Syst Biol 6. doi:10.1038/msb.2010.53
Vaughn K, Lehnen G (1991) Mitotic disrupter herbicides. Weed Sci 39:450–457
Verkest A, Weinl C, Inzé D, De Veylder L, Schnittger A (2005) Switching the cell cycle. Kip-related proteins in plant cell cycle control. Plant Physiol 139:1099–1106
Acknowledgements
A portion of this work was supported through a grant from the 1,4-Group to MAC.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
XLSX 11,838 kb
Rights and permissions
About this article
Cite this article
Campbell, M.A., Gleichsner, A., Hilldorfer, L. et al. The sprout inhibitor 1,4-dimethylnaphthalene induces the expression of the cell cycle inhibitors KRP1 and KRP2 in potatoes. Funct Integr Genomics 12, 533–541 (2012). https://doi.org/10.1007/s10142-011-0257-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-011-0257-9