Abstract
We describe here the isolation of a novel gene, designated AlSAP, from A. littoralis in a first step to exploit the potential of this halophyte grass as a genetic resource to improve salt and drought tolerance in plants and, particularly, in cereals. The Aeluropus genome contains a single AlSAP gene which has an intron at its 5’UTR. Sequence homology analysis showed that the AlSAP protein is characterized by the presence of two conserved zinc-finger domains A20 and AN1. AlSAP is induced not only by various abiotic stresses such as salt, osmotic, heat and cold but, also by abscisic acid (ABA) and salicylic acid (SA). Tobacco plants expressing the AlSAP gene under the control of the duplicated CaMV35S promoter exhibited an enhanced tolerance to abiotic stresses such as salinity (350 mM NaCl), drought (soil Relative Water Content (RWC) = 25%), heat (55°C for 2.5 h) and freezing (−20°C for 3 h). Moreover, under high salt and drought conditions, the transgenic plants were able to complete their life cycle and to produce viable seeds while the wild-type plants died at the vegetative stage. Measurements of the leaf RWC and of the root and leaf endogenous Na+ and K+ levels in AlSAP transgenic lines compared to wild-type tobacco, showed an evident lower water loss rate and a higher Na+ accumulation in senescent-basal leaves, respectively. Finally, we found that the steady state levels of transcripts of eight stress-related genes were higher in AlSAP transgenic lines than in wild-type tobacco. Taken together, these results show that AlSAP is a potentially useful candidate gene for engineering drought and salt tolerance in cultivated plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
As plants have a sessile nature, they are constantly exposed to various biotic and abiotic (e.g., drought, salinity, high and low temperature and light intensity) stresses, which have a great impact on their growth and productivity. To cope with these stresses, plants trigger a network of events. These events start with stress signal perception, followed by transduction cascades that eventually lead to the expression of target genes which participate in stress tolerance (Shinozaki and Yamguchi-Shinozaki 2000) by promoting morphological, biochemical and physiological changes (Pastori and Foyer 2002).
The “Stress Associated Protein” (SAP) gene family is composed of genes encoding proteins containing A20/AN1 zinc-finger domains. Their number ranges from 19 in Populus trichocarp, 18 in Oryza sativa (OsSAP) and Sorghum bicolour, 14 in Arabidopsis thaliana (AtSAP), 11 in Zea mays, 10 in Physcomitrella patens and Vitis vinifera, to 3 in Chlamydomonas reinhardtii (Jin et al. 2007; Vij and Tyagi 2008). The A20/AN1 protein family has been divided into 2 types. Type I harbours the traditional pattern Cx2Cx9−12Cx1−2Cx4Cx2Hx5HxC. Type II has an expanded pattern Cx4Cx9−12Cx1−2Cx4Cx2Hx5HxC (Jin et al. 2007). Moreover, most of type I members lack intron and have an A20 domain while most of type II members have a single intron but do not exhibit an A20 domain. It has been demonstrated that most rice type I SAP genes are induced by one or more abiotic stresses and perform functions in a stress-specific or tissue-specific manner (Jin et al. 2007; Vij and Tyagi 2006). The term ‘zinc finger’ represents the sequence motifs in which cysteines and/or histidines coordinate a zinc atom(s) to form local peptide structures required for their specific functions (Takatsuji 1998). The zinc finger domain enables different proteins to interact with or bind to DNA, RNA or other proteins. Some classes of zinc-finger motifs such as TFIIIA- and GATA-types (in most cases) are part of the DNA-binding domains of transcription factors. However, other classes such as LIM- and RING-finger types are mostly implicated in protein–protein interactions (Takatsuji 1998).
In animals, the family of proteins containing A20/AN1 zinc-finger domain is well-characterized and plays a central role in regulating the immune response by acting as a negative regulator of inflammation and apoptosis (Heyninck and Beyaert 2005; Hishiya et al. 2006; Huang et al. 2004). The A20 zinc-finger domain characterized by multiple Cys2/Cys2 finger motifs was first identified in the TNFα-inducible protein of the human endothelial cells (Dixit et al. 1990; Opipari et al. 1990). On the other hand, one putative zinc-finger domain AN1 was identified in the proteins encoded by the Xenopus laevis animal hemisphere 1 (AN1) maternal RNA (Linnen et al. 1993). This AN1 zinc-finger domain is usually associated with the A20 zinc-finger. Such type of proteins is present in all eukaryotes (Evans et al. 2004).
Recently, the A20/AN1 zinc-finger domain proteins have been shown to be involved in stress response in plants. However, their cyto-protective molecular mechanism remains elusive (De Valck et al. 1999; Evans et al. 2004; Lee et al. 2000). Yet, Vij and Tyagi (2008) suggested “these proteins most probably belong to the regulatory class of proteins in the stress signalling cascade as they lack any typical nuclear localization signal”. In animals, such proteins have already been shown to function in the cytosol. These proteins may use their zinc-finger domains for protein–protein interactions as shown in the case of the human ZnF216 protein (Scott et al. 1998). The indica rice OsSAP1 was identified as the first plant protein containing both zinc-finger domains A20 (present at the N-terminus) and AN1 (present at the C-terminus). It was found that the OsSAP1 transcript is induced in response to different types of stresses including cold, desiccation, salt, submergence, heavy metals, wounding and ABA (Mukhopadhyay et al. 2004). It was also shown to be an intron-less and present as a single copy gene in the rice genome. Furthermore, overexpression of this gene in transgenic tobacco plants increased their tolerance to cold, salt, and dehydration stresses (Mukhopadhyay et al. 2004; Vij and Tyagi 2006). More recently, it has been shown that expression of OsSAP8, a member of the SAP gene family in rice, is induced by various abiotic stress treatments like OsSAP1. Sub-cellular localization of OsSAP8-GFP fusion protein indicated that OsSAP8 is a cytoplasmic protein. Yeast two-hybrid analysis indicated that A20 and AN1 zinc-finger domains of OsSAP8 interact with each other. Taken together, these data prove that SAP gene family products are cytoplasmic proteins and might carry out their functions via protein–protein interactions mediated by A20 and AN1 zinc-finger domains. Overexpression of OsSAP8 in transgenic rice and tobacco plants conferred tolerance to high salt (800 mM NaCl), drought (water deficit for 23 days during anthesis) and cold (4 ± 1°C for 4 days) stresses (Kanneganti and Gupta 2008). On the other hand, overexpression of the rice zinc finger A20/AN1 gene ZFP177 (identical to OsSAP9) in tobacco plants led not only to an increased tolerance to both high and low temperature and H2O2 stresses but also to an over sensitivity to dehydration and salt stresses (Huang et al. 2008).
In an objective of improving drought and salt tolerance in cereals, especially wheat and barley, we investigate the potential of A. littoralis to isolate genes involved in key mechanisms of environmental stress response. A. littoralis is a perennial, monocotyledonous halophyte (can endure up to 600 mM NaCl) growing in dry salty areas or marshes (Li and Liu 1994; Gulzar et al. 2003). It is also a C4 photosynthesis plant, salt-secreting and rhizomatous. This plant is diploid (2n = 2X = 14) and has a relative small genome around 342 Mb (Zouari et al. 2007). Thus, A. littoralis has the potential to become a precious genetic resource, not only for understanding the molecular mechanisms of stress-responses in monocots but also for improving tolerance to abiotic stresses in economically important crops. To understand the genetic basis of salt tolerance mechanisms in A. littoralis at the genomic level, we have isolated, sequenced and annotated 492 transcripts with a size ranging from ESTs (Expressed Sequence Tag) to full length cDNAs (Zouari et al. 2007). Among these, we report here the isolation and characterization of a gene (AlSAP, DQ885218) encoding for an A20/AN1 zinc- finger protein induced by salt, drought, cold, heat, ABA and SA. By stress assays, we have found that the overexpression of the AlSAP gene in tobacco improved tolerance to continuous salt and drought stresses under greenhouse conditions.
Materials and methods
Plant materials
Aeluropus littoralis and Nicotiana tabacum var. Xanthi were used in this study. Seeds of an A. littoralis ecotype were collected from salt marshes near “Sfax”, a coastal town in the middle of Tunisia.
AlSAP gene isolation
The AlSAP cDNA was isolated from an A. littoralis root cDNA library (cloned in pDNR-LIB vector, CLONTECH) prepared by using plants stressed for 15 days with 300 mM NaCl as described previously (Zouari et al. 2007). The total cDNA sequence of 707 bp was obtained by end sequencing (ABI 3100 automatic DNA sequencer, APPLIED BIOSYSTEMS) of the cDNA clone using M13F and M13R (supplementary Table 1) primers. The sequence was deposited to the Genbank with the accession number DQ885218 (UniProtKB: A1YAQ3). Bioinformatics analysis has revealed significant homology with different A20/AN1 zinc-finger proteins that belong to the Stress Associated Protein (SAP) gene family and that are isolated from different plants. Therefore, the gene was designated as AlSAP (A. littoralis Stress Associated Protein). The primers AlF5’ and AlR3’ (supplementary Table 1) were used to amplify the genomic clone of AlSAP gene. The generated fragment of 2,230 bp was cloned in the pGEM T-easy vector (PROMEGA) and three different positive clones were sequenced using SP6 and T7 primers.
Stress assay in Aeluropus littoralis
Surface sterilized seeds of A. littoralis (1% sodium hypochlorite solution for 15 min, followed by six washings with autoclaved Milli-Q water) were germinated in Eppendorf tubes containing 500 μl half strength MS (Murashige and Skoog 1962) solid medium under 16 h photoperiod at 25°C. The tubes containing the seedlings were perforated and their caps were cut when the plants’ roots reached their bottom. Later, the seedlings were transferred to a nutrient solution as described by Zouari et al. (2007) and grown for 2 months before treating them with different stress factors: high salinity (300 mM NaCl), high osmotic pressure (10% PEG 8000), low temperature (4°C) and high temperature (37°C). The plants were also treated with 100 μM abscisic acid (ABA) and 10 mM salicylic acid (SA) for hormonal stress. Plants were sampled at 0, 1, 2, 3 and 6 days after each treatment, frozen in liquid nitrogen and stored at −80°C for RNA extraction.
Semi-quantitative RT-PCR
Total RNA was isolated from the A. littoralis plants subjected to various stress treatments using Trizol reagent (INVITROGEN) according to the manufacturer’s protocol. The RNA was treated with DNase I (MBI, FERMENTAS) at 37°C for 15 min in order to remove the remaining genomic DNA. For semi-quantitative RT-PCR analysis, 5 μg of treated total RNA was reverse-transcribed using the oligo-dT (18 mer) primer and SuperScript reverse transcriptase (INVITROGEN) according to the manufacturer’s instructions. The PCR amplification of AlSAP gene was performed using 2 μl of 1/10th dilution cDNA as a template and two specific primers Al5’UTR and Al3’UTR (supplementary Table 1). The reaction included an initial 3 min of denaturation at 94°C, then 30 cycles of 30 s at 94°C, 40 s at 55°C and 1 min at 72°C, and finally 10 min extension at 72°C. As an internal control, a fragment of actin gene (380 bp) was amplified using the following primers ACTF and ACTR (supplementary Table 1). To ensure reproducibility, experiments were repeated three times with similar results.
The transcript accumulation was monitored for eight stress-related genes in transgenic lines and wild-type tobacco plants. These genes encode catalase (CAT1, U93244.1) (Takahashi et al. 1997), manganese superoxide dismutase (MnSOD, AB093097.1), ascorbate peroxidase (APX, U15933.1) (Orvar and Ellis 1995), osmotin (M2979) (Singh et al. 1989) and four encoding group 2 LEA proteins NtERD10A (AB049335.1), NtERD10B (AB049336.1), NtERD10C (AB049337.1), NtERD10D (AB049338.1) (Kasuga et al. 2004). Total RNA was extracted from wild-type tobacco and transgenic lines and then reverse-transcribed using oligo-dT (18 mer) as described above to generate first strand cDNA. The amplification of stress-related genes was performed using 2 μl of 1/10th dilution cDNA as a template and two specific primers with 30 PCR cycles. This reaction included an initial 3 min denaturation at 94°C, then 30 cycles of 30 s at 94°C, 40 s at 60°C, 45 s at 72°C, and finally 10 min extension at 72°C. The following gene-specific primers were used: CAT1 (cat1F, cat1R), MnSOD (MnsodF, MnsodR), APX (apxF, apxR), osmotin (osmF, osmR), NtERD10A (NtEAF, NtEAR), NtERD10B (NtEBF, NtEBR), NtERD10C (NtECF, NtECR) and NtERD10D (NtEDF, NtEDR) (supplementary Table 1). RT-PCR experiments were repeated three times to validate the results. To determine if equal amounts of total RNA were used in the RT-PCR reactions among samples, the RT-PCR for the house-keeping actin gene was performed under the same above-mentioned conditions.
Overexpression of AlSAP in Saccharomyces cerevisiae and stress-tolerance assays
For overexpression of AlSAP gene in S. cerevisiae, strain W303 (MATa ade2 ura3 leu2 his3 trp1), the cDNA fragment was released from the plasmid pDNR-LIB by digestion with EcoRI and XbaI. To obtain the plasmid pYES2-AlSAP, the resulting product was inserted into the EcoRI/XbaI sites of pYES2 vector (INVITROGEN) which is a 2-μm-based multi-copy yeast plasmid and contains the URA3 gene and the Gal1 promoter for selection and expression in yeast. Yeast cells were grown overnight in YPD medium (1% yeast extract; 2% peptone and 2% dextrose, DIFCO) at 30°C in a rotary shaker (180 rpm) to mid-exponential phase. They were then transformed with 1 μg of pYES2 empty vector or pYES2-AlSAP constructs using the EZ-transformation kit (Q-BIOGENE) according to the manufacturer protocol. The recombinants colonies were selected on Yeast Nitrogen Base plates lacking uracil (YNBUra−). This medium is the same as for YNB except that uracil-lacking CSM was added instead of the CSM mix. For the stress-tolerance assays, positive colonies were grown overnight to mid-exponential phase in YNBUra− medium, adjusted to an OD600 = 1 and serially diluted (10−2, 5 × 10−2 or 10−3) with fresh medium. Finally, 5 μl aliquots of each dilution were cultivated onto YNBUra−Gal 2% solid or liquid medium supplemented with NaCl (1.5 M), LiCl (0.1 M) KCl (1.5 M) or Mannitol (1.5 M) and incubated at 30°C. The growth rate was evaluated visually or by measuring the OD600 for solid or liquid medium respectively. The galactose at 2% was added to YNBUra− plates to induce the expression of AlSAP gene under the control of Gal1 promoter. For quantitative RT-PCR, yeast cells were cultivated in YNBUra− liquid medium containing 2% galactose at 30°C. After the cell growth reached the exponential growth phase (OD600 = about 1), cells were harvested by centrifugation, frozen in liquid nitrogen immediately and stored at −80°C until the isolation of total RNA samples. Total RNA was extracted using Trizol reagent (INVITROGEN) according to the manufacturer’s protocol. Total RNA (10 μg) were treated with DNase I (MBI FERMENTAS) for 30 min at 37°C and further incubated at 65°C for 10 min. The treated total RNA was reverse-transcribed using the oligo-dT (18 mer) primer and SuperScript reverse transcriptase (INVITROGEN) according to the manufacturer’s instructions. The AlSAP gene was amplified using 2 μl of 1/10th dilution of cDNA as a template and two specific primers Al5’UTR and Al3’UTR. As an internal control, a fragment of 18S rRNA (600 pb) was amplified using the following primers: 18SF and 18SR (supplementary Table 1).
Construction of binary vector and transformation of tobacco
The plasmid pDNR-LIB was digested with XbaI and SmaI to release the cDNA fragment of AlSAP gene which was purified of the gel and finally cloned in the binary vector pCAMBIA 2300 (CAMBIA, Canberra, Australia) under the control of the CaMV35S promoter and CaMV35S terminator. The obtained construct pCAMBIA2300-AlSAP was then mobilized into Agrobacterium tumefaciens strain LBA4404 (Hoekema et al. 1983) by freeze–thaw transformation method (Chen et al. 1994). Agrobacterium-mediated leaf disc transformation of tobacco was carried out per standard protocol (Horsch et al. 1988). Transformants (T0) were selected on MS agar medium containing 250 mg/1 kanamycin. The integration and expression of AlSAP was ascertained by Southern and northern blot analysis, respectively (see below).
Southern and northern blot analyses
Genomic DNA was isolated using the CTAB (N-acetyl-N, N, N-trimethylammonium bromide) method (Murray and Thompson 1980). For Southern blot analysis, 20 μg of genomic DNA extracted from PCR-positive transgenic tobacco lines or from A. littoralis were digested with HindIII and with EcoRI, BamHI, BglII, HindIII respectively. The digested DNA was separated by electrophoresis on a 1% agarose gel and then transferred onto Hybond-N+ nylon membrane (AMERSHAM-PHARMACIA). The AlSAP cDNA fragment amplified by PCR with a pair specific primer (Al5’UTR, Al3’UTR) and labeled with [α-32P]dCTP (AMERSHAM-PHARMACIA) was used as a probe. After hybridization at 65°C, the membrane was washed once with 2XSSC plus 0.1% SDS at 65°C for 20 min and then twice with 1XSSC plus 0.1% SDS at 65°C for 30 min. For RNA blot analysis, total RNA was extracted from 4-week-old seedlings of transgenic tobacco using Trizol reagent (INVITROGEN). About 20 μg of total RNA samples were resolved on a 1.5% formaldehyde gel and blotted onto a Hybond-N+ membrane (AMERSHAM). Hybridization and washing conditions were identical to the above-mentioned DNA blot. Hybridization was detected by autoradiography. The RNA also served for RT-PCR analyses, as described in the semi-quantitative RT-PCR section.
Evaluation of transgenic tobacco plants for abiotic stress tolerance
In vitro assays: The AlSAP transgenic tobacco plants (T9, T16 and T22) of homozygous T2 generation were used in the subsequent abiotic stress assays. Seeds of WT and transgenic plants were surface-sterilized and plated on MS0, MS0 m (MS0 plus 300 mM mannitol) and MS0 s (MS0 plus 200 mM NaCl) culture medium plates. The plates were placed in a growth chamber under a 16 h light/8 h dark cycle at 25°C. Germination rates were scored after 4 weeks. To evaluate the growth rate under osmotic and salt stress conditions, 10 day-old transgenic and wild-type seedlings were transferred to MS0, MS0 m, MS0 s and to MS0sm (MS0 plus 300 mM mannitol and 200 mM NaCl). Plates were held vertically. After 1 month of incubation, root length was monitored. For ionic stress tolerance analysis, 15 day-old seedlings of WT and transgenic lines were transferred on MS medium supplemented with 50 or 100 mM LiCl. After incubation in growth chamber for 10 days, the fresh and dry weights of the plants were determined. For freezing and heat stresses, 1 month-old seedlings of transgenic and WT plants grown in peat under greenhouse conditions were incubated at −20°C for 3 h or at 55°C for 2.5 h, respectively. The plant’s fresh and dry weights were determined after 1 week of recovery in greenhouse.
Leaf disk floating assays: One cm diameter leaf disks (5 disks per treatment) were prepared from leaves of identical development stage of both wild-type and transgenic plants and floated on 0, 300 and 800 mM NaCl solution for 72 h. The total chlorophyll content in each sample was calculated after extraction in aqueous 80% acetone using the following formulae (Arnon 1949) which express [Chl a], [Chl b] and [Chls a + b] in μg·ml−1: [Chl a] = 12.70·A663–2.69·A645, [Chl b] = 22.90·A645–4.68·A663, [Chls a + b] = 20.21·A645 + 8.02·A663. The A663 and A645 represent absorbance values read at 663 and 645 nm wavelengths, respectively.
Greenhouse assays: To monitor the effects of drought and salt tratments on AlSAP overexpressing plants under greenhouse conditions, seeds of three homozygous transgenic lines (T9, T16 and T22) and WT plants were germinated and grown on MS medium supplemented with 250 mg/l kanamycin for 1 month. The seedlings were then transferred to pots filled with a 3:1 mixture of soil and peat and grown in a greenhouse for two more weeks before exposure to stress treatments. Transgenic and WT plants were irrigated with tap-water using a program that maintained the RWC of soil at 75% under normal conditions. For salt stress treatment, the same irrigation program was used for watering the plants with a solution containing 350 mM NaCl until the end of plant cycle. This NaCl concentration was increased gradually from 200 to 350 mM within the first 15 days of stress treatment. Leaves (young (top) and old (bottom)) and roots were collected from salt stressed treated plants and then dried at 80°C for 24 h. Finally, the dried material was incubated in 0.5% HNO3 for a week. The Na+ and K+ contents were analyzed in the filtrate using atomic absorption spectrophotometry.
For drought stress, plants were watered with tap-water using a program that maintained the RWC of the soil at 25%. The RWC of the leaves was estimated every 6 days for the whole period (30 days) of withholding water according to the method of Turner (1981): RWC(%) = (FW−DW)/(TW−DW) × 100. Leaves of uniform position were taken from WT and transgenic lines and were immediately weighed (fresh weight: FW), hydrated to full turgidity for 12 h at 4°C by floating them on distilled water in a closed Petri dish. They were also weighed to obtain the turgid weight (TW). Finally, the leaves were oven-dried at 80°C for 24 h and were weighed to determine the dry weight (DW). For all treatments, plant height, root length, number of fertile pods and yield of seeds were determined.
Sequence analysis
We performed the BLAST search of the AlSAP gene at NCBI (http://www.ncbi.nlm.nih.gov/Blast). Various tools from Expasy (http://www.expasy.org/tools) were used to deduce the translated product and compute theoretical pI and molecular weight. The putative domains were identified using the InterProscan search (http://www.ebi.ac.uk/interproscan). The multiple sequence alignments and the degree of amino acid sequence identity were determined by the use of the biological sequence editor software BioEdit 7.0.0. Finally, to predict the phylo-genomic relationships between AlSAP gene and O. sativa/A. thaliana genes, we have used the Greenphyl Orthologs Search Tool (GOST, http://greenphyl.cines.fr/cgi-bin/gost.cgi).
Results
Analysis of the AlSAP sequence
A cDNA clone, AlSAP, was isolated from roots of salt treated A. littoralis plants (300 mM for 15 days). Sequence analysis using the web-based annotation tool linked to BioEdit software revealed that AlSAP cDNA was 707 bp long, including a complete open reading frame of 477 bp with 5’-UTR and 3’-UTR regions of 118 and 112 bp, respectively (supplementary Fig. 1a). AlSAP encodes a predicted polypeptide of 159 amino acids with a molecular weight of 17.69 kDa and a pI of 8.01. The amino acid sequence analysis by NetPhos 2.0 server showed the presence of some predicted phosphorylation sites with a high score (>0.9) at serine (S-5, S-63, S-64, S-97, S-101), threonine (T-70, T-102) and tyrosine (Y-136) (supplementary Fig. 1a). It has been shown that the protein phosphorylation at these residues affects a multitude of cellular signaling processes (Blom et al. 1999). In addition, one kinase-specific phosphorylation site PKC (protein kinase C) was predicted at threonine (T-102, score of 0.9) (supplementary Fig. 1a) by NetPhosK 1.0 server (Blom et al. 2004).
The conserved domain search of AlSAP amino acid sequence in NCBI and the InterProScan in EMBL-EBI server predicted the presence of two zinc-finger domains as a characteristic of the SAP gene family in plants (Fig. 1a). The first one was located at the N-terminus region between amino acids 16 and 40 and was similar (E-value of 0.007) to the zf-A20 domain (smart00259) of the human A20 protein with a consensus of Cx2−4Cx11Cx2C, where x represents any amino acid. The second domain was found at C-terminus of the protein, covering the region from amino acid 100–139, which was similar (E-value = 4 × 10−11) to the zf-AN1 domain (Pfam01428) having a consensus sequence of Cx2−4Cx9−12Cx1−2Cx4Cx2Hx5HxC.
Sequence analysis of the AlSAP protein highlighting the A20/AN1 conserved domains and the prediction of phylogenomic relationships between AlSAP gene and O.sativa/A.thaliana SAP genes. a The amino acids at N-terminal A20 and the C-terminal AN1 type zinc-finger domains are conserved in the AlSAP protein. Conserved cysteines and histidines are indicated in bold type. b Phylogenetic tree of rice and Arabidopsis SAP gene families encoded A20/AN1 zinc-finger proteins. The unrooted tree was generated using the Greenphyl Orthologs Search Tool (GOST, http://greenphyl.cines.fr/cgi-bin/gost.cgi) which predicts the phylogenomic relationships (percentage of orthology) between AlSAP gene and O. sativa/A. thaliana SAP genes
Using blastp in NCBI, the homology searches run with the translated full-length amino acid sequences of the AlSAP cDNA clone revealed a significant similarity to the A20/AN1 zinc-finger proteins isolated from plants, human (ZNF216 and AWP1) and animals. The prediction of orthologs to the AlSAP gene in the Greenphyl database using the GOST tools (GOST, http://greenphyl.cines.fr/cgi-bin/gost.cgi) revealed the presence of two orthologs in O. sativa, OsSAP9 (Os07g0168800) and OsSAP6 (Os03g0792900) with a high percentage of 87 and 52% orthology, respectively (Fig. 1b). AlSAP showed the highest identity values at the amino acid levels with the two rice SAP proteins, OsSAP9 (Q7Y1W9, 79%) and OsSAP6 (Q852K5, 63%) (Vij and Tyagi 2006), whereas, for other SAPs from rice, Arabidopsis and maize the identity values were much lower, ranging from 25 to 40%. Thus, AlSAP showed 37 and 41% of identity to OsSAP1 (Mukhopadhyay et al. 2004) and OsSAP8 (Kanneganti and Gupta 2008), respectively. These authors have also shown that OsSAP1, OsSAP6, OsSAP8 and OsSAP9 are members of the SAP gene family characterized by the presence of zf-A20 and zf-AN1 zinc-finger domains in their putative encoded proteins. Apart from the plant homologues, the AlSAP showed 35% identity to the human ZNF216 and AWP1 proteins, concentrated in the zf-A20 and zf-AN1 zinc finger domains.
Comparison of the AlSAP full-length cDNA with a genomic fragment (2,230 bp) generated by PCR amplification and sequencing, revealed the presence of two exons of 83 and 624 bp separated by one intron of 1,523 bp (supplementary Fig. 1b).This long intron is located in the 5’UTR region while the coding sequence is continuous. In addition, Southern blot analysis revealed that AlSAP is a single copy gene (Fig. 2). A RAP-DB database-search (http://rapdb.dna.affrc.go.jp) showed that AlSAP gene has the same genomic structure than its two rice orthologous genes, OsSAP9 and OsSAP6, located on rice chromosome 7 and 3 respectively. The OsSAP9 and OsSAP6 genes also harbour an intron of of 1,537 and 1,430 bp length, respectively in their 5’UTR. It was also reported that OsSAP8 possesses two introns (176 and 739 bp) located in the 5’UTR region (Kanneganti and Gupta 2008).
Genomic organization of AlSAP gene. The AlSAP gene is present as a single copy in the genome of A. littoralis. Total genomic DNA from A. littoralis plant was digested with HindIII (H), BamHI (Ba), EcoRI (E) (do not cut in the sequence of AlSAP gene) and BglII (Bg) (cut once at position 2,115), and analyzed by Southern blot using the full-length cDNA of AlSAP as a probe. The positions of the molecular size standards are indicated
Stress-induced transcription of AlSAP gene
We have investigated the accumulation of AlSAP transcripts in response to multiple abiotic stresses (salt, osmotic, heat and cold) as well as to hormonal stresses (ABA and SA). The transcript levels increased gradually as a function of exposure time of A. littoralis plants to PEG, heat and cold stresses and remained at elevated levels for 6 days (Fig. 3b, d and e). In the case of salt stress, transcript accumulation reached a high level after a 2 day treatment and then gradually declined back to its basal level from days 3–6 (Fig. 3a). A comparable pattern of transcript accumulation was observed during the ABA treatment, except that the mRNA level remained high both on days 2 and 3 following the treatment (Fig. 3c). For SA treatment, the transcripts started to accumulate within 1 day of treatment, reached the maximum level on the 2nd and 3rd days and then declined on the 6th day (Fig. 3f).
Expression pattern of the AlSAP gene in response to different stresses applied to A. littoralis plants. RT-PCR analysis was performed with AlSAP specific primers using the RNA isolated from the A. littoralis plants subjected to normal conditions (C), a 300 mM NaCl, b heat at 37°C, c 100 μM ABA, d 10% PEG, e Cold at 4°C and f 10 mM SA. Actin amplification was used as an internal control (lower panel).1–6 day represents the duration of treatment in days. (-RT) represents the amplification of the RNA without RT-reaction. (H2O) represents the amplification in the absence of templates
Heterologous expression of AlSAP gene in yeast cells
The fact that the AlSAP gene is induced in response to multiple abiotic stresses, led us to further determine whether the expression of this gene can protect cells against environmental stresses. In that aim, we used yeast (Saccharomyces cerevisiae) as a fast heterologous model system. Wild-type yeast strain W303 was transformed with the plasmid pYES2 containing the AlSAP gene driven by the galactose inducible promoter GAL1 or by the empty pYES2 vector. The cells overexpressing AlSAP (as confirmed by RTPCR, Fig. 4a) exhibited a better growth than control cells under NaCl, LiCl, KCl and mannitol stresses (Fig. 4b). The number of transgenic AlSAP cells was two-fold and six-fold higher than control cells under LiCl and NaCl stresses, respectively (Fig. 4c). Finally, in hyperosmotic liquid medium containing KCl or mannitol at 1.5 M, the growth rate of AlSAP cells was two-fold higher than that of control cells (Fig. 4c). These findings suggest that overexpression of AlSAP in yeast increased cell tolerance to ionic and osmotic stresses. Based on these results, we further investigated whether AlSAP can operate the same role in transgenic tobacco plants.
Abiotic stress tolerance of yeast cells overexpressing the AlSAP gene. a The overexpression of AlSAP in yeast, grown in liquid YNB-Ura−/Gal2% medium, was confirmed by RT-PCR analysis. The 18S rRNA used as an internal control. b A culture with an OD600 = 1 of transformed cells with empty plasmid pYES2 as control or pYES2 containing AlSAP was serially diluted to 10−2, 5 × 10−2 or 10−3. 5 μl of each dilution were plated onto solid YNB-Ura/Gal2% plates (control) or supplemented with different concentrations of NaCl (1.2 M), LiCl (0.1 M), KCl (1.5 M) or Mannitol (1.5 M). Colonies were photographed after 5–6 days of incubation at 30°C. cYeast cells containing pYES2-AlSAP and pYES2 were incubated in liquid YNB-Ura/Gal2% medium at 30°C till OD600 reached 0.2, then the stresses were started by adding different concentrations of NaCl (1.5 M), KCl (1.5 M), LiCl (0.1 M) or Mannitol (1.5 M) to the cultures and finally the yeast cell growth was monitored by measuring the OD600 at different periods
Heterologous expression of AlSAP gene in transgenic tobacco plants
Generation and characterization of AlSAP tobacco plants
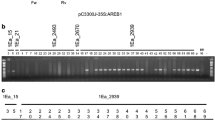
To investigate the physiological function of AlSAP, we generated 7, independent transgenic tobacco lines overexpressing AlSAP under the control of the duplicated cauliflower mosaic virus 35S promoter (Fig. 5a). The copy number of the integrated AlSAP gene was determined by Southern blot analysis (Fig. 5b). The constitutive expression of AlSAP in the selected transgenic lines was established by northern blot and semi-quantitative RT-PCR analyses (Fig. 5c, d). The absence of obvious phenotypic difference between AlSAP-expressing plants and WT plants under normal growth conditions indicates that ectopic expression of AlSAP does not affect the overall plant morphology.
Analysis of transgenic tobacco lines overexpressing the AlSAP gene. a Schematic map of the T-DNA inserted in the binary vector pCAMBIA2300: AlSAP, used for tobacco transformation. b Southern blot analysis of transgenic and wild type tobacco plants determining the T-DNA copy number. Genomic DNA was digested with HindIII, separated by electrophoresis on a 1% agarose gel and transferred onto a nylon membrane. The transferred DNA was hybridized with the [α- 32P]dCTP labeled AlSAP full-length cDNA probe. c Northern blot analysis of AlSAP transcript levels in wild-type tobacco and in the transgenic lines. RNA was transferred onto a nylon membrane and hybridized with the [α-32P]dCTP labeled AlSAP full-length cDNA probe. Equal loading in each lane was confirmed by ethidium bromide staining (lower panel). d Expression pattern of the AlSAP gene in three transgenic lines (9, 16 and 22) used for tolerance assays. RT-PCR analysis was performed with AlSAP specific primers using the RNA isolated from the three transgenic lines and WT plants. Actin amplification was used as an internal control (lower panel)
Evaluation of AlSAP tobacco for stress tolerance under in vitro conditions
To evaluate the level of stress tolerance of the generated events, we selected 3 transgenic lines, T9, T22 and T16 that harboured 1, 2 or 3 copies of the AlSAP gene respectively (Fig. 5b). Homozygous T2 seedlings were first evaluated in vitro for their tolerance to salt (NaCl), ionic (LiCl) and osmotic (mannitol) stresses.
Seeds of Wild-type tobacco and of the three homozygous transgenic lines T9, T16 and T22 were germinated on different culture media -MS0 (control), MS0 s (salt) and MS0 m (osmotic). Germination rates of wild-type and transgenic seeds cultured on the control MS0 medium (Fig. 6a) were similar, indicating that AlSAP gene expression has no detrimental effect on seed germination. On the other hand, germination of wild-type seeds was significantly impaired under stress conditions when compared to transgenic seeds. Whereas only 29% of the wild-type seeds germinated on MS0 s medium supplemented with 200 mM NaCl after 1 month,.seeds of the T16, T22 and T9 lines germinated at a rate of 96, 89 and 80%, respectively (Fig. 6b). Along the same line, 55% of the wild-type seeds germinated on MS0 m medium supplemented with 300 mM mannitol while 74, 95 and 98% of the T9, T22 and T16 transgenic seeds, respectively, did so (Fig. 6b). The transgenic lines harboring more than one copy of AlSAP gene (T16 and T22) showed a higher germination rate under salt and osmotic stresses than the transgenic line carrying a single copy (T9). This could be related to the higher level of accumulation of AlSAP transcripts in lines 16 and 22 than in line 9 (Fig. 5d). In addition, the germination experiments revealed that the wild-type seedlings stopped their growth, producing small (osmotic stress) or yellow (salt stress) leaves and less root biomass than transgenic seedlings. Interestingly, shoots and roots of all the transgenic ssedlings continued to grow after germination under these conditions (Fig. 6a). To further investigate the effect of salt and mannitol on the growth rate, 10-day-old wild-type and transgenic seedlings were transferred to MS0 s medium (NaCl 200 mM), MS0 m (mannitol 300 mM) or a combination of the two chemicals. After another month, root and leaf growth of wild-type seedlings were strongly retarded, whereas, growth inhibition was less pronounced in transgenic lines (Fig. 7a). Root lengths in wild-type plants exhibited severe reduction of 78, 88 and 89% under osmotic, salinity and a combination of the two stresses respectively, compared to control conditions. On the other hand, in transgenic lines, root elongation inhibition reached 35% for the two types of stresses (osmotic or salinity) and 62% for the combination of the two stresses (Fig. 7b). More importantly, we noticed that the root length reduction observed under osmotic or salt stress in transgenic plants was compensated by an increase in root number when compared to control conditions (Fig. 7a). Transgenic seedlings were able to produce a fourth pair of leaves during the stress treatment whereas WT seedlings had produced only the second pair of leaves.
Effect of salt (NaCl 200 mM) and osmotic (mannitol 300 mM) stresses on seed germination and plant phenotype in transgenic tobacco plants overexpressing the AlSAP gene. a Photographs were taken 4 weeks (control and mannitol) and 2 months (salt) after seed germination. b Percentage of seed germination in the case of WT and T2 homozygous transgenic plants (T9, T16 and T22) growing on MS0 medium (control), MS0 s (NaCl 200 mM) or MS0 m (mannitol 300 mM). Results are presented as means and standard errors from three independent experiments (150 seeds of each line were sown for each experiment)
Effect of salt (NaCl 200 mM), osmotic (mannitol 300 mM) and the combined stresses on shoot and root growth of WT plants and three transgenic AlSAP tobacco lines. a Photographs were taken 4 weeks after incubation in vitro. b Root length expressed in cm in the case of WT and transgenic plants (T9, T16 and T22) grown on MS medium (control) or on MS medium supplemented with NaCl (200 mM), mannitol (300 mM) or the two chemicals. Results are presented as means and standard errors from three independent experiments (10 plants of each line were grown for each experiment)
Finally, the alteration in chlorophyll content under salt stress was evaluated using a detached leaf disk assay. This assay can be taken as a reliable index of the damage to the photosynthesis apparatus under stress. When leaf disks were floated on a salt solution containing 300 or 800 mM NaCl for 72 h, disks of WT plants bleached more intensely than those of transgenic lines (Fig. 8a). This was later found consistent with the measurement of the chlorophyll content which dropped dramatically in WT plants (4–6%) while the AlSAP transgenic lines retained almost 30–50% of their chlorophyll under similar conditions (Fig. 8b). Taken together, these results demonstrate that transgenic tobacco plants overexpressing AlSAP have a better ability to tolerate salinity stress than wild-type plants. To investigate if this tolerance to salinity was due to osmotic and/or ionic mechanisms, 15 day-old WT and transgenic seedlings were challenged with a strictly ionic agent such as 50 or 100 mM LiCl. Stress tolerance was evaluated by monitoring the plant fresh and dry weights, 10 days following stress application. As shown in Fig. 9a, a significant difference in root and leaf growth rates was observed between the transgenic and WT seedlings. In fact, 10 days after stress application (50 mM LiCl), the transgenic seedlings reached the 4th leaf pair stage whereas the WT seedlings were at the 3rd leaf pair stage. In addition, the biomass production of WT plants, measured by fresh and dry weights, was four-fold to five-fold lower than that of transgenic lines (Fig. 9b, c). These results indicate that expression of the AlSAP gene conferred not only osmotic but also ionic stress tolerance.
Leaf disk floating assays as an evaluation of salinity tolerance in transgenic AlSAP tobacco lines (T2 generation). a Representative picture that shows phenotypic differences in leaf disks. b % of chlorophyll retention from leaf segments of WT and two transgenic lines plants (T9 and T22) after incubation in 300 and 800 mM NaCl solution. Leaf disks floating in water served as experimental control. After 72 h of incubation, the % of chlorophyll retention in the leaf discs was measured
Effect of strict ionic stress treatments on growth rate of WT and transgenic seedlings. Tobacco seedlings grown in MS medium for 15 days were transferred to culture plates containing 50 or 100 mM LiCl. a The photograph was taken 10 days after transplantation. b The fresh and c dry weight of WT and three transgenic plants (T9, T16 and T22) were determined
Expression of stress-associated genes in AlSAP tobacco plants
Semi-quantitative RT-PCR was used to compare expression of some stress-related genes between wild-type and AlSAP overexpressing tobacco plants under normal conditions. Transcript accumulation of the eight stress-related genes tested (CAT1, MnSOD, APX, osmotin and NtERD10 A, B, C or D) were higher in AlSAP transgenic plants than in wild-type plants in the absence of stress (Fig. 10a, b). Based on these results, it could be concluded that AlSAP might act as an upstream activator of some stress-related genes, which could lead to the tolerance to salt and drought stresses observed in transgenic lines. The induction of CAT1, MnSOD and APX genes in AlSAP transgenic lines is probably related to an increase of the plant capacity to eliminate the reactive oxygen species (ROS) generated by salt and drought stresses. In addition, the upregulation of the osmotin gene (a dehydrin) and the four genes of group 2 LEA protein (NtERD10 A, B, C or D) in AlSAP transgenic lines is another element that could explain the tolerance observed to drought and salt stresses.
Semi-quantitative RT-PCR assay of stress-related genes in WT and transgenic AlSAP tobacco lines grown under normal conditions. Total RNA was extracted from wild-type tobacco and transgenic lines (9, 16 and 22) and then reverse-transcribed using oligo-dT (18 mer) as described in materials and methods to generate first strand cDNA. The amplification of stress-related genes was performed using 2 μl of 1/10th dilution cDNA as a template and two specific primers. Actin amplification was used as an internal control. The stress-related genes used for the test are as follows: a genes encoding for ROS scavenging enzymes: catalase (CAT1, U93244.1), manganese superoxide dismutase (MnSOD, AB093097.1), ascorbate peroxidase (APX, U15933.1). b Genes encoding for protecting proteins: osmotin (M2979) and group 2 LEA proteins NtERD10A (AB049335.1), NtERD10B (AB049336.1), NtERD10C (AB049337.1) and NtERD10D (AB049338.1)
Evaluation of AlSAP tobacco for stress tolerance under greenhouse conditions
Because AlSAP expression in transgenic plants was found to allow their growth in vitro under salt and osmotic stresses, it was crucial to investigate its ultimate effect on seed production and yield. For this purpose, we performed salt and drought tolerance experiments with WT and the three T2 generation transgenic lines (T9, T22, T16) grown in soil in the greenhouse. Plants were grown either under optimal water supply (control conditions: RWC of the soil was maintained at 75%), under drought stress (the RWC of the soil was maintained at 25%) or continuous salt stress using NaCl (350 mM) for irrigation until the end of the plant cycle. As an indicator of salinity and drought stress tolerance in T2 transgenic lines, several critical growth parameters (plant fresh weight, root and shoot length, leaf and fertile pod numbers, time to flowering and seed weight) were scored (Table 1). The T2 generation transgenic plants behaved similarly to the WT counterparts for all these parameters when grown under non-stress conditions, confirming that AlSAP overexpression does not induce any growth or yield penalty in transgenic plants (Table 1; Fig. 11-control). In contrast, we have noticed a slight increase in the root and shoot length and in leaf number in transgenic lines when compared to WT plants (Table 1). This constrasts with the almost 50% yield penalty observed in transgenic rice lines overexpressing OsSAP8 under normal growth conditions (Kanneganti and Gupta 2008).
Relative salt tolerance of WT and T2-AlSAP-overexpressing transgenic tobacco lines (events T9, T16 and T22) at the adult stage under greenhouse conditions. WT and transgenic plants were grown under either normal condition, a continuous presence of 350 mM NaCl or a continuous drought stress (soil RWC of 25%) until the end of the plant cycle. Note that WT plants could not sustain growth under these stress conditions while transgenic plants were able to produce viable seeds (upper panel). Photographs show the root growth of WT plants and T16 plants grown under control, salt and drought conditions at the end of their life cycle
Under salt and drought stresses, the transgenic plants clearly performed differently than the WT. They indeed continued to grow, reached maturity, flowered and set seeds while the WT plants showed chlorosis, a stunted phenotype, were not able to produce viable seeds and ultimately died (Fig. 11). It should be noted that the biggest difference between transgenic and WT plants was observed in seed production. Thus, the three lines overexpressing the AlSAP gene produced under salt and drought stresses, between 55 and 75% of their respective seed yield observed under control conditions (Table 1). This could be related to the fact that the transgenic plants exposed to the continuous salt or drought stresses showed a relatively normal growth (Fig. 11) reflected by average plant weights, shoot and root lengths, leaf and fertile pod numbers remaining not deeply altered compared to control conditions (Table 1). Conversely, growth of WT plants was severely affected by a continuous exposure to salinity or drought stress, and their flowers were not able to produce any seed (Table 1; Fig. 11). Importantly, the yield of transgenic plants was more affected by the drought stress than by the salt stress. While the flowering time in transgenic plants remained unchanged under salt stress, flowers appeared with a delay of nearly 20 days under drought stress compared to control conditions. On the other hand, WT flowered 12 days earlier under both stresses than under control conditions. Such behaviour is considered as a normal response of sensitive plants to escape stress conditions.
As the transcription of the AlSAP gene is induced by high- and low-temperature stress (Fig. 3b, e), we also evaluated T2 plants of T9, T16 and T22 transgenic lines along with WT plants, for freezing (−20°C for 3 h) and heat (55°C for 2.5 h) tolerance. After a recovery period of 1 week all WT seedlings were wilted, whereas all transgenic seedlings recovered following the two types of stresses (Fig. 12a, b). Analysis of the seedling fresh weight after 1 week recovery period revealed that the transgenic seedlings had a three- and eight-fold higher fresh weight than WT seedlings following heat and freezing stresses, respectively (Fig. 12c). Additionally, the dry weights of transgenic seedlings was two- and four- fold higher than those of WT in the case of heat and freezing stresses, respectively (Fig. 12d).
Overexpressing AlSAP gene in transgenic tobacco confers tolerance to freezing (−20°C for 3 h) and heat (55°C for 2.5 h) stresses under greenhouse conditions. One-month-old seedling of WT plants and three AlSAP transgenic tobacco lines (T9, T16 and T22) were grown a at 55°C for 2.5 h or b at −20°C for 3 h and then transferred back to the normal condition for recovery. Photographs were taken before stress treatments and after 1 week of recovery. The AlSAP transgenic lines recovered better and growth of new leaves with elongated internodes was observed. c The fresh and d dry weights of WT and three AlSAP transgenic plants (T9, T16 and T22) were measured 1 week after recovery. Results are presented as means and standard errors from three independent experiments (10 plants of each line were grown for each experiment)
Characterization of the physiological status of AlSAP plants under stress treatment
To examine the basic mechanism conferring the salt-tolerant phenotype of AlSAP-overexpressing lines, we have measured the endogenous Na+ and K+ levels in leaves (young and old) and roots of plants grown in the absence or presence of NaCl for 60 days. Under normal conditions, no significant difference is observed in Na+ or K+ accumulation and partitioning for young/old leaves and roots of WT and transgenic plants (Fig. 13a, b). However, a dramatic change is observed under stress conditions, especially, in the partitioning of Na+ and K+. Thus, in transgenic plants, the Na+ concentration in old leaves was 3.5 fold higher than in young leaves (Fig. 13a). Conversely, in WT plants, the Na+ content in the young leaves was at least twice that of old leaves. In addition, Na+ accumulation in WT roots was three-fold higher than in those of transgenic plants. Moreover, K+ accumulation was ten-fold higher in transgenic roots compared to WT ones (Fig. 13b). All these findings suggest that transgenic plants were able to maintain a higher selectivity of K+ over Na+ in roots and sequestered almost 65% of the Na+ uptake in senescent leaves, thereby keeping young leaves and roots protected from Na+ ion toxicity. This could be one factor explaining the relatively normal growth of transgenic plants and their ability to produce viable seeds under salt stress. On the other hand, WT plants sequestered 70 and 20% of the Na+ uptake in roots and young leaves, respectively.
Physiological characterization of WT and transgenic AlSAP tobacco plants grown under normal and stress conditions. a Time-course of relative water content (RWC) during plant dehydration. b Na+ and c K+ content (calculated as mg of ions per g of dry weight (DW) of the tissue) in young (Y) and old (O) leaves and roots of the WT and AlSAP transgenic plants (WT, T16) grown under the continued presence of 350 mM NaCl or in H2O. In the histogram for each determination, roots, old leaf (bottom), young leaf (top), and roots were collected from three different plants of each type. Values are the mean ± SD (n = 3). Each value represents the average of independent measurements in three plants. Error bars represent SD
To validate and dissect the physiological mechanisms of drought stress tolerance afforded by AlSAP overexpression in tobacco plant, the RWC of leaves (WT and T16 line) was estimated every 6 days after withholding water for 30 days. The leaves tested were of similar size and developmental stage. After stopping irrigation for 12 days and until the end of stress, the RWC of detached leaves overexpressing AlSAP was almost double than that of WT leaves (Fig. 13c). These results clearly showed that the water content of transgenic leaves was higher than that of WT plants. Water loss in plants occurs via stomata and cuticula, and under well-watered conditions, stomata remain open. Most of the water loss is due to stomatal transpiration. Under stress conditions, stomata are closed and then cuticular transpiration rate becomes the most important component for water loss and consequently for plant survival (Savé et al. 1993). The results obtained on RWC of detached leaves suggest that the constitutive expression of AlSAP gene in plant tissues may alter stomatal behaviour or increase water uptake. In order to explain the difference in the water loss rate between the WT and transgenic plants, key parameters (water use efficiency (WUE), mean transpiration rate (MTR), stomatal conductance and net carbon assimilation rate (NAR)) must be determined.
Discussion
AlSAP belongs to the SAP gene family, is a single gene and has no ortholog in dicots
We reported here the isolation of the AlSAP gene, a single copy gene of the halophyte grass A. littoralis and demonstrated that it belongs to SAP gene family. This family is composed of genes encoding proteins containing A20/AN1 zinc-finger domains. AlSAP contains both A20 and AN1 domains at the N-terminus and C-Terminus regions of the protein, respectively. In animals, the A20 protein has been characterized as an inhibitor of both apoptotic and necrotic cell death (De Valck et al. 1996). These authors also suggest that self-association of A20 is mediated by its zinc finger domain. The AN1 domain was found at the C-terminus of AN1, an ubiquitin-like protein in Xenopus laevis (Linnen et al. 1993). In humans, two A20/AN1 proteins, ZNF216 and AWP1 (associated with PRK1) have been widely studied. In animals, the A20/AN1 zinc-finger domain-containing family of proteins is well-characterized and plays a central role in regulating the immune response and inflammation as well as in inhibiting apoptosis (Heyninck and Beyaert 2005; Hishiya et al. 2006; Huang et al. 2004). In plants, the A20/AN1 zinc-finger domain proteins have been shown to be involved in stress responses (Huang et al. 2008; Jin et al. 2007; Kanneganti and Gupta 2008; Mukhopadhyay et al. 2004; Vij and Tyagi 2006). Using the GOST tools in Greenphyl database, we found that the most likely rice ortholog of AlSAP is the OsSAP9 gene (87% of orthology). No ortholog was identified in dicots. Jin et al. (2007) recently reported that most zf-AN1 genes expanded in a monocot or eudicot-specific manner according to their phylogenetic tree. This suggests that SAP gene expansion could have occurred after the divergence of monocots and dicots and could explain the absence of orthologs to AlSAP in A. thaliana.
AlSAP is induced in response to a range of stresses
We have shown that AlSAP gene was induced by salt, drought, cold, heat, ABA and SA. It was reported that all members of the rice SAP family are also induced by at least one type of abiotic stress, indicating that the OsSAP genes are a key component of stress response in rice (Vij and Tyagi 2006). The OsSAP1 and OsSAP8 genes were found to be induced by salt, drought, cold, desiccation, submergence, wounding, ABA and heavy metals (Kanneganti and Gupta 2008; Mukhopadhyay et al. 2004). An early accumulation of transcripts for both genes was observed within 30 min after stress treatment in 1 week-old seedlings of indica rice but it declined rapidly within 24 h later. This time course transcription profile differs from that observed for the AlSAP gene. The induction of AlSAP gene started later after application of the stress (1–2 days) but remained at a high level for a longer period (1–6 days). The OsSAP9 (ZFP177) which is the nearest ortholog to the AlSAP gene (87% of orthology and 79% of amino acid identity) was demonstrated to be induced by heat and cold stresses but down-regulated by salt stress in japonica rice (Huang et al. 2008). These authors have also shown that following drought stress, OsSAP9 is upregulated as early as 2 h after treatment and then down-regulated at 6 h. Our results, along with those reported by Huang et al. (2008), show differential regulation of AlSAP and its close rice ortholog OsSAP9, indicating that evolutionary changes in promoter structure occur between closely-related glycophytic (rice) and halophytic (A. littoralis) species and may have contributed to develop a salt tolerance mechanism in A. littoralis. Many evolutionary biologists consider that evolution often proceeds by changes in the spatial and temporal patterns of gene expression (Doebley and Lukens 1998). A detailed analysis of the transcripts accumulation under abiotic stresses of AlSAP in A. littoralis along with its ortholog OsSAP9 in rice can throw more light on the importance of the temporal expression patterns of these genes in stress tolerance. Indeed, it has been hypothesized that differences in salt tolerance mechanisms between salt-sensitive A. thaliana and salt-tolerant Thellungiella halophila result from a differential regulation of the same basic set of genes involved in salt tolerance (Xiong and Zhu 2002; Zhu 2000, 2001).
Heterologous expression of the AlSAP gene in yeast cells and tobacco enhance their tolerance to abiotic stresses
The overexpression of AlSAP gene in yeast cells or in transgenic tobacco conferred tolerance to ionic and osmotic stress under in vitro conditions. In addition, the expression of AlSAP gene conferred freezing- and heat-stress tolerance in transgenic tobacco. This contrasts with the salt resistance conferred by the transgenic expression of Rab17 in Arabidopsis which was due to a mechanism of osmotic rather than ionic tolerance (Figueras et al. 2004). Similarly, the transcript of PDH45 (Pea DNA helicase 45) was shown to accumulate following Na+ but not Li+ treatments (Sanan-Mishra et al. 2005). The increased ability to grow under NaCl stress of the transgenic plants overexpressing this gene could likely be explained by an osmotic tolerance mechanism. The dual osmotic and ionic tolerance conferred by the AlSAP strongly supports the proposition of this candidate gene for improving salt and drought tolerance in cereals.
Under greenhouse conditions, AlSAP transgenic tobacco exposed to continuous salt or drought stress showed a higher level of tolerance. Indeed, these plants under salt or drought stress were able to maintain 75 and 55% of the yield observed under unstressed conditions, respectively. Under salinity stress, WT plants sequestered 70 and 20% of the Na+ uptake in roots and young leaves, respectively while AlSAP plants sequestered Na+ in senescent basal leaves. Similar differential Na+ sequestration between old and young leaves was observed in tobacco and rice plants overexpressing glyoxalase (Singla-Pareek et al. 2003, 2008).
To our knowledge, experimental evidence for conferring abiotic stress tolerance has so far been reported in three genes belonging to the SAP gene family: OsSAP1, OsSAP8 and OsSAP9 (Mukhopadhyay et al. 2004; Huang et al. 2008; Kanneganti and Gupta 2008). In these reports, tolerance evaluation was performed either under in vitro conditions or using the recovery system in a greenhouse. By contrast, in our study, along with in vitro tolerance assays, we carried out experiments under greenhouse conditions applying a continuous stress during the whole plant cycle in order to mimic the conditions found in semi-arid and arid regions. By comparing our results and those reported by Mukhopadhyay et al. (2004) and Kanneganti and Gupta (2008), it is clear that overexpressing of AlSAP, OsSAP1 or OsSAP8 in tobacco conferred tolerance to drought and salt stresses. However, it is impossible to answer if transgenic tobacco plants expressing AlSAP are more or less tolerant than those transformed with OsSAP1 or OsSAP8. In fact, the experimental procedures used for stress tolerance evaluation are different in the three cases. Furthermore, the three genes have a weak amino acid identity around 35% (no orthology relation). Finally, the OsSAP1 and OsSAP8 transcripts levels increased within 30 min after subjecting seedlings to various stresses while AlSAP transcript peaked within 2 days. This could suggest that the products of OsSAP1 and OsSAP8 are required in the early stage of stress response while AlSAP could be needed later. In contrast to these two above-mentioned genes (OsSAP1, OsSAP8), OsSAP9 (Huang et al. 2008) showed 79% of amino acid identity and 87% of orthology with AlSAP (according to the phylo-genomic tree generated by GOST tools). It has been demonstrated that the overexpression of OsSAP9 in transgenic tobacco conferred tolerance to both low and high temperature stresses but increased sensitivity to salt and drought stresses. The contrasting results about tolerance level to drought and salt stresses conferred by the two nearest orthologs genes AlSAP and OsSAP9 could be due to either sequence/structural differences between these two proteins or to their origins (halophyte and glycophyte monocots). Our results show that AlSAP and its ortholog OsSAP9 act differently in transgenic tobacco despite their amino acid identity. In addition, the expression patterns of AlSAP in A. littoralis and of OsSAP9 in rice are different in response to salt and drought stresses. For this purpose, further studies to clone the promoter of AlSAP gene and compare it with the one of its ortholog (OsSAP9) in rice are required.
We have shown that the steady state level of transcripts of some stress associated genes encoding proteins involved in anti-oxidative and protection activities are higher in unstressed AlSAP tobacco than in WT plants. It is well known that antioxidant enzyme activity is increased in plants in response to various environmental and chemical stresses (Allen 1995; Baek et al. 2006). ‘Late Embryogenesis Abundant’ or LEA proteins and osmotin are thought to protect macromolecules and membranes under stress conditions (Grover et al. 2001). In contrast to our findings, Huang et al. (2008) found that overexpression of OsSAP9 in transgenic tobacco inhibited the expression of stress-related genes such as osmotin, NtERD10 A, C or D. Taken together, these results suggest that AlSAP from the halophyte grass (A. littoralis) and its nearest ortholog from rice OsSAP9 act differently in tobacco plants.
A proposed role for AlSAP
The AlSAP protein has predicted serine, threonine, tyrosine and PKC phosphorylation sites, together with the presence of zf-A20 and zf-AN1 domains. It was suggested (as revealed by yeast two hybrid analyses) that A20 interacts not only with itself but also with the AN1 domain while the AN1 does not interact with itself (Kanneganti and Gupta 2008). Moreover, it was also suggested that OsSAP1 and OsSAP8 were cytoplasmic proteins and might carry out their functions via protein–protein interactions aided by their A20 and AN1 zinc finger domains (Kanneganti and Gupta 2008; Mukhopadhyay et al. 2004). It is therefore possible that AlSAP may interact with proteins needed for the phosphorylation cascade that targets proteins directly involved in cellular protection or transcription factors controlling specific sets of stress-regulated genes. This model is supported by several arguments based on mammalian A20/AN1 zinc finger data. Indeed, it was demonstrated that the human A20/AN1 zinc finger protein AWP1 interacts with a serine/threonine protein kinase PRK1 and thus may play a regulatory role in mammalian signal transduction pathways (Duan et al. 2000).
References
Allen RD (1995) Dissection of oxidative stress tolerance using transgenic plants. Plant Physiol 107:1049–1054
Arnon DI (1949) Copper enzymes in isolated chloroplasts polyphenoloxidase in Beta, vulgaris. Plant Physiol 24:1–15
Baek K, Skinner DZ, Ling P, Chen X (2006) Molecular structure and organization of the wheat genomic manganese superoxide dismutase gene. Genome 49:209–218
Blom N, Gammeltoft S, Brunak S (1999) Sequence- and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol 294:1351–1362. doi:10.1006/jmbi.1999.3310
Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S (2004) Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 4:1633–1649
Chen H, Nelson RS, Sherwood JL (1994) Enhanced recovery of transformants of Agrobacterium tumifaciens after freeze–thaw transformation and drug selection. Biotechniques 16:664–668
De Valck D, Heyninck K, Van Criekinge W, Contreras R, Beyaert R, Fiers W (1996) A20, an inhibitor of cell death, self-associates by its zinc finger domain. FEBS Lett 384:61–64. doi:10.1016/0014-5793(96)00283-9
De Valck D, Jin DY, Heyninck K, Van de Craen M, Contreras R, Fiers W, Jeang KT, Beyaert R (1999) The zinc finger protein A20 interacts with a novel anti-apoptotic protein which is cleaved by specific caspases. Oncogene 18:4182–4190
Dixit VM, Green S, Sarma V, Holzman LB, Wolf FW, O’Rourke K, Ward PA, Prochownik EV, Marks RM (1990) Tumor necrosis factor-alpha induction of novel gene products in human endothelial cells including a macrophage-specific chemotaxin. J Biol Chem 265:2973–2978
Doebley J, Lukens L (1998) Transcriptional regulators and the evolution of plant form. Plant Cell 10:1075–1082
Duan W, Sun B, LiT Wei, Tan B, Lee M, Teo TS (2000) Cloning and characterization of AWP1, a novel protein that associates with serine/threonine kinase PRK1 in vivo. Gene 256:113–121
Evans PC, Ovaa H, Hamon M, Kilshaw PE, Hamm S, Bauer S, Ploegh HL, Smith TS (2004) Zinc-finger protein A20, a regulator of inflammation and cell survival, has de-ubiquitinating activity. Biochem J 378:727–734. doi:10.1042/BJ20031377
Figueras M, Pujal J, Saleh A, Savé R, Pagès M, Goday A (2004) A Maize Rab17 overexpression in Arabidopsis plants promotes osmotic stress tolerance. Ann Appl Biol 44:251–257
Grover A, Kapoor A, Lakshmi OS, Agarwal S, Sahi CH, Katiyar-Agarwal S, Agarwal M, Dubey H (2001) Understanding molecular alphabets of the plant abiotic stress responses. Curr Sci 80:206–221
Gulzar S, Khan MA, Ungar IA (2003) Effects of salinity on growth, ionic content and plant-water status of Aeluropus lagopoides. Commun Soil Sci Plant Anal 34:1657–1668
Heyninck K, Beyaert R (2005) A20 inhibits NF-kB activation by dual ubiquitin-editing functions. Trends Biochem Sci 30:1–4. doi:10.1016/j.tibs.2004.11.001
Hishiya A, Iemura S, Natsume T, Takayama S, Ikeda K (2006) A novel ubiquitinbinding protein ZNF21 functioning in muscle atrophy. EMBO J 25:554–564. doi:10.1038/sj.emboj.7600945
Hoekema A, Hirsch PR, Hooykaas PJJ, Schilperoort RA (1983) A binary plant vector strategy based on separation of the vir and T-region of the Agrobacterium tumefaciens Ti plasmid. Nature 303:179–180. doi:10.1038/303179a0
Horsch RB, Fry J, Hoffmann N, Neidermeyer J, Rogers SG, Fraley RT (1988) Leaf disc transformation. In: Gelvin SB, Schilperoort RA (eds) Plant Molecular Biology Manual. Kluwer Academic Publishers, Dordrecht, pp 1–9
Huang J, Teng L, Li L, Liu T, Li L, Chen D, Xu LG, Zhai Z, Shu HB (2004) ZNF216 is an A20-like and IkappaB kinase gammainteracting inhibitor of NF kappaB activation. J Biol Chem 279:16847–16853. doi:10.1074/jbc.M309491200
Huang J, Wang MM, Jiang Y, Bao YM, Huang X, Sun H, Xu DQ, Lan HX, Zhang HS (2008) Expression analysis of rice A20/AN1-type zinc finger genes and characterization of ZFP177 that contributes to temperature stress tolerance. Gene 420:135–144. doi:10.1016/j.gene.2008.05.019
Jin Y, Wang M, Fu J, Xuan N, Zhu Y, Lian Y, Jia Z, Zheng J, Wang G (2007) Phylogenetic and expression analysis of ZnF-AN1 genes in plants. Genomics 90:265–275. doi:10.1016/j.ygeno.2007.03.019
Kanneganti V, Gupta AK (2008) Overexpression of OsiSAP8, a member of stress associated protein (SAP) gene family of rice confers tolerance to salt, drought and cold stress in transgenic tobacco and rice. Plant Mol Biol 66:445–462. doi:10.1007/s11103-007-9284-2
Kasuga M, Miura S, Shinozaki K, Yamaguchi-Shinozaki K (2004) A combination of the Arabidopsis DREB1A gene and stress-inducible rd29A promoter improved drought- and low-temperature stress tolerance in tobacco by gene transfer. Plant Cell Physiol 45:346–350
Lee EG, Boone DL, Chai S, Libby SL, Chien M, Lodolce JP, Ma A (2000) Failure to regulate TNF-induced NF-κB and cell death responses in A20-deficient mice. Science 289:2350–2354. doi:10.1126/science.289.5488.2350
Li MY, Liu YJ (1994) Halophytes of yellow river delta in north shandong province of China. J Qufu Normal Univ 125–133
Linnen JM, Bailey CP, Weeks DL (1993) Two related localized mRNAs from Xenopus laevis encode ubiquitin-like fusion proteins. Gene 128:181–188. doi:10.1016/0378-1119(93)90561-G
Mukhopadhyay A, Vij S, Tyagi AK (2004) Overexpression of a zinc-finger protein gene from rice confers tolerance to cold, dehydration, and salt stress in transgenic tobacco. Proc Natl Acad Sci USA 101:6309–6314. doi:10.1073/pnas.0401572101
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Plant Physiol 15:15473–15497. doi:10.1111/j.1399-3054.1962.tb08052.x
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4325
Opipari AW, Boguski MS, Dixit VM (1990) The A20 cDNA induced by tumor necrosis factor alpha encodes a novel type of zinc finger protein. J Biol Chem 265:14705–14708
Orvar BL, Ellis BE (1995) Isolation of a cDNA encoding cytosolic ascorbate peroxidase in tobacco. Plant Physiol 108:839–840
Pastori GM, Foyer CH (2002) Common components, networks, and pathways of cross-tolerance to stress. The central role of “Redox” and abscisic acid-mediated controls. Plant Physiol 129:460–468. doi:10.1104/pp.011021
Sanan-Mishra N, Hoi Pham X, Sopory SK, Tuteja N (2005) Pea DNA helicase 45 overexpression in tobacco confers high salinity tolerance without affecting yield. Proc Natl Acad Sci USA 102:509–514. doi:10.1073/pnas.0406485102
Savé R, Alegre L, Pery M, Terradas J (1993) Ecophysiology of after-fire resprouts of Arbutus unedo L. Orsis 8:107–119
Scott DA et al (1998) Identification and mutation analysis of a cochlearexpressed, zinc finger protein gene at the DFNB7/11 and dn hearing-loss-loci on human chromosome 9q and mouse chromosome19. Gene 215:461–469. doi:10.1016/S0378-1119(98)00316-3
Shinozaki K, Yamguchi-Shinozaki K (2000) Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr Opin Plant Biol 3:217–223. doi:10.1016/S1369-5266(00)00067-4
Singh NK, Nelson DE, Kuhn D, Hasegawa PM, Bressan RA (1989) Molecular cloning of osmotin and regulation of its expression by ABA and adaptation to low water potential. Plant Physiol 90:1096–1101
Singla-Pareek SL, Reddy MK, Sopory SK (2003) Genetic engineering of the glyoxalase pathway in tobacco leads to enhanced salinity tolerance. Proc Natl Acad Sci USA 100:14672–14677. doi:10.1073/pnas.2034667100
Singla-Pareek SL, Yadav SK, Pareek A, Reddy MK, Sopory SK (2008) Enhancing salt tolerance in a crop plant by overexpression of glyoxalase II. Transgenic Res 17:171–180. doi:10.1007/s11248-007-9082-2
Takahashi H, Chen Z, Du H, Liu Y, Klessig DF (1997) Development of necrosis and activation of disease resistance in transgenic tobacco plants with severely reduced catalase levels. Plant J 11:993–1005
Takatsuji H (1998) Zinc-finger transcription factors in plants. Cell Mol Life Sci 54:582–596. doi:10.1007/s000180050186
Turner NC (1981) Techniques and experimental approaches for the measurement of plant water status. Plant Soil 58:339–366. doi:10.1007/BF02180062
Vij S, Tyagi AK (2006) Genome-wide analysis of the stress associated protein (SAP) gene family containing A20/AN1 zinc-finger(s) in rice and their phylogenetic relationship with Arabidopsis. Mol Genet Genomics 276:565–575. doi:10.1007/s00438-006-0165-1
Vij Sh, Tyagi AK (2008) A20/AN1 zinc-finger domain-containing proteins in plants and animals represent common elements in stress response. Funct Integr Genomics 8:301–307. doi:10.1007/s10142-008-0078-7
Xiong L, Zhu JK (2002) Salt tolerance. In: Somerville CR, Meyerowitz EM (eds) The Arabidopsis Book. American Society of Plant Biologists, Rockville. doi:10.1199/tab.0048
Zhu JK (2000) Genetic analysis of plant salt tolerance using Arabidopsis. Plant Physiol 124:941–948
Zhu JK (2001) Plant salt tolerance. Trends Plant Sci 6:66–72. doi:10.1016/S1360-1385(00)01838-0
Zouari N, Ben Saad R, Legavre Th, Azaza J, Sabau X, Jaoua M, Masmoudi K, Hassairi A (2007) Identification and sequencing of ESTs from the halophyte grass Aeluropus littoralis. Gene 404:61–69
Acknowledgments
Special thanks are extended to C. Périn and M. Conte (Plant Development and Genetic Improvement (DAP) unit of CIRAD) as well as to Kh. Belhaj (CBS), A. Price (University of Aberdeen, UK) for their critical review of the manuscript. The authors are also grateful to S. Abid, a teacher of English, for the English revision. Part of this work was conducted under the REFUGE platform funded by Agropolis Fondation, Montpellier France. This study was supported by a grant from Ministry of Higher Education Scientific Research and Technology of Tunisia (contrat programme 2006–2010) and by the European project CEDROME (INCO-CT-2005-015468).
Author information
Authors and Affiliations
Corresponding author
Additional information
R. Ben Saad and A. Hassairi should be considered as first authors, as they contributed equally to this work.
The AlSAP sequence was deposited to the Genbank with the accession number DQ0885218 (UniProtKB: A1YAQ3).
Electronic supplementary material
Below is the link to the electronic supplementary material.
11103_2009_9560_MOESM1_ESM.tif
Supplementary Fig. 1 The nucleotide (707 bp) and the deduced amino acid sequence (159 aa) of AlSAP gene and its genomic structure. (a)The position of 5’UTR (118 bp) and 3’UTR (112 bp) are marked by line. The position of start (atg) and the stop (tga) codons are indicated by *. The A20 and AN1 domains of the peptide are indicated by dark and light grey shading respectively. The predicted serine (S), threonine (T) and tyrosine (Y) phosphorylation sites by NetPhos 2.0 Server (http://www.cbs.dtu.dk/services/NetPhos/) with a score > 0.8 are indicated by a circle. The predicted of the threonine kinase specific protein phosphoylation site (score 0.9) by NetPhosK 1.0 Server is indicated by a square. (b) The nucleotide sequence of AlSAP gene showing the presence of an intron (italic letters, 1,523 bp) in the 5’UTR (light grey shading), the ORF and the 3’UTR (dark grey shading) (TIFF 800 kb)
Rights and permissions
About this article
Cite this article
Ben Saad, R., Zouari, N., Ben Ramdhan, W. et al. Improved drought and salt stress tolerance in transgenic tobacco overexpressing a novel A20/AN1 zinc-finger “AlSAP” gene isolated from the halophyte grass Aeluropus littoralis . Plant Mol Biol 72, 171–190 (2010). https://doi.org/10.1007/s11103-009-9560-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-009-9560-4