Abstract
We characterized transcriptional responses of potato plants to multiple abiotic stresses and used this information to identify potential mechanisms through which over-expression of the stress related transcription factor CBF1 from Arabidopsis thaliana (AtCBF1) confers multiple stress tolerance. Most transcriptional changes were specific to each condition, but genes involved in phenyl-propanoid biosynthesis were affected by all abiotic stresses evaluated. Interestingly, over-expression of AtCBF1 in potato plants not only conferred tolerance to low temperatures, as previously reported, but also to high-light conditions at 22 °C, suggesting that it confers multiple stress tolerance by enhancing the ability of plants to cope with an excess of radiant energy. Finally, we found that transcriptional changes triggered by abiotic stress were much larger than those resulting from AtCBF1 over-expression in potato, revealing that overexpression of an heterologous transcription factor causes minor alterations in the plant transcriptome in comparison to transcriptional changes triggered by abiotic stresses.
Resumen
Caracterizamos respuestas transcripcionales de plantas de papa a múltiples estreses abióticos y utilizamos esta información para identificar mecanismos potenciales a través de los cuales la sobreexpresión del factor de transcripción CBF1 relacionado con agobio de Arabidopsis thaliana (AtCBF1) confiere tolerancia múltiple al estrés. La mayoría de los cambios transcripcionales fueron específicos para cada condición, pero se afectaron los genes involucrados en la biosíntesis de fenil-propanoides por todos los estreses abióticos evaluados. Interesantemente, la sobreexpresión del AtCBF1 en plantas de papa no solo confirieron tolerancia a bajas temperaturas, como se ha reportado previamente, sino también a condiciones de alta luminosidad a 22 °C, lo que sugiere múltiple tolerancia al estrés mediante el aumento de la habilidad de las plantas para hacer frente a un exceso de energía radiante. Finalmente, encontramos que los cambios transcripcionales disparados por el agobio abiótico fueron mayores que aquellos que resultaron de la sobreexpresión de AtCBF1 en papa, revelando que la sobreexpresión de un factor heterólogo de transcripción causa alteraciones menores en el transcriptoma de la planta en comparación a cambios transcripcionales disparados por estreses abióticos.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The increase in food demand caused by the continuous growth of the population, along with the absence of additional areas with optimal environmental conditions for crop production, is leading to an expansion of agricultural activities in marginal regions (Khush 2001; Rosegrant and Cline 2003; Tilman et al. 2002). In addition, man-induced global climate changes are leading to repeated episodes of drought or extreme temperatures in geographical regions where those abiotic stresses were uncommon. Therefore, understanding the molecular and physiological mechanisms underlying the adaptive responses of plants to abiotic stress conditions is crucial. This knowledge is required not only to facilitate the development of novel cultivars with higher levels of stress tolerance, but also to evaluate and minimize potential environmental and health related risks associated with these biotechnological innovations.
The mechanisms leading to stress tolerance are extremely complex. Some species are adapted to live in environments where intense stress is always present, so they have constitutive adaptations (Bohnert et al. 1995). Others display physiological and molecular adaptations which allow them to tolerate harsh conditions only if they were previously exposed to anticipatory stress-associated signals (generally milder stress conditions). This process, known as acclimation, involves homeostatic alterations that contribute to improving the adjustment of organisms to changes in external conditions (Thomashow 2001). Thus, an acclimation period can confer tolerance to extreme environmental conditions that might otherwise be lethal, without the cost of expressing constitutive adaptive responses.
Acclimation to both drought and low temperature conditions is mediated through specific as well as common signaling pathways, which result in a large reprogramming of the plant transcriptome (Shinozaki and Yamaguchi-Shinozaki 2000; Shinozaki et al. 2003). In addition to increasing the expression of genes encoding hydrophilic proteins that prevent membrane damage, and molecular chaperons that stabilize proteins against freezing or dehydration-induced denaturation, low temperatures and water deficits also induce the expression of genes encoding scavengers of reactive oxygen species (ROS) (Cushman and Bohnert 2000; Mittler 2002; Zhu 2002). These molecules are formed when electrons transport chains become fully reduced as a consequence of an excess of radiant energy that cannot be processed through the photosynthetic apparatus. This can be due to a decrease in photosynthetic activity caused by drought induced stomatal closure, or to a reduction in enzymatic activity associated with low temperature conditions. Indeed, ROS scavengers are critical to avoid photo-oxidative damage to membranes in response to abiotic stress (Foyer and Allen 2003).
Physiological and molecular responses to drought and low temperatures are mediated by both ABA-dependent as well as by ABA-independent signaling pathways (Qin et al. 2011). An important regulatory element found in the promoters of drought and cold regulated genes is the sequence known as CRT/DRE (C-repeat/Dehidratation Responsive Element), composed of a core CCGAC element (Baker et al. 1994; Yamaguchi-Shinozaki and Shinozaki 1994). The DREB family of transcription factors bears homology to AP2/ERF DNA-binding proteins and recognizes CRT/DRE sequences. Members of the DREB2 family are expressed in response to drought, and confer tolerance to it (Liu et al. 1998). On the other hand, DREB1b, DREB1c, and DRE·B1a, also known as CBF1, CBF2 and CBF3, are induced in response to cold exposure and activate the expression of cold-regulated genes (COR) upon binding to CRT/DRE elements present in their promoters. Interestingly, over-expression of CBF1 induces constitutive expression of COR genes and enhances freezing tolerance in Arabidopsis thaliana in non-acclimated plants (Jaglo-Ottosen et al. 1998). In addition, over-expression of any member of the DREB1 family of transcription factors from Arabidopsis thaliana, induces tolerance to multiple abiotic stresses in other species (Khan 2011). For example, expression of Arabidopsis CBF1 confers tolerance to chilling, oxidative, and drought stress in tomato (Hsieh et al. 2002a, b).
Solanum tuberosum, a cultivated tuber-bearing potato plant, is sensitive to low temperature conditions as well as to drought (Chen and Li 1980), and an important biotechnological aim is to develop transgenic potato plants capable of tolerating multiple stresses. The first aim of the work described here was to identify the genes whose expression is affected by different abiotic stress conditions in potato (i.e. to define the “potato abiotic stress transcriptome”), in order to determine convergent targets of different stress associated signaling pathways that could be manipulated to trigger multiple stress tolerance in this species. Several studies that analyze the potato transcriptome in response to drought, cold as well as other stresses such as high temperatures and salt, have already been conducted (Carvallo et al. 2011; Gangadhar et al. 2014; Hancock et al. 2014; Kondrak et al. 2012; Rensink et al. 2005a, b; Vasquez-Robinet et al. 2008; Zhang et al. 2014). Here we analyzed the potato transcriptome in response to an additional environmental stress, high light conditions at warm temperatures (22 °C), and compared the effect of this stress to that of drought and cold conditions.
In addition, several groups have recently reported that over-expression of A. thaliana CBF1 and CBF3 genes in potato plants confers tolerance to salt stress (Behnam et al. 2006), freezing (Movahedi et al. 2012; Pino et al. 2007, 2008)), high temperatures (Dou et al. 2015) and drought (Movahedi et al. 2012), but the physiological and molecular mechanisms through which CBF genes confer abiotic stress tolerance in potato plants are not well understood. Potato plants have three CBF genes in tandem, which are orthologues of Arabidopsis CBF1, CBF2 and CBF3 and, indeed, the potato CBF1 orthologue is also induced in response to cold treatments (Pennycooke et al. 2008). Interestingly, overexpressing the AtCBF3 gene in potato plants results in the upregulation of some genes that are part of the Arabidopsis CBF regulon, but other genes induced by AtCBF3 in potato encode proteins whose ortholgues have not been identified as part of the Arabidopsis CBF regulon (Carvallo et al. 2011). Whether these genes responding to the overexpression of the AtCBF3 gene in potato but not in Arabidopsis also respond to other stress treatments in potato is not known. Therefore, a second aim of this project was to determine whether a “potato stress-transcriptome” could be used to obtain clues on the mechanisms underlying multiple stress tolerance in transgenic potato plants over-expressing an AtCBF1 gene.
Finally, genetically modified organisms (GMOs) are subject to intense monitoring by governmental regulatory agencies, both in relation to environmental bio-safety, as well as in terms of potential effects on human health. Therefore, the characterization of potentially unintended effects associated with the transgenic event analyzed is likely to be an important step of the regulatory process. In this particular case, we considered “unintended” effects all those changes in the expression of genes whose transcripts levels are affected by overexpression of a stress related transcription factor, but do not respond to stress treatments in wild type plants. Recent work conducted to characterize “unintended” effects associated with the over-expression of an abiotic-transcriptional regulator suggests that this strategy may not necessarily cause extensive “unintended” transcriptional alterations (Abdeen et al. 2010). However, this result might be biased given that a later study evaluated the effect of over-expressing an A.thaliana transcription factor in A.thaliana. Here we used a combination of approaches to characterize the effect of overexpressing Arabidopsis thaliana CBF1 in a heterologous species. This approach allowed us to narrow down the transcriptional changes that are likely to be considered “unintended” effects to a small subset of genes, which should facilitate the evaluation of the potential risks associated with these transgenic plants. In addition, we also observed that the transcriptional changes induced by different abiotic stresses were much larger than those caused by AtCBF1 over-expression in potato. This indicates that overexpression of an heterologous transcription factor in a crop species only causes limited effects on the plant transcriptome, limiting the extent of potential “unintended” effects.
Materials and Methods
Plant Material and Growth Conditions
Potato plants (Solanum tuberosum cv. Désirée) were grown under sterile conditions in glass bottles containing Murashige and Skoog (MS, Sigma) medium solidified with 0.8 % phytoagar (Sigma). Plants were propagated in vitro and grown under 16 h light (50 μmol.m−2.s−1 cool-white fluorescent illumination) / 8 h dark cycles, at 22 °C. The analysis of physiological and molecular responses of plants to different stress treatments was conducted on plants transferred to plastic pots containing a mixture of perlite:vermiculite:peat moss (1:1:1), and acclimated for 1 month under the same light and temperature conditions in which the treatments were applied.
Stress Treatments
To analyze the effect of low temperature conditions, plants were first grown under long days (16 h light/8 h darkness) with fluorescent illumination (50 μmol.m−2.s−1) at 22 °C. After 1 month, half of the plants were exposed to 4 °C under similar light conditions, while the remaining plants were kept at 22 °C. The effect of high light stress was evaluated on plants grown first under long day conditions in a growth chamber illuminated with high pressure sodium lamps at 50 μmol.m−2.s−1, at 22 °C. After 1 month, half of the plants were exposed to 800 μmol.m−2.s−1 at 22 °C, while the remaining plants were kept under 50 μmol.m−2.s−1 at 22 °C. For drought stress, plants were grown under natural radiation (1450 μmol.m−2.s−1 at midday) in a greenhouse in 3 l pots supplemented with daily irrigation until water soil capacity was saturated. After 1 month under this condition, irrigation was interrupted for half of the plants while the remaining plants continued with the previous daily irrigation regime.
Anthocyanin and Chlorophyll Measurement
For anthocyanin determination, petioles from the third completely expanded leaf were harvested and placed in 1.5 ml plastic tubes containing 1 ml of 1 % HCl methanol, and kept in darkness for 2 days at 4 °C. The amount of anthocyanin was determined spectrophotometrically, measuring absorbance at 530 nm and correcting for chlorophyll absorption (Mancinelli et al. 1991). For chlorophyll determination, leaf discs of 20 mm2 were placed in 1.5 ml plastic tubes containing 1 ml of N,N-dimethylformamide and incubated for 3 days in darkness at 4 °C. Chlorophyll levels were measured spectrophometrically, determining absorbance at 647 and 660 nm, and calculating chlorophyll concentration according to Moran and Porath (Moran and Porath 1980).
Leaf Area, Chlorophyll Fluorescence, Photosynthetic Rate and Leaf Conductance Measurements
Leaf area was determined using a Li-3100C Area Meter (Li-Cor Inc., Lincoln, NE, USA). Chlorophyll fluorescence measurements were determined using the Fluorescence Monitoring System 2 (FMS-2, Hansatech Instruments Ltd, Kings Lynn, UK), and Fv/Fm ratios were measured in dark adapted leaves. This parameter is known to correlate with the number of functional PSII reaction centers, so that it can be used to quantify photoinhibition (Somersalo and Krause 1989). Maximum photosynthetic rates were measured using a Li-6200 Portable Photosynthesis System (Li-Cor, Lincoln, NE, USA), with a 0.25-L chamber attached to a regulated portable power (QB1 205LI-670, Quantum Devices Inc., Barneveld, WI) at 1500 μmol m-2.s-1 of red light. Leaf conductance was measured using the Leaf Porometer Model SC-1 (Decagon Devices Inc., Pullman, WA, USA).
Transpiration and Desiccation Tolerance Measurements
Transpiration was estimated from water consumption measurements, which were based on changes in pot weight recorded on a daily basis. The response to short term desiccation was measured analyzing changes in fresh weight of detached leaves (Verslues et al. 2006) that were kept under continuous illumination (50 μmol.m−2.s−1) at 22 °C for 24 h.
Development of AtCBF1 Overexpressing Potato Plants
The coding sequence of the Arabidopsis thaliana CBF1 gene was amplified by PCR and ligated into the binary vector pCHF3 (Fankhauser et al. 1999). The resulting plasmid pCHF3.AtCBF1, which contains the coding sequence under the control of CaMV 35S promoter and the Rubisco terminator, was transformed into Agrobacterium tumefaciens strain GV3101 by electroporation. Potato plants (Solanum tuberosum Cv. Désirée) were transformed with the pCHF3. AtCBF1 as previously described (Beaujean et al. 1998).
Molecular and Phenotypic Characterization of Transgenic Plants
To confirm the presence of the transgene, 11 independent lines were transferred to plastic pots containing a mixture of perlite:vermiculite:peat moss (1:1:1), and allowed to grow for 3 weeks. DNA was isolated using the CTAB method and was used to confirm the presence of the transgene by PCR. The primers used for genotyping were: 1224 (5′-CGCCAGGGTTTTCCCAGTCACGAC-3′) and 1233 (5′-AGCGGATAACAATTTCACACAGGA-3′), which recognizes the promoter and terminator regions of the pCHF3. The expression of AtCBF1 transgene was then measured in PCR confirmed lines. For this, RNA was isolated from mature leaves using the RNeasy® Plant Mini Kit (Qiagen). Reverse transcription was done using SuperScriptTM III Reverse Transcripatase (Invitrogen®). Expression was evaluated using semi-quantitative RT-PCR. The actin gene was used as a housekeeping gene to normalize expression levels across samples.
For phenotypic analysis, AtCBF1 over-expressing and WT potato plants were grown in 300 ml plastic pots under long day conditions for 3 weeks. Total height, as well as length of specific internodes, was measured with a ruler. Potato yield was also measured in plants grown for 4 months in 3 l plastic pots kept under the same conditions. For the later we assessed tuber number as well as tuber weight per plant.
Expression Profiling
Total RNA extracted from young leaves belonging to two different potato plants were pooled for each biological replicate, and two biological replicates were obtained and characterized for each experimental condition. Five micrograms of RNA from each sample were processed and hybridized to GeneChip® Tomato Genome Arrays (Affymetrix, Inc.), according to the manufacturer’s instructions. Microarray signals were made comparable by scaling the average overall signal intensity of all probe sets to a target signal of 250. Data were analyzed using MAS5. Microarray data was analyzed using different statistical approaches described below. Microarray results were confirmed for a subset of genes and conditions using qRT-PCR. For this, total RNA was isolated using the RNeasy® Plant Mini Kit (Qiagen). Then, RNA samples were subjected to a DNAse treatment with RQ1 RNase-Free Dnase (Promega), and to retrotranscription using SuperScriptTM III Reverse Transcripatase (Invitrogen®) and oligo-dT. Synthesized cDNAs were amplified with FastStart Universal SYBR Green Master (Roche) using the Mx3000P® QPCR System (Stratagene). Data were analyzed by MxPro™ QPCR Software (Stratagene). We quantified the relative amount of RNA of the CHS, PAL1, and PORb genes using the Ef1 gene as housekeeping control (Nicot et al. 2005). The list of the primers used is detailed in the ESM_18.
ANOVA
One way ANOVA was performed on the resulting data to identify genes regulated by different abiotic stress factors. ANOVA was also used to identify genes whose expression was up or down regulated in transgenic plants overexpressing AtCBF1 compared to non transgenic WT controls. Multiple testing was taken into account by converting p-values to q-values (Storey and Tibshirani 2003). Genes considered for further analysis had a q-value of less than 0.1 and a 2 fold change in expression compared to their corresponding controls, and had a present call in all samples from at least one experimental condition. BLASTP was used to determine the identity of each gene and, whenever possible, the closest Arabidopsis homologue was established.
Hierarchical Clustering
Global expression analysis of AtCBF1-OX plants exposed to abiotic stress was performed using the statistical programming framework R (Gentleman et al. 2004). In order to produce a complete-linkage hierarchical clustering dendrogram, a metric based on absolute correlation levels between sample expression profiles was considered. The obtained ordination of samples was robust against the use of different linkage methods. Moreover, sample profiles built upon the whole set of transcripts or just a non-specific filtered subset of genes produced identical results.
Linear Model Analysis
To analyze transcriptional responses of AtCBF1-OX plants exposed to abiotic stress, the Bioconductor Limma package was used to fit a linear model to the expression value for each gene, and to assess the significance of differential expression between different experimental conditions (Smyth 2004). The Benjamini and Hochberg’s method was used to control the false discovery rate (fdr) at a 0.05 level (Benjamini and Hochberg 1995). Additionally, a minimal fold change level of two was required for a gene to be dubbed as differentially expressed.
Template Matching Analysis
A feature selection method based on a method known as template matching (Pavlidis and Noble 2001) was implemented in R scripts, in order to identify genes with patterns of expression strongly correlated to ideal responders to genetic background, temperature or light stress conditions main effects in AtCBF1-OX plants exposed to multiple stresses. The number of genes associated to each one of these templates, with a correlation larger than a given absolute R threshold value, was used as a proxy for the extent of global transcriptional changes induced by the corresponding condition, at that statistical significance level.
Multi Response Permutation Procedure
The MRPP analysis (Reiss et al. 2010) was performed using the R package Vegan (Dixon 2009). The MRPP analysis assess for group mean differences, testing for unusually low average intra-group dissimilarities compared against random group assignment of sampling units. In the same spirit of analysis of variance procedures, if two groups are different, the average of the within-group dissimilarities ought to be less than the average dissimilarities between random collections of samples. The statistic of the test, δ, was calculated as a weighted sum of within-group average distances. Its associated statistical significance was estimated through random shuffling group memberships. Finally, the effect size parameter A = 1−δobserved/expected, allowed us to establish a quantitative comparison between the magnitude of the transcriptional changes induced by differences in genomic backgrounds, and those associated with environmental stress conditions.
Results and Discussion
Global Analysis of the Potato Transcriptome to Multiple Abiotic Stresses Reveals Convergent Effects on the Phenyl-Propanoid Biosynthetic Pathway
To identify common as well as specific molecular mechanisms underlying responses of potato plants to low temperatures, drought, and high light stress, we conducted a global analysis of the potato transcriptome. For this, we used Affymetrix tomato arrays that have already been successfully used to evaluate gene expression in potato (Bagnaresi et al. 2008) and conducted the experiments using tissue culture potato plantlets transferred to plastic pots containing a mixture of perlite:vermiculite:peat moss. The plants were grown for 1 month in a growth chamber under a 16 h light (50 μmol.m−2.s−1)/ 8 h dark cycle at 22 °C and then one group of plants was exposed for 1 day to a sharp decrease in temperature from 22 to 4 °C keeping the same light conditions, while another group of plants was exposed to an increase in light intensity from 50 to 800 μmol.m−2.s−1 at warm temperatures (22 °C). Then, the transcriptome of both groups of plants was compared to that of plants remaining under control conditions (i.e. low light and warm temperatures). In addition, we also analyzed the transcriptome of potato plants that were grown in the greenhouse with daily irrigation, and compared it to the transcriptome of plants that were grown identically for 1 month but received no additional water for the last 2 days before they were harvested.
Out of approximately 10,000 genes present in the array, 7982 genes showed signal intensities above background levels in at least one environmental condition, revealing that Affymetrix tomato arrays were indeed useful to evaluate the transcriptome of potato plants. Using a one way analysis of variance we identified 1427 genes with a statistically significant stress treatment effect (q-value < 0.1), and a three-fold (or more) increase or decrease in expression under at least one condition, compared to expression levels observed in plants not exposed to stressful conditions (Fig. 1, ESM_1). The number of stress regulated genes increased to 3138 if we also considered genes whose expression increased or decreased at least two fold. Thus, this analysis indicates that up to 40 % of the potato genes evaluated show significant changes in expression under the abiotic stress conditions tested here. Many of the gene families identified as stress regulated in our experiments have previously been identified as stress regulated in studies conducted with field grown potato plants, but we should be cautious in extrapolating the results obtained here with plants derived from tissue cultured plantlets to those taking place in field grown plants under conventional conditions.
Genes preferentially up-regulated in response to a decrease in water availability included several members of the homeodomain, NAC, bZIP, zinc finger and MYB families of transcription factors (ESM_1 and ESM_2), which have previously been implicated in transcriptional responses to drought in general (Ariel et al. 2007; Golldack et al. 2011; Nakashima et al. 2012; Uno et al. 2000) as well as specifically in potato (Kondrak et al. 2012; Vasquez-Robinet et al. 2008). In addition to transcription factors, the list of strongly drought up-regulated genes included genes encoding metabolic enzymes such as the proline dehydrogenase ERD5, or signaling factors such as phosphatases, known to mediate ABA signaling in other species (ESM_1 and ESM_2). Genes down-regulated under drought were mostly involved in protein translation (ESM_1 and ESM_2). In addition, several genes associated with cell wall elongation and chloroplast function were also down-regulated in these water stressed plants (ESM_1 and ESM_2).
A significant overlap was found between genes up-regulated in response to drought, and those up-regulated in response to low temperature, which was twice the overlap expected by chance (p = 1.6 × 10−28, hypergeometric distribution) (Fig. 1a). Some of these genes encoded transcriptional regulators of the b-ZIP gene family, which are likely involved in ABA signaling (ESM_3). In addition, genes involved in sugar metabolism, mainly starch degradation and sucrose synthesis, were also found among those simultaneously up-regulated in response to both stress treatments (ESM_1 and ESM_3). Interestingly, a common consequence of plant exposure to drought or low temperature conditions is a reduction in leaf water potential and, therefore, the enhanced expression of genes involved in starch degradation and sucrose synthesis might have an osmotic function helping to maintain cell turgor under water limiting conditions. Interestingly, we found an aquaporin related gene whose expression was strongly up-regulated in response to low temperatures but was strongly down-regulated in response to drought (ESM_4). The opposite behavior of this gene most likely reflects the different adaptive strategies activated by these two stresses to overcome a similar physiological problem. Indeed, while reduced leaf water potential under drought is caused by limited water availability, dehydration under low temperature conditions is triggered by reduced rates of water transport into cells. Therefore, the down-regulation of this aquaporin could be involved in minimizing water loss from cells under drought, while its up-regulation in response to low temperatures most likely facilitates water flow into cells, helping to maintain high water potentials within cells in both cases.
On the other hand, more than 200 genes were affected by the cold treatment but not (or to a much lesser extent) by water deficit (Fig. 1a). An interesting subset of genes in this class included LATE ELONGATED HYPOCOTYL 1 (LHY) and several of its homologs, such as REVEILLE1 (RVE1) and RVE8/LCL5 (ESM_5). LHY is a single MYB transcription factor that is part of the circadian clock in plants (Schaffer et al. 1998). LHY acts by directly promoting or repressing the expression of core-clock components, as well as many clock output genes (Carre and Kim 2002). Interestingly, LHY has recently been shown to control the circadian regulation of CBF1-3 expression and to mediate the cold induction of their expression in Arabidopsis. Furthermore, double mutants of LHY and its closest homolog CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) display increased sensitivity to freezing temperatures (Dong et al. 2011). Thus, the observation of increased expression of LHY and several of its homologs in response to cold suggests that this gene family may have an important role mediating temperature responses in the plant kingdom, helping to keep the cold transcriptome under circadian control.
Among genes down-regulated in response to low temperature conditions we found many associated with mitochondrial electron transport. This might be associated with increased electron flow through the alternative oxidase pathway, which might function to minimize the production of reactive oxygen species under low temperature conditions. Indeed, an almost ten-fold increase in expression was observed for a gene encoding an alternative oxidase homologue (AOX) in response to the low temperature treatment (see probe ID Les.4223.1.S1_at S1). Interestingly, AOX enzymes function to prevent the formation of an excess reactive oxygen species in the mitochondria under stressful environmental conditions, but may also have more pervasive effects, including some that are extramitochondrial (Fiorani et al. 2005).
As expected, all the above mentioned transcriptional changes were associated with rapid changes in physiological processes. Both drought and low temperature conditions triggered a reduction in photosynthetic activity (Fig. 2a). This response was associated with a reduction in chlorophyll levels in plants exposed to low temperatures but not in those exposed to drought (Fig. 2b). On the other hand, plants exposed to drought (but not those under low temperature conditions) displayed reduced leaf conductance (Fig. 2c). Both treatments triggered a reduction in the Fv/Fm ratio (Fig. 2d), which indicates photo-oxidative damage most likely generated by absorbed light that exceeds the photosynthetic capacity of plants exposed to these abiotic stresses.
Physiological parameters measured in potato plants exposed to drought and low temperature stresses. a Photosynthetic rate. b Leaf chlorophyll content. c Stomatal conductance (d) Fv/Fm. In (a) data are mean ± SD of four biological replicates. In (b) and (d) data are mean ± SD of three biological replicates. In (c) data are mean ± SD of five biological replicates. Asterisk indicates significant differences between treatments (p < 0.05)
Since both low temperature and drought conditions increase oxidative stress leading to photo-inhibition, we expected to find a large overlap between genes induced by high light stress, compared to those induced in response to drought and low temperatures. To specifically evaluate the effect of an excess of radiant energy, potato plants grown in a growth chamber under low light conditions (50 μmol.m−2.s−1) for 1 month were transferred to an identical growth chamber under high light conditions (800 μmol.m−2.s−1), keeping them at warm temperatures (22 °C) and well irrigated. Interestingly, a strong and fast hyponastic response was observed in plants exposed to high light conditions (Fig. 3a), which most likely contributed to minimize light exposure. In spite of this light avoidance response, this treatment caused a decrease in the Fv/Fm ratio (Fig. 3c) as well as in photosynthetic activity (Fig. 3b). However, no changes in overall chlorophyll levels were observed during the first 24 h under high light conditions (Fig. 3d). Thus, these data shows both convergent as well as stress specific responses to high light conditions compared to those seen in response to drought and cold stress. Interestingly, from the 1427 genes whose expression increased or decreased more than three-fold under stress conditions, only 17 had a simultaneous response in the same direction (12 up-regulated and 5 down-regulated) under the three stress treatments evaluated (Fig. 1). Among the 12 genes that were up-regulated, five were associated with secondary metabolism, particularly with the production of phenolic compounds such as anthocyanins and flavonoids (Table 1). Anthocyanins are colored pigments, which can protect plants against ultraviolet light (Steyn et al. 2002). Among the genes associated with key enzymes in this pathway that were up-regulated more than three fold in all stress treatments, we found a gene encoding a chalcone synthase (CHS), an observation that was confirmed by RT-qPCR (Fig. 4a). Extending the analysis to those genes that showed at least a two-fold change in expression in the same direction under all conditions revealed a similar enrichment in genes involved in phenylpropanoid biosynthesis (10 out of 30 genes) (Table 1). Another key rate limiting enzyme in this pathway that was up-regulated more than two fold under all three stress treatments is phenyl alanine ammonia lyase (PAL) (Fig. 4b). Although the statistical significance of the high-light effect was slightly above the threshold for this particular gene (ESM_1), the effect was confirmed by RT-qPCR. Thus, these results suggest that the synthesis of protective pigments is likely to be the main common mechanism underlying adaptation to different abiotic stresses in potato plants. Consistent with this hypothesis, we found an increase in the amount of anthocyanins in response to the three stresses evaluated in this work (Fig. 4c).
Physiological parameters measured in potato plant under high light stress. a Angle between the third leaf and the stem. b Fv/Fm. c Photosynthetic rate. d Chlorophyll content. In (a) and (b) data are mean ± SD of three biological replicates. In (c) and (d) data are mean ± SD of five biological replicates. Asterisk indicates significant differences between treatments (p < 0.05)
Effect of abiotic stress on anthocyanin biosynthesis, CHS and PAL expression. a Chalcone Synthase expression analyzed by microarrays and qRT-PCR under three different abiotic stresses in potato plants. b Expression level of Phenyl Ammonio Lyase analyzed by microarrays and qRT-PCR under three different abiotic stresses. c Anthocyanin content in petioles of plants exposed to drought and low temperature stresses. Data are mean ± SD of five biological replicates. Asterisk indicates significant differences between treatments (p < 0.05)
A subset of genes involved in maintaining redox homeostasis was strongly induced under high light stress but not in response to the other stress treatments (ESM_1 and ESM_6). This finding suggests that plants exposed to drought and low temperature conditions only had a moderate oxidative stress and, thus, the increment in the synthesis of protective pigments was likely an anticipatory response to protect plants from further oxidative damage rather than an immediate response triggered by strong oxidative damage.
In addition to the subset of genes that were commonly up-regulated by all three stresses, five genes were down-regulated at least three-fold under all stress conditions (Table 2). This group increased to 19 genes if we considered those genes down-regulated at least two-fold in response to all stresses. A distinct functional category was not identified among these genes, but we found genes encoding an expansin, an aquaporin and a cell wall remodeling enzyme, which might be associated with a reduction in cell elongation and leaf area normally observed in response to these stresses (ESM_7). Finally, light signals acting through specific photoreceptors, such as phytochromes and cryptochromes, have been shown to modulate responses to drought, low temperature and high light stresses (Carvalho et al. 2011; Yu et al. 2010). Interestingly, the three abiotic stresses evaluated here triggered a reduction in the expression of CRYPTOCHROME INTERACTING bHLH 1 (CIB1) (ESM_8), a bHLH transcription factor shown to mediate light regulation of flowering time in Arabidopsis (Liu et al. 2008). Thus, it will be interesting to test whether CIB1 plays a role modulating stress responses in potato or other plants.
Potato Plants Overexpressing the Arabidopsis thaliana CBF1 Gene Showed Enhanced Tolerance to High Light Conditions at Warm Temperatures
Overexpression of the CBF family of transcription factors (DREB1s) has been shown to confer tolerance to multiple abiotic stresses in several species (Khan 2011). In potato in particular, AtCBF1 overexpression has been shown to confer freezing, drought and salinity stress tolerance (Behnam et al. 2007; Movahedi et al. 2012), while over-expression of AtCBF3 confers heat stress tolerance (Dou et al. 2015). Since low temperature conditions and drought affect plant growth enhancing susceptibility to photoinhibition, we hypothesized that tolerance to multiple stresses in plants overexpressing CBF1 could be associated with enhanced tolerance to high light stress. To test this possibility we developed transgenic potato plants overexpressing the Arabidopsis thaliana CBF1 (AtCBF1) gene, and characterized tolerance of these plants to low temperatures, drought and high light stress.
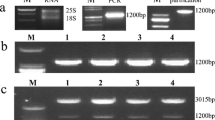
Eleven independent transgenic lines were obtained, which were positive for a PCR reaction detecting the AtCBF1 transgene (Fig. 5a). These 11 independent lines also displayed detectable levels of the AtCBF1 mRNA by RT-PCR (Fig. 5b). Two lines, AtCBF1-OX-6 and AtCBF1-OX-11, were selected for further studies. Transgenic and wild type (WT) plants were grown in a growth chamber under long day conditions, and plant height, internode length, and tuber yield were measured (ESM_9). Previous work has shown that ectopic expression of AtCBF1 in tomato or potato produces growth retardation and dwarfism (Pino et al. 2007; Zhang et al. 2004). In agreement with this observation, these plants showed a strong (AtCBF1-OX_6) or intermediate (AtCBF1-OX_11) dwarf phenotype (Fig. 5c and ESM_9). In contrast, although it was reported that the constitutive expression of the AtCBF1 gene causes a reduction in tuber yield (Pino et al. 2007), we found that tuber yield was similar in WT and in our two transgenic lines, at least under the growth conditions we used for this evaluation (ESM_9).
To evaluate the response of these plants to low temperature stress, WT and transgenic plants overexpressing AtCBF1 were transferred from 22 to 4 °C. The Fv/Fm ratio decreased after 1 day at 4 °C in both genotypes, but the reduction was significantly less severe in the transgenic plants overexpressing AtCBF1, revealing an increased tolerance to low temperature conditions in these plants (ESM_10). Indeed, after exposure to low temperature conditions for 2 weeks, chlorotic spots and wilting were observed in WT plants, while transgenic plants did not show any of these symptoms (ESM_10).
To evaluate desiccation tolerance, leaves from transgenic and WT plants were cut and kept under continuous illumination at 22 °C, while fresh weight was evaluated during a 75 h period (ESM_11). During the first hours, leaves from WT plants lost more water than leaves from transgenic plants, indicating enhanced drought sensitivity, which was coupled to a slower stomatal closure in comparison to the transgenic plants. We also evaluated drought tolerance with another assay that mimics more realistic drought stress conditions. For this, WT and AtCBF1 transgenic plants were grown for 1 month with daily irrigation in a growth chamber under long day conditions at 22 °C. After 1 month, irrigation was interrupted for half of the plants. Notoriously, 10 days after interruption of irrigation, leaf wilting was observed in WT plants but not in AtCBF1-OX transgenic plants (ESM_11d). This differential response to drought was associated with an earlier stomatal closure in the transgenic plants with respect to the WT plants, which allows them to maintain a better plant water status as well as to keep more water in the soil for a longer period of time (ESM_11). In addition, transgenic plants over-expressing AtCBF1 seemed to be more tolerant to drought stress as a consequence of reduced photo-inhibition. As shown in ESM_11e, drought caused a reduction in Fv/Fm in both genotypes, but this reduction was less intense in the AtCBF1 transgenic plants. This was also associated with a milder decrease in photosynthetic activity in the AtCBF1 transgenic compared to WT plants in response to drought (ESM_11).
Finally, to test if some of the enhanced tolerance to drought and low temperature was due to an improved ability to tolerate an excess of radiant energy, we conducted experiments where the only environmental variable modified was light intensity. The Fv/Fm ratio decreased similarly in all genotypes in plants transferred for 24 h from low (50 μmol.m−2.s−1) to high light conditions (800 μmol.m−2.s−1) (Fig. 6a). However, after a few days, chlorotic spots were detected in WT but not in AtCBF1-OX plants (Fig. 6f). This response was associated with a decrease in photosynthetic rate (Fig. 6b), which was stronger in WT than in transgenic plants. In addition, while WT plants showed a strong hyponastic response to high light stress, this response was strongly attenuated in the transgenic plants (Fig. 6c). WT plants showed a reduction in leaf area after 30 days compared to the transgenic plants grown under the same conditions (Fig. 6d), most likely as a consequence of a decrease in the amount of intercepted radiation in WT plants, together with a decrease in photosynthetic rate (Fig. 6b). Anthocyanin content was constitutively higher in the transgenic plants under control conditions, allowing transgenic plants to overcome the stress imposed by the high light treatment better than WT plants (Fig. 6e). This difference in anthocyanin content between genotypes was maintained under the stress treatment (Fig. 6e).
Tolerance to high light stress in AtCBF1-OX transgenic potato plants. a Fv/Fm. b Photosynthetic rate and chlorophyll content in stress treated plants relative to control plants. c High light avoidance measured as the angle between the third leaf and the stem. One month old WT and transgenic potato plants were transferred from 50 μmol.m−2s−1 to 800 μmol.m−2.s−1 for 1 day before measurements. d Leaf area of 2 month old plants. e Anthocyanin content in petioles of plants exposed to high light stress. f Leaves from transgenic and WT plants exposed to high light conditions. In d, e and f, plants were grown under control or high light conditions (800 μmol.m−2.s−1) for 1 month. * and ** indicate significant differences between AtCBF1-OX plants and WT controls under each condition (p < 0.05 and p < 0.01 respectively)
Transcriptional Changes Associated with AtCBF1 Overexpression in Potato Plants
To better understand the molecular mechanisms behind the multiple stress tolerance observed in AtCBF1 transgenic plants, we compared the transcriptional changes triggered by AtCBF1 overexpression with those induced in response to low temperature, high light, and drought stress in potato plants. For this, we first identified genes whose expression was either up or down regulated by AtCBF1 under control conditions, i.e. in well watered plants grown at warm temperatures, under moderate/low light intensity conditions. We found 138 genes that showed at least a two-fold increase in expression, and 187 genes with a two-fold decrease in mRNA levels in AtCBF1 over-expressing plants compared to WT plants (ESM_12). Interestingly, 45 of the 138 genes up-regulated by AtCBF1 over-expression were also up-regulated in response to at least one of the stress treatments analyzed before. In addition, 34 of the 187 genes down-regulated in AtCBF1 transgenic plants were also affected by at least one of the stress treatments analyzed. This observation suggests that over-expression of AtCBF1 confers multiple stress tolerance by modulating the expression of a subset of abiotic stress-responsive genes.
Several genes that could be responsible for the enhanced stress tolerance observed in the potato AtCBF1 transgenic plants were identified. One of the genes that was strongly up-regulated by ectopic overexpression of AtCBF1 was a homolog of AN1, a transcription factor recently shown to confer enhanced abiotic stress tolerance (Ben Saad et al. 2010; Huang et al. 2008). In addition, genes that are known components of the CBF1 regulon, such as ERD10 and LEA 14, were also up-regulated in the potato AtCBF1 transgenics, suggesting that the stress tolerance phenotypes are linked to known activities of CBF1 (Carvallo et al. 2011; Gilmour et al. 2004) (ESM_13). Interestingly, a gene that could explain some of the enhanced tolerance to high light stress is that encoding NADPH:PROTOCHLOROPHYLLIDE OXIDOREDUCTASE B (PORB). Reduced levels of the PORB gene product are usually associated with photoinhibition caused by chlorophyll degradation (Frick et al. 2003). Expression of this gene was strongly reduced in WT plants in response to high light, but this reduction was attenuated in the transgenic plants (ESM_14), and observation that was confirmed by RT-qPCR (ESM_14).
Finally, potato plants have CBF1 orthologue, and therefore it was interesting to determine if the expression of this gene was responding to the stress treatments evaluated here and/or if overexpression of AtCBF1 had any effect on the expression of the potato CBF1 orthologue. Unfortunately, expression levels of the potato CBF1 orthologue in our microarray data were below the threshold required to consider expression data as reliable for further statistical analysis and, therefore this gene was not present in our lists of expressed genes. However, an inspection of the raw data revealed that the signal intensity of the microarray probe associated with the potato CBF1 gene indeed increased more than ten times in wild type as well as in potato plants expressing the AtCBF1 gene in response to cold conditions (data not shown), as previously reported (Pennycooke et al. 2008). Under warm conditions, however, expression of the potato CBF1 orthologue was extremely low and it was therefore not possible to determine whether the expression of the endogenous gene was affected to any significant extent by overexpression of its Arabidopsis orthologue.
Transcriptional Profiles as an Aid for Risk Assessment of Transgenic Plants Expressing a Heterologous Transcription Factor
The first generation of transgenic plants authorized for cultivation in the field were genetically modified with single genes controlling simple traits, such as herbicide tolerance, or enhanced resistance to different pests (Shah et al. 1986; Vaeck et al. 1987). In contrast, the next generation of transgenic plants includes those that over-express transcription factors that can simultaneously affect, directly or indirectly, many different traits, contributing to increasing crop yield by modulating developmental programs or tolerance to abiotic stresses (Preuss et al. 2012). An important issue that needs to be addressed by the governmental regulatory agencies controlling the cultivation of genetically modified (GM) plants in the field is how to assess the environmental and health related risks associated with this next generation of transgenic plants, and which risk management procedures should be implemented (Ricroch et al. 2011).
The demonstration of “substantial equivalence” with their non-genetically modified counterparts is a key regulatory requirement for commercialization of GM plants (Craig et al. 2008). Non-targeted omic approaches have been useful to evaluate this issue (Ricroch et al. 2011). In particular, many studies have shown that first generation GM plants are usually substantially equivalent to their non-GM counterparts in terms of their chemical composition, analyzed with metabolomic approaches, and also in terms of their transcriptome, analyzed through DNA microarrays (Ricroch et al. 2011). However, GM plants over-expressing regulatory transcription factors are likely to target many different genes and pathways and, therefore, differences in their transcriptional profiles compared to their non-GM counterparts are expected. Then, it is important to evaluate the extent of those changes both quantitatively and qualitatively in order to obtain adequate information to estimate and evaluate environmental and/or health risks associated with this generation of GM plants.
To further evaluate the overlap between AtCBF1 and stress regulated genes in potato plants we analyzed the effect of AtCBF1 not only under control conditions, but also the transcriptome of WT and AtCBF1 overexpressing plants exposed to low temperatures or high light conditions (Fig. 7). We performed a global analysis of transcriptional profiles to quantitatively compare the magnitude of changes caused by over-expression of the AtCBF1 transcription factor in relation to those induced by changes in environmental conditions normally experienced by plants on a daily and/or seasonal basis. The dendrogram structure resulting from hierarchical clustering of sample expression profiles revealed that biological variability, dissimilar genotypic backgrounds, differences in control conditions and, finally, high light stress and low temperature induced transcriptional changes present an increasing level of global dissimilarity (Fig. 7a). Particularly relevant to this study was the finding that the magnitude of changes associated with environmental stress treatments was greater than the one linked to genotypic effects. This was in line with the observation that a linear model analysis of differential expression reported 290, 848, and 1067 differentially expressed genes (adjusted p-value q = 0.01) associated to genomic background, light, and temperature main effect factors respectively (Fig. 7b).
Transcriptome analysis of wild type and AtCBF1-OX transgenic plants exposed to abiotic stresses. a Global transcriptomic analysis using gene expression heatmaps. The transcriptional profiles of a non-specifically filtered subset of 2184 genes were considered. Sample names were built using the following keywords: WT, CBF1, for wild type and AtCBF1-OX genotype samples respectively, CHL, HL for control high-light, and high-light samples respectively, CT, LT for control temperature and low-temperature samples respectively. Numbers 1 and 2 indicated for the CT and LT samples, as well as the letters A and B indicated for the CHL and HL samples represent the two different biological replicates obtained for each treatment. Complete-linkage hierarchical clustering was used to reorder samples and genes according to relative similarities to highlight global patterns of gene expression. b Venn diagram for gene sets differentially modulated (Benjamini-Hochberg adjusted p-values below the 0.01 level and a fold-change greater than 2) under the three main effects considered in this work: genomic background, light, and temperature treatments
Similar results were also obtained using a template matching analysis of expression profiles. For a given statistical significance level, light stress treatment committed the largest set of genes, which was approximately 20 % larger than the one induced by temperature stress, and up to three times larger than the one associated with genetic background differences (ESM_15). Finally the multi response permutation test procedure (MRPP)- a multivariate non-parametric procedure for testing the hypothesis of no difference between two or more groups -, indicated that the transcriptional changes triggered by low temperature and high light stresses were approximately five times larger than those associated with AtCBF1 over-expression (ESM_16). These results, taken together, strongly indicate that over-expression of a key stress-related transcription factor has modest effects on the transcriptome compared to the effect triggered by fluctuations in environmental conditions.
Along a quantitative assessment of induced global transcriptional changes, an extremely important step needed for risk assessment of transgenic plants over-expressing a transcription factor is to identify unintended effects associated with this genetic modification. Given that the aim of over-expressing AtCBF1 is to enhance abiotic stress tolerance, those genes that were regulated by AtCBF1 but not by abiotic stress constitute a cluster of unintended target genes. To further evaluate the overlap between AtCBF1 and stress regulated genes in potato plants we analyzed the effect of AtCBF1 not only under control conditions, but also the transcriptome of WT and AtCBF1 over-expressing plants exposed to low temperatures or high light conditions. An overlap analysis of the set of genes showing statistically significant differential changes under the three main effects was used to identify genes regulated by AtCBF1 but not by abiotic stresses, genes regulated by high light and/or low temperature but not by AtCBF1, as well as genes regulated by AtCBF1, high light and low temperatures (Fig. 7b and ESM_17). The cluster of genes regulated only by AtCBF1 included 198 genes. Although some of these genes might be regulated by abiotic stress under conditions not evaluated here, or at different times during the course of exposure to stress, a significant proportion of this subset of genes may represent true non-stress related targets of AtCBF1 activity. Therefore, this subset of genes should be the focus of the analysis aimed at evaluating potential environmental or health related hazards associated with transgenic plants over-expressing AtCBF1, before these or similarly GM plants are authorized for cultivation in the field and eventually for commercial release.
Conclusions
The use of a non-targeted genomics approach allowed us to identify genes simultaneously regulated by different abiotic stresses in potato plants. This data set can help identifying novel candidate genes that could be used to develop stress tolerant potato plants. In addition, our results show that comparison of an “abiotic stress transcriptome” with that of GM plants over-expressing a stress related transcription factor is a useful strategy to determine potential mechanisms through which particular transcription factors confer stress tolerance. Indeed, in this case we provide strong evidence that tolerance to multiple abiotic stresses in potato plants over-expressing AtCBF1 is due, at least in part, to an increased ability to tolerate radiant energy in excess of the maximal amount that can be channeled through the photosynthetic process. Since this tolerance to multiple abiotic stresses may be due to increased anthocyanins levels, and these metabolites are produced at the expense of diverting carbon away from TCA intermediates, it will be important to evaluate carbon metabolism and sugar composition in the tubers of these or similar transgenic plants before they can be used for commercial purposes (Payyavula et al. 2012).
Finally, we also show that a comparative genomics strategy can be used to identify unintended targets, i.e. genes affected by the over-expression of a stress related transcription factor, which are not regulated by stress in wild type potato plants. This approach should help narrow down the list of genes on which bio-safety risk assessments should focus before this or similar GM plants are authorized for cultivation in the field.
References
Abdeen, A., J. Schnell, and B. Miki. 2010. Transcriptome analysis reveals absence of unintended effects in drought-tolerant transgenic plants overexpressing the transcription factor ABF3. BMC Genomics 11: 69.
Ariel, F.D., P.A. Manavella, C.A. Dezar, and R.L. Chan. 2007. The true story of the HD-Zip family. Trends in Plant Science 12: 419–426.
Bagnaresi, P., A. Moschella, O. Beretta, F. Vitulli, P. Ranalli, and P. Perata. 2008. Heterologous microarray experiments allow the identification of the early events associated with potato tuber cold sweetening. BMC Genomics 9: 176.
Baker, S.S., K.S. Wilhelm, and M.F. Thomashow. 1994. The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Molecular Biology 24: 701–713.
Beaujean, A., R.S. Sangwan, A. Ledercadonnel, and B.S. Sangwan-Norreel. 1998. Agrobacterium-mediated transformation of three economically important potato cultivars using sliced internodal explants: an efficient protocol of transformation. Journal of Experimental Botany 49: 1589–1595.
Behnam, B., A. Kikuchi, S. Yamanaka, M. Kasuga, K. Yamaguchi-Shinozaki, and K. Watanabe. 2006. The Arabidopsis DREB1A gene driven by the stress-inducible rd29A promoter increases salt-stress tolerance in proportion to its copy number in tetrasomic tetraploid potato (Solanum tuberosum). Plant Biotechnology 23: 169–177.
Behnam, B., A. Kikuchi, F. Celebi-Toprak, M. Kasuga, K. Yamaguchi-Shinozaki, and K. Watanabe. 2007. Arabidopsis rd29A::DREB1A enhances freezing tolerance in transgenic potato. Plant Cell Reports 26: 1275–1282.
Ben Saad, R., N. Zouari, W. Ben Ramdhan, J. Azaza, D. Meynard, E. Guiderdoni, and A. Hassairi. 2010. Improved drought and salt stress tolerance in transgenic tobacco overexpressing a novel A20/AN1 zinc-finger AISAP gene isolated from the halophyte grass Aeluropus littoralis. Plant Molecular Biology 72: 171–190.
Benjamini, Y., and Y. Hochberg. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society: Series B Methodological 57: 289–300.
Bohnert, H.J., D.E. Nelson, and R.G. Jensen. 1995. Adaptations to environmental stresses. The Plant Cell 7: 1099.
Carre, I.A., and J.Y. Kim. 2002. MYB transcription factors in the Arabidopsis circadian clock. Journal of Experimental Botany 53: 1551–1557.
Carvalho, R.F., M.L. Campos, and R.A. Acevedo. 2011. The role of phytochrome in stress tolerance. Journal of Integrative Plant Biology 53: 920–929.
Carvallo, M.A., M.T. Pino, Z. Jeknic, C. Zou, C.J. Doherty, S.H. Shiu, T.H. Chen, and M.F. Thomashow. 2011. A comparison of the low temperature transcriptomes and CBF regulons of three plant species that differ in freezing tolerance: Solanum commersonii, Solanum tuberosum, and Arabidopsis thaliana. Journal of Experimental Botany 62: 3807–3819.
Chen, H.-H., and P.H. Li. 1980. Characteristics of cold acclimation and deacclimation in tuber-bearing Solanum species. Plant Physiology 65: 1146–1148.
Craig, W., M. Tepfer, G. Degrassi, and D. Ripandelli. 2008. An overview of general features of risk assessments of genetically modified crops. Euphytica 164: 853–880.
Cushman, J.C., and H.J. Bohnert. 2000. Genomic approaches to plant stress tolerance. Current Opinion in Plant Biology 3: 117–124.
Dixon, P. 2009. VEGAN, a package of R functions for community ecology. Journal of Vegetation Science 14: 927–930.
Dong, M.A., E.M. Farre, and M.F. Thomashow. 2011. CIRCADIAN CLOCK-ASSOCIATED 1 and LATE ELONGATED HYPOCOTYL regulate expression of the C-REPEAT BINDING FACTOR (CBF) pathway in Arabidopsis. Proceedings of the National Academy of Sciences 108: 7241–7246.
Dou, H., K. Xv, Q. Meng, G. Li, and X. Yang. 2015. Potato plants ectopically expressing Arabidopsis thaliana CBF3 exhibit enhanced tolerance to high-temperature stress. Plant, Cell & Environment 38: 61–72.
Fankhauser, C., K.C. Yeh, J.C. Lagarias, H. Zhang, T.D. Elich, and J. Chory. 1999. PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science 284: 1539–1541.
Fiorani, F., A.L. Umbach, and J.N. Siedow. 2005. The alternative oxidase of plant mitochondria is involved in the acclimation of shoot growth at low temperature. A study of Arabidopsis AOX1a transgenic plants. Plant Physiology 139: 1795–1805.
Foyer, C.H., and J.F. Allen. 2003. Lessons from redox signaling in plants. Antioxidants & Redox Signaling 5: 3–5.
Frick, G., Q. Su, K. Apel, and G.A. Armstrong. 2003. An Arabidopsis porB porC double mutant lacking light-dependent NADPH:protochlorophyllide oxidoreductases B and C is highly chlorophyll-deficient and developmentally arrested. The Plant Journal 35: 141–153.
Gangadhar, B.H., J.W. Yu, K. Sajeesh, and S.W. Park. 2014. A systematic exploration of high-temperature stress-responsive genes in potato using large-scale yeast functional screening. Molecular Genetics and Genomics 289: 185–201.
Gentleman, R., V. Carey, D. Bates, B. Bolstad, M. Dettling, S. Dudoit, B. Ellis, L. Gautier, Y. Ge, J. Gentry, K. Hornik, T. Hothorn, W. Huber, S. Iacus, R. Irizarry, F. Leisch, C. Li, M. Maechler, A. Rossini, G. Sawitzki, C. Smith, G. Smyth, L. Tierney, J. Yang, and J. Zhang. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biology 5: R80.
Gilmour, S., S. Fowler, and M. Thomashow. 2004. Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Molecular Biology 54: 767–781.
Golldack, D., I. Lüking, and O. Yang. 2011. Plant tolerance to drought and salinity: stress regulating transcription factors and their functional significance in the cellular transcriptional network. Plant Cell Reports 30: 1383–1391.
Hancock, R.D., W.L. Morris, L.J. Ducreux, J.A. Morris, M. Usman, S.R. Verrall, J. Fuller, C.G. Simpson, R. Zhang, P.E. Hedley, and M.A. Taylor. 2014. Physiological, biochemical and molecular responses of the potato (Solanum tuberosum L.) plant to moderately elevated temperature. Plant, Cell and Environment 37: 439–450.
Hsieh, T.-H., J.-t. Lee, Charng Y-y, and M.-T. Chan. 2002a. Tomato plants ectopically expressing Arabidopsis CBF1 show enhanced resistance to water deficit stress. Plant Physiology 130: 618–626.
Hsieh, T.-H., J.-T. Lee, P.-T. Yang, L.-H. Chiu, Charng Y-y, Y.-C. Wang, and M.-T. Chan. 2002b. Heterology expression of the Arabidopsis C-repeat/dehydration response element binding factor 1 gene confers elevated tolerance to chilling and oxidative stresses in transgenic tomato. Plant Physiology 129: 1086–1094.
Huang, J., M.-M. Wang, Y. Jiang, Y.-M. Bao, X. Huang, H. Sun, D.-Q. Xu, H.-X. Lan, and H.-S. Zhang. 2008. Expression analysis of rice A20/AN1-type zinc finger genes and characterization of ZFP177 that contributes to temperature stress tolerance. Gene 420: 135–144.
Jaglo-Ottosen, K.R., S.J. Gilmour, D.G. Zarka, O. Schabenberger, and M.F. Thomashow. 1998. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280: 104–106.
Khan, M.S. 2011. The role of DREB transcription factors in abiotic stress tolerance of plants. Biotechnology and Biotechnology Equipment 25: 2433–2442.
Khush, G.S. 2001. Challenges for meeting the global food and nutrient needs in the new millennium. Proceedings of the Nutrition Society 60: 15–26.
Kondrak, M., F. Marincs, F. Antal, Z. Juhasz, and Z. Banfalvi. 2012. Effects of yeast trehalose-6-phosphate synthase 1 on gene expression and carbohydrate contents of potato leaves under drought stress conditions. BMC Plant Biology 12: 74.
Liu, Q., M. Kasuga, Y. Sakuma, H. Abe, S. Miura, K. Yamaguchi-Shinozaki, and K. Shinozaki. 1998. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10: 1391–1406.
Liu, H., X. Yu, K. Li, J. Klejnot, H. Yang, D. Lisiero, and C. Lin. 2008. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322: 1535–1539.
Mancinelli, A.L., F. Rossi, and A. Moroni. 1991. Cryptochrome, phytochrome, and anthocyanin production. Plant Physiology 96: 1079–1085.
Mittler, R. 2002. Oxidative stress, antioxidants and stress tolerance. Trends in Plant Science 7: 405–410.
Moran, R., and D. Porath. 1980. Chlorophyll determination in intact tissues using N,N-Dimethylformamide. Plant Physiology 65: 478–479.
Movahedi, S., B.E. Sayed Tabatabaei, H. Alizade, C. Ghobadi, A. Yamchi, and G. Khaksar. 2012. Constitutive expression of Arabidopsis DREB1B in transgenic potato enhances drought and freezing tolerance. Biologia Plantarum 56: 37–42.
Nakashima, K., H. Takasaki, J. Mizoi, K. Shinozaki, and K. Yamaguchi-Shinozaki. 2012. NAC transcription factors in plant abiotic stress responses. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms 1819: 97–103.
Nicot, N., J.-F. Hausman, L. Hoffmann, and D. Evers. 2005. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. Journal of Experimental Botany 56: 2907–2914.
Pavlidis, P., and W.S. Noble. 2001. Analysis of strain and regional variation in gene expression in mouse brain. Genome Biology 2: 0042.0041–0042.0015.
Payyavula, R., D. Navarre, J. Kuhl, A. Pantoja, and S. Pillai. 2012. Differential effects of environment on potato phenylpropanoid and carotenoid expression. BMC Plant Biology 12: 39.
Pennycooke, J., H. Cheng, S. Roberts, Q. Yang, S. Rhee, and E. Stockinger. 2008. The low temperature-responsive, SolanumCBF1 genes maintain high identity in their upstream regions in a genomic environment undergoing gene duplications, deletions, and rearrangements. Plant Molecular Biology 67: 483–497.
Pino, M.-T., J.S. Skinner, E.-J. Park, Z. Jeknić, P.M. Hayes, M.F. Thomashow, and T.H.H. Chen. 2007. Use of a stress inducible promoter to drive ectopic AtCBF expression improves potato freezing tolerance while minimizing negative effects on tuber yield. Plant Biotechnology Journal 5: 591–604.
Pino, M.-T., J.S. Skinner, Z. JekniĆ, P.M. Hayes, A.H. Soeldner, M.F. Thomashow, and T.H.H. Chen. 2008. Ectopic AtCBF1 over-expression enhances freezing tolerance and induces cold acclimation-associated physiological modifications in potato. Plant, Cell & Environment 31: 393–406.
Preuss, S.B., R. Meister, Q. Xu, C.P. Urwin, F.A. Tripodi, S.E. Screen, V.S. Anil, S. Zhu, J.A. Morrell, G. Liu, O.J. Ratcliffe, T.L. Reuber, R. Khanna, B.S. Goldman, E. Bell, T.E. Ziegler, A.L. McClerren, T.G. Ruff, and M.E. Petracek. 2012. Expression of the Arabidopsis thaliana BBX32 gene in soybean increases grain yield. PLoS ONE 7: e30717.
Qin, F., K. Shinozaki, and K. Yamaguchi-Shinozaki. 2011. Achievements and challenges in understanding plant abiotic stress responses and tolerance. Plant and Cell Physiology 52: 1569–1582.
Reiss, P.T., M.H. Stevens, Z. Shehzad, E. Petkova, and M.P. Milham. 2010. On distance-based permutation tests for between-group comparisons. Biometrics 66: 636–643.
Rensink, W., A. Hart, J. Liu, S. Ouyang, V. Zismann, and C.R. Buell. 2005a. Analyzing the potato abiotic stress transcriptome using expressed sequence tags. Genome 48: 598–605.
Rensink, W.A., S. Iobst, A. Hart, S. Stegalkina, J. Liu, and C.R. Buell. 2005b. Gene expression profiling of potato responses to cold, heat, and salt stress. Functional and Integrative Genomics 5: 201–207.
Ricroch, A.E., J.B. Berge, and M. Kuntz. 2011. Evaluation of genetically engineered crops using transcriptomic, proteomic, and metabolomic profiling techniques. Plant Physiology 155: 1752–1761.
Rosegrant, M.W., and S.A. Cline. 2003. Global food security: challenges and policies. Science 302: 1917–1919.
Schaffer, R., N. Ramsay, A. Samach, S. Corden, J. Putterill, I.A. Carre, and G. Coupland. 1998. The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93: 1219–1229.
Shah, D.M., R.B. Horsch, H.J. Klee, G.M. Kishore, J.A. Winter, N.E. Tumer, C.M. Hironaka, P.R. Sanders, C.S. Gasser, S. Aykent, N.R. Siegel, S.G. Rogers, and R.T. Fraley. 1986. Engineering herbicide tolerance in transgenic plants. Science 233: 478–481.
Shinozaki, K., and K. Yamaguchi-Shinozaki. 2000. Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Current Opinion in Plant Biology 3: 217–223.
Shinozaki, K., K. Yamaguchi-Shinozaki, and M. Seki. 2003. Regulatory network of gene expression in the drought and cold stress responses. Current Opinion in Plant Biology 6: 410–417.
Smyth, G.K. 2004. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistical Applications in Genetics and Molecular Biology 3: 3.
Somersalo, S., and G.H. Krause. 1989. Photoinhibition at chilling temperature. Planta 177: 409–416.
Steyn, W.J., S.J.E. Wand, D.M. Holcroft, and G. Jacobs. 2002. Anthocyanins in vegetative tissues: a proposed unified function in photoprotection. New Phytologist 155: 349–361.
Storey, J.D., and R. Tibshirani. 2003. Statistical significance for genomewide studies. Proceedings of the National Academy of Sciences of the United States of America 100: 9440–9445.
Thomashow, M.F. 2001. So what’s new in the field of plant cold acclimation? Lots! Plant Physiology 125: 89–93.
Tilman, D., K.G. Cassman, P.A. Matson, R. Naylor, and S. Polasky. 2002. Agricultural sustainability and intensive production practices. Nature 418: 671–677.
Uno, Y., T. Furihata, H. Abe, R. Yoshida, K. Shinozaki, and K. Yamaguchi-Shinozaki. 2000. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proceedings of the National Academy of Sciences of the United States of America 97: 11632–11637.
Vaeck, M., A. Reynaerts, H. Höfte, S. Jansens, M. de Beuckeleer, C. Dean, M. Zabeau, M.V. Montagu, and J. Leemans. 1987. Transgenic plants protected from insect attack. Nature 328: 33–37.
Vasquez-Robinet, C., S.P. Mane, A.V. Ulanov, J.I. Watkinson, V.K. Stromberg, D. De Koeyer, R. Schafleitner, D.B. Willmot, M. Bonierbale, H.J. Bohnert, and R. Grene. 2008. Physiological and molecular adaptations to drought in Andean potato genotypes. Journal of Experimental Botany 59: 2109–2123.
Verslues, P., M. Agarwal, S. Katiyar-Agarwal, J. Zhu, and J.-K. Zhu. 2006. Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. The Plant Journal 45: 523–539.
Yamaguchi-Shinozaki, K., and K. Shinozaki. 1994. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6: 251–264.
Yu, X., H. Liu, J. Klejnot, and C. Lin. 2010. The cryptochrome blue light receptors. Arabidopsis Book 8: e0135.
Zhang, X., S.G. Fowler, H. Cheng, Y. Lou, S. Rhee, E.J. Stockinger, and M.F. Tomashow. 2004. Freezing-sensitive tomato has a functional CBF cold response pathway, but a CBF regulon that differs from that of freezing-tolerant <i>Arabidopsis</i> The Plant Journal 39: 905–919.
Zhang, N., B. Liu, C. Ma, G. Zhang, J. Chang, H. Si, and D. Wang. 2014. Transcriptome characterization and sequencing-based identification of drought-responsive genes in potato. Molecular Biology Reports 41: 505–517.
Zhu, J.-K. 2002. Salt and drought stress signal transduction in plants. Annual Review of Plant Biology 53: 247–273.
Acknowledgments
This work was supported by a fellowship from the Argentinean National Research Council to L.S., and by grants from Agencia Nacional de Promoción Científica y Tecnológica to M.J.Y.
Authors’ Contributions
LS performed most of the experiments in this study with technical assistance from EP, MLR, GGS. AC performed the statistical analysis. LS, CEH, RJS, JJC and MJY provided input in designing experiments and in the preparation of the manuscript and LS, CEH and MJY wrote the paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Founding Information
These experiments were funded by the Argentinean National Research Council (CONICET).
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
Complete lists of genes regulated by drought, low temperature and high-light stress in potato plants. Tab1. Genes up-regulated under drought stress. Tab2. Genes down-regulated under drought stress. Tab3. Genes up-regulated under low temperature stress. Tab4. Genes down-regulated under low temperature stress. Tab5. Genes up-regulated under high light stress. Tab6. Genes down-regulated under high light stress. (XLS 1118 kb)
ESM 2
Examples of genes regulated under drought stress. (a) Up-regulated genes. (b) Down-regulated genes (GIF 35 kb)
ESM 3
Examples of genes up-regulated under both drought and low temperature stresses (GIF 21 kb)
ESM 4
Contrasting regulation of an aquaporin gene by drought and low temperatures (GIF 15 kb)
ESM 5
Expression of circadian clock associated genes from the MYB family of transcription factors under low temperature, drought and high light (GIF 21 kb)
ESM 6
Regulation of genes involved in redox homeostasis by high light stress (GIF 20 kb)
ESM 7
Down regulation of genes associated with the control of cell elongation and leaf area in response to abiotic stress (GIF 23 kb)
ESM 8
Regulation of CIB1 expression by abiotic stress in potato (GIF 13 kb)
ESM 9
Phenotypic characterization of AtCBF1-OX transgenic potato plants. (a) Total height and 3rd internode length of two independent AtCBF1-OX transgenic lines compared to wild-type potato plants. (b) Tuber yield. In (a) data are mean ± S.E. of six replicates. In (b) data are mean ± S.E. of nine replicates. * indicates significant differences between treatments (p<0.05). (GIF 47 kb)
ESM 10
Cold stress tolerance in AtCBF1-OX transgenic potato plants. (a) Fv/Fm measured in the leaves of WT and AtCBF1-OX transgenic plants under control and low temperature conditions. Data are mean ± S.E. of three replicates. Different letters indicate statistically significant differences (p<0.05). (b) Representative leaf of WT and AtCBF1-OX transgenic potato plants after prolonged cold exposure. (c) WT and AtCBF1-OX transgenic plants exposed for one week to low temperature conditions. (GIF 139 kb)
ESM 11
Tolerance to drought stress in AtCBF1-OX transgenic potato plants. (a) Water loss in detached leaves of WT and AtCBF1-OX plants during three days. (b) Leaf transpiration rate during drought treatment. (c) Transpiration rate measured as water loss from plants grown in pots during a drought treatment. (d) Photograph of WT and AtCBF1OX transgenic potato plants after ten days of no water supply. (e) Fv/Fm after ten days of drought. (f) Photosynthesis rate and chlorophyll content of stress treated relative to control plants after 10 days of drought. Data in (a) are mean ± S.E. of twelve leaves. Data in (b), (c) and (d) are mean ± S.E. of three plants. Data in F are mean ± S.E. of four plants. * indicates significant differences between treatments (p<0.05). (GIF 114 kb)
ESM 12
Genes regulated by AtCBF1 under control conditions and in WT plants under stress conditions. Tab1. Genes up-regulated by AtCBF1 under control growth conditions. Tab2. Genes down-regulated by AtCBF1 under control growth conditions. Tab3. Genes up-regulated by AtCBF1 under control growth conditions and under any of the abiotic stresses analyzed before. Tab4. Genes down-regulated by AtCBF1 under control growth conditions and by any of the abiotic stresses analyzed before. (XLS 149 kb)
ESM 13
Expression of cold regulated genes in WT and AtCBF1-OX transgenic plants under control conditions (GIF 17 kb)
ESM 14
Expression of PORB in WT and AtCBF1-OX transgenic plants in response to high light stress (GIF 18 kb)
ESM 15
Template Matching Analysis. Number of genes significantly correlated to each one of these templates, as a function of the considered significance level. For any given statistical cutoff level, light stress treatments committed the largest set of genes (~20% bigger than the one induced by temperature stress, and up to 300% bigger than the one associated to genetic background differences). (GIF 35 kb)
ESM 16
Multi Response Permutation Procedure for gene expression analysis. MRPP results obtained for the low temperature and high-light stress conditions are reported in the first and second row respectively. A, δ, <δ>, and pv, are the effect-size, the observed and expected weighted mean of intra group distances, and the associated p-value, respectively. (XLS 25 kb)
ESM 17
Lists of genes regulated by AtCBF1-OX under control or abiotic stress conditions in potato plants. 1 indicates regulated by the treatment being evaluated, 0 indicates not regulated by the treatment being evaluated. Tab1. Genes regulated by temperature stress but not by light or genetic background. Tab2. Genes regulated by light but not by low temperature conditions or genetic background. Tab3. Genes regulated by genetic background but not by low temperatures or high-light stress. Tab4. Genes regulated by low temperatures and by high-light but not by genetic background. Tab5. Genes regulated by temperature stress and genetic background, but not by high-light. Tab6. Genes regulated by genetic background and by high-light, but not by low temperatures. Tab7. Genes regulated by genetic background, high-light and low temperatures. (XLS 1621 kb)
ESM 18
Primers used for qRT-PCR validation of microarray data (XLS 24 kb)
Rights and permissions
About this article
Cite this article
Storani, L., Hernando, C.E., Staneloni, R.J. et al. AtCBF1 Overexpression Confers Tolerance to High Light Conditions at Warm Temperatures in Potato Plants. Am. J. Potato Res. 92, 619–635 (2015). https://doi.org/10.1007/s12230-015-9476-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12230-015-9476-2