Abstract
Haemophilus parasuis (H. parasuis) is associated with meningitis, polyserositis, polyarthritis and bacterial pneumonia. At present, its prevention and control is difficult because of the lack of suitable subunit vaccines. Nowadays, high-throughput methods, immunoproteomics, are available to screen for more vaccine candidates. A protein extraction method for H. parasuis and two-dimensional electrophoresis (2-DE) were optimized to provide high-resolution profiles covering pH 3 to 10. Twenty immunoreactive spots were excised from gels after strict comparison between 2-DE Western blot membranes and the relevant gels. Matrix-assisted laser desorption/ionization-time of flight-mass spectrometry (MALDI-TOF-MS) and MALDI-TOF–TOF-MS successfully identified 16 different proteins. Fifteen of them were reported as immunoreactive proteins in H. parasuis for the first time. In addition, recombinant HP5-7 (ABC transporter, periplasmic-binding protein) showed immunoreactivity both with hyperimmune rabbit serum and convalescent swine serum. Four recombinants of the 14 successfully expressed genes showed immunoreactivity with hyperimmune rabbit serum.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
H. parasuis is a Gram-negative bacterium that belongs to the Pasteurellaceae family. Its clinical infection signs are termed Glässer disease (Riley et al. 1977). There are 15 distinct serotypes, while 15–41 % of the isolates were reported as non-typable. Serotypes 1, 5, 10, 12, 13 and 14 are the most virulent (Angen et al. 2004; Oliveira et al. 2003). The most prevalent serotypes in China are 4 and 5 (Cai 2006; Zhou et al. 2010).
Abuse of antimicrobials in farmed animals is a hazard to humans, so vaccination is the preferred method. However, pigs immunized with monovalent vaccines are protected against challenge with the homologous serotype strains but not with other heterologous ones (Takahashi et al. 2001). Three ABC-type transporters (OppA, YfeA, and PlpA) and one curli protein assembly (CsgG) showed cross-reactivity when tested with sera raised against serovars 4 and 5 of H. parasuis (Hong et al. 2011). The purified recombinants of outer membrane protein (OMP) P2 and OmpP5 (also known as OmpA, PalA, P2, D15, HPS-06257) provided partial protection against H. parasuis infection in mice (Ahn et al. 2012; Sturgill et al. 2013; Tang et al. 2010; Zhou et al. 2009). However, some reports indicated that recombinant OmpP5 could not provide satisfactory protection in mice after bacterial challenge (Zhou et al. 2009). Recently, it was reported that recombinant virulence-associated trimeric autotransporters (VtaA) could provide partial protection against H. parasuis infection in colostrum-deprived piglets (Olvera et al. 2011). Thus, in contrast to other diseases of similar importance, there are few effective subunit vaccines.
The whole H. parasuis genome sequences SH0165 (serotype 5) (Yue et al. 2009) and 29755 (serotype 5) (Mullins et al. 2011) have been completed. In the post-genomics era, immunoproteomics has emerged as a high-throughput method for screening novel vaccine candidates. All of these developments lay a solid foundation for our research.
In this paper, we used an immunoproteomic approach to identify immunogenic antigens according to the immune response in H. parasuis serotype 5-immunized rabbits.
Materials and methods
Bacterial strains and culture conditions
H. parasuis strain GD10 (serotype 5) was isolated from a diseased pig in Guangdong Province in China. It was kindly provided by Dr. Guiping Wang (Guangdong Modern Agriculture Research Institute) and cultured in tryptic soy broth (TSB; Sigma-Aldrich Co. LLC., MO, USA) with 5 % newborn calf serum (Tian Jin Hao Yang Biological Manufacture Co., Ltd., Tian Jin, China) and 20 μg/mL NAD (Sinopharm Chemical Reagent Co., Ltd., China). Cultures were incubated at 37 °C in a rotary incubator (180 rpm) until the late stage of the exponential phase (Cai 2006).
Hyperimmune sera preparation
Polyclonal antibodies were raised in rabbits immunized with formaldehyde-inactivated GD10 bacteria after the rabbits were determined to be negative for GD10 antibodies by whole-cell enzyme-linked immunosorbent assay (ELISA). Three doses of 1.0 × 108 cells per rabbit were administered by subcutaneous injections at 2-week intervals (Cai 2006).
Convalescent sera screening
The sera of apparently healthy swine that had not been immunized with any H. parasuis vaccine were screened by ELISA using recombinant (OMP) P2, whose sequence is conserved among H. parasuis but specific to other bacteria, especially Pasteurella multocida (PM) and Actinobacillus pleuropneumoniae (APP). A swine that tested positive was considered to have been infected by H. parasuis and to have recovered.
Protein sample extraction
Proteins were extracted according to the approach of Zhang et al. with slight modifications (Zhai et al. 2012). Trichloroacetic acid (TCA, Sinopharm Chuan Kang Pharmaceutical Co., Ltd. China) was added to the sample preparation solution extract at a final concentration of 5 %.
Isoelectric focusing (IEF)
IEF was performed according to the approach of Zhang et al. with slight modifications (Zhai et al. 2012) and was carried out at 20 °C for 10.5 h (max. voltage 8,000 V; max. current 50 μA per IPG strip; total 28,000 V/h).
Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis
After SDS-PAGE, one gel was stained with Coomassie Brilliant Blue (CBB-G250), while the duplicate gel was subjected to Western blot analysis. After blocked, the membrane was incubated with anti-GD10 sera from immunized rabbits (1:2,000 dilution) at 37 °C for 2 h. Three replicates were run for each sample.
Image processing
Imagemaster 7.0 (GE Healthcare) was used to analyze the images scanned from the PVDF membrane and the corresponding two-dimensional electrophoresis (2-DE) gels.
MALDI-TOF-MS and database searches
Spots that identified as immunoreactive were excised and analyzed by MALDI-TOF-MS/MALDI-TOF–TOF-MS (NanJing Steed BioTechnologies Co. Ltd., Nanjing, China). The MASCOT server (http://www.matrixscience.com) was used to analyze the peptide mass fingerprinting (PMF) data.
Bioinformatic analysis
The compute pI/Mw server (http://expasy.org/tools/pi_tool.html) was used to calculate isoelectric point (pI) and molar mass (MW). The TMHMM Server v.2.0 (http://www.cbs.dtu.dk/services/TMHMM/) was used to predict transmembrane helices. The PSORT server (http://www.psort.org/) was used to predict the subcellular location and the SignalP 3.0 server (http://www.cbs.dtu.dk/servicess/SignalIP) identified potential signal peptides.
Characterization of recombinant immunoreactive proteins
Primer Premier 5.0 and Oligo 6.24 were used to design PCR primers (Table 1). Gene sequences of the 16 identified proteins were cloned into the pET-32a vector and expressed in Escherichia coli BL21 (DE3). Whole-cell extracts were subjected to SDS-PAGE with CBB-R250 staining and Western blotting using the anti-GD10 hyperimmune rabbit sera, swine convalescent sera and their corresponding negative sera.
Results
2-DE profiles of H. parasuis bacterial proteins and Western blot analysis
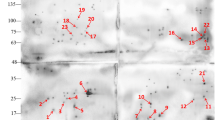
IEF separated over a pH range of 3–10 and 13-cm length in the first direction and a separation using SDS-PAGE in the vertical dimension identified 375 spots. Most proteins had a molar mass between 10 and 150 kDa (Fig. 1a). The repeat gels showed the corresponding immunoreactive spots (Fig. 1b).
The numbers on the left (M) indicate the molar mass of standards (kDa). The numbers in the figure represent different independent spots. Arrows indicate the corresponding proteins. 2-DE and Western blotting of whole-cell proteins from GD10. a CBB-G250-stained 2-DE gel of whole-cell proteins from GD10 (pH 3–10, 13 cm). b Western blot of corresponding gels of whole-cell proteins from GD10 using immunized anti-GD10 hyperimmune sera
Identification of immunoreactive spots
Twenty immunoreactive spots (HP5-1 to HP5-20) were excised from repeated 2-DE gels and analyzed by MALDI-TOF-MS/MALDI-TOF–TOF-MS followed by PMF searches. Finally, the 20 protein spots were identified as belonging to 16 different proteins (Table 2).
Bioinformatics analysis
The prediction results of subcellular locations, transmembrane helices and signal peptide cleavage sites of all identified proteins are listed in Table 3.
Characterization of recombinant immunoreactive proteins gene
Without HP5-3 and HP5-14/HP5-15, 4 out of the 14 successfully expressed proteins could react with hyperimmune rabbit sera raised against GD10 (Fig. 2). The four proteins were a double-stranded DNA (dsDNA)-mimic protein, Holliday junction DNA helicase B, an ABC transporter and periplasmic-binding protein/LacI regulator. The ABC transporter also reacted with the swine convalescent sera (Fig. 3).
The numbers on the left (M) indicate the molar mass of standards (kDa). The numbers in the figure represent different independent spots. Arrows indicate the corresponding proteins. CBB-R250-stained gel and Western blotting of the four recombinant immunoreactive proteins from cell lysates. a CBB-R250-stained gel of the four recombinant immunoreactive proteins in cell lysates CBB-R250-stained gel of induced 1 recombinant HP5-1, 2 recombinant HP5-2, 3 recombinant HP5-7, 4 recombinant HP5-17, and 5 pET-32a vector. b Western blot analysis of the four recombinant immunoreactive proteins in cell lysates. Western blot of induced 1 recombinant HP5-1, 2 recombinant HP5-2, 3 recombinant HP5-7, 4 recombinant HP5-17, and 5 pET-32a vector reacted with anti-GD10 hyperimmune sera. Western blot of induced 6 recombinant HP5-1, 7 recombinant HP5-2, 8 recombinant HP5-7, 9 recombinant HP5-17, and 10 pET-32a vector reacted with negative rabbit sera
The numbers on the left (M) indicate the molar mass of standards (kDa). The numbers in the figure represent different independent spots. Arrows indicate the corresponding proteins. CBB-R250-stained gel and Western blot analysis of the recombinant immunoreactive proteins from cell lysates. CBB-R250-stained gel of induced 1 recombinant HP5-7 and 2 pET-32a vector. Western blot analysis of induced 3 recombinant HP5-7 and 4 pET-32a vector reacted with convalescent swine serum. Western blot analysis of induced 5 recombinant of HP5-7 and 6 pET-32a vector reacted with negative swine sera
Discussion
We obtained 2-DE profiles with better resolution, over a broader pH range and produced clearer Western blots with the modification of protein sample preparation, compared with other proteomics studies in H. parasuis (Zhou et al. 2009).
The 16 identified immunoreactive proteins could be divided into three categories. The first category comprised proteins that have already been studied as subunit vaccines in H. parasuis. HP5-7, HP5-8, HP5-10, and HP5-20 were identified as the same protein: ABC transporter, a ubiquitous membrane protein (Schmitt and Tampe 2002) in all species, mediating the uptake and efflux of a diverse array of compounds (Grote et al. 2009; Locher 2004), including the non-classical secretion of signaling molecules and toxins. Three other ABC-type transporters (OppA, YfeA, and PlpA), which show amino acid identity of 34, 47, and 32 %, respectively, with the protein represented by HP5-7, showed cross-reactivity when tested with sera raised against serotypes 4 and 5 of H. parasuis (Hong et al. 2011).
The second category was proteins whose homologues in other bacteria were reported to be immunogenic. HI1450 may function as a dsDNA-mimic (HP5-1) to recognize, inhibit, or regulate an as yet unidentified dsDNA-binding protein. It is an immunoreactive protein against hyperimmune rabbit sera when expressed in E. coli (Parsons et al. 2004, 2005).
Elongation factor-Tu (EF-Tu, HP5-4) plays a central role during the selection of the correct amino acids during the elongation phase of translation (Kavaliauskas and Knudsen 2012). Many studies have verified the immunoreactivity of EF-Tu (Vergauwen et al. 2010).
Heme-binding protein A (HbpA, HP5-5) was identified as a virulence determinant in a model of Haemophilus influenzae invasive disease constructed by an insertional mutation of hbpA in a type b and a non-typable H. influenzae strain (Morton et al. 2005, 2009). HbpA of APP grown under iron-restricted conditions generated immunoreactivity in an immunoproteomic analysis (Chung et al. 2012).
The acquisition of iron from transferrin by Gram-negative bacterial pathogens is dependent on a periplasmic ferric-ion-binding protein, FbpA (HP5-11). It was highly antigenic in mice and showed intraspecies and interspecies antigenic homogenicity and specific anti-FbpA antibodies are fully cross-reactive (Ferreiros et al. 1999).
The third category included proteins with no previous report of immunogenicity. The smallest subunit of E. coli RNA polymerase is termed omega (HP5-9) (Gentry and Burgess 1986). A deletion mutation in this gene of Mycobacterium smegmatis (M. smegmatis) caused reduced sliding motility and defective biofilm formation. This resulted from a deficiency in generation of the extracellular matrix and the mutant bacterium failed to synthesize the short-chain mycolic acids that are characteristic of biofilm growth in M. smegmatis (Mathew et al. 2006).
Indolepyruvate ferredoxin oxidoreductase (IOR, HP5-12) from hyperthermophilic archaeon Pyrococcus kodakaraensis KOD1 catalyzes the oxidative decarboxylation of arylpyruvates by forming a heterooligomeric complex (alpha2beta2) (Siddiqui et al. 1998).
Most of the AraC family (HP5-13) are thought to be transcriptional activators that regulate genes related to carbon metabolism, stress responses, or pathogenesis (Egan 2002; Zeng and Spiro 2013). The Mycobacterium tuberculosis mutant strain disrupted in the AraC homologue Rv1931c exhibited reduced survival both in macrophages and in a mouse infection model (Frota et al. 2004).
Purine nucleoside phosphorylase (PNP, HP5-18) is a key enzyme to transfer glycosyl residues to acceptor bases. It has potential applications in the synthesis of nucleoside analogs used in the treatment of antiviral infections and in anticancer chemotherapy (Martins et al. 2011).
Pyridoxine kinase (HP5-19), which also phosphorylates pyridoxal (PL) and pyridoxamine (PM) in vitro, functions solely in the vitamin B6 salvage pathway. In E. coli, it contains an additional PL kinase associated with biosynthesis of pyridoxal 5′-phosphate (Yang and Winkler 1996).
The crystal structure of the Holliday junction DNA helicase B (HP5-2) bound to a single E. coli RuvA tetramer at 3.1-Å resolution has been solved.
Periplasmic-binding proteins (PBPs, HP5-6) are essential components of bacterial transport systems and are necessary for bacterial growth and survival (Shi et al. 2009).
The LacI regulator (HP5-17) is involved in the adaptive response of Streptococcus pneumoniae via its control of competence, adherence, and virulence (Chapuy-Regaud et al. 2003).
No details of oligopeptide permease ABC transporter membrane protein (HP5-3), hypothetical protein HPS_10240 (HP5-14/HP5-15), or putative solute/DNA competence effector (HP5-16) are available.
In conclusion, we obtained clear 2-DE and Western blot profiles of immunogenic proteins from H. parasuis. We identified 16 immunoreactive proteins, 15 of which are novel in H. parasuis. These data represent the basis for developing promising subunit vaccines.
References
Ahn J, Hong M, Yoo S, Lee E, Won H, Yoon I, Jung JK, Lee H (2012) Soluble expression of OmpA from Haemophilus parasuis in Escherichia coli and its protective effects in the mouse model of infection. J Microbiol Biotechnol 22:1307–1309
Angen O, Svensmark B, Mittal KR (2004) Serological characterization of Danish Haemophilus parasuis isolates. Vet Microbiol 103:255–258
Cai XW (2006) Isolation, identification, diagnosis and inactivated vaccine research of Haemophilus parasuis.
Chapuy-Regaud S, Ogunniyi AD, Diallo N, Huet Y, Desnottes JF, Paton JC, Escaich S, Trombe MC (2003) Regr, a global laci/galr family regulator, modulates virulence and competence in Streptococcus pneumoniae. Infect Immun 71:2615–2625
Chung JW, Kuster-Schock E, Gibbs BF, Jacques M, Coulton JW (2012) Immunoproteomic analyses of outer membrane antigens of Actinobacillus pleuropneumoniae grown under iron-restricted conditions. Vet Microbiol 159:187–194
Egan SM (2002) Growing repertoire of arac/xyls activators. J Bacteriol 184:5529–5532
Ferreiros C, Criado MT, Gomez JA (1999) The neisserial 37 kda ferric binding protein (fbpa). Comp Biochem Physiol B Biochem Mol Biol 123:1–7
Frota CC, Papavinasasundaram KG, Davis EO, Colston MJ (2004) The arac family transcriptional regulator rv1931c plays a role in the virulence of Mycobacterium tuberculosis. Infect Immun 72:5483–5486
Gentry DR, Burgess RR (1986) The cloning and sequence of the gene encoding the omega subunit of Escherichia coli RNA polymerase. Gene 48:33–40
Grote M, Polyhach Y, Jeschke G, Steinhoff HJ, Schneider E, Bordignon E (2009) Transmembrane signaling in the maltose abc transporter malfgk2-e: periplasmic malf-p2 loop communicates substrate availability to the atp-bound malk dimer. J Biol Chem 284:17521–17526
Hong MAJ, Yoo S, Hong J, Lee E, Yoon I, Jung JK, Lee H (2011) Identification of novel immunogenic proteins in pathogenic Haemophilus parasuis based on genome sequence analysis. Vet Microbiol 148:89–92
Kavaliauskas DNP, Knudsen CR (2012) The busiest of all ribosomal assistants: elongation factor tu. Biochemistry 51:2642–2651
Locher KP (2004) Structure and mechanism of abc transporters. Curr Opin Struct Biol 14:426–431
Martins NH, Meza AN, Santos CR, de Giuseppe PO, Murakami MT (2011) Molecular cloning, overexpression, purification, crystallization and preliminary x-ray diffraction analysis of a purine nucleoside phosphorylase from Bacillus subtilis strain 168. Acta Crystallogr Sect F: Struct Biol Cryst Commun 67:618–622
Mathew R, Mukherjee R, Balachandar R, Chatterji D (2006) Deletion of the rpoz gene, encoding the omega subunit of RNA polymerase, results in pleiotropic surface-related phenotypes in Mycobacterium smegmatis. Microbiology 152:1741–1750
Morton DJ, Madore LL, Smith A, Vanwagoner TM, Seale TW, Whitby PW, Stull TL (2005) The heme-binding lipoprotein (hbpa) of Haemophilus influenzae: role in heme utilization. FEMS Microbiol Lett 253:193–199
Morton DJST, Bakaletz LO, Jurcisek JA, Smith A, VanWagoner TM, Whitby PW, Stull TL (2009) The heme-binding protein (hbpa) of Haemophilus influenzae as a virulence determinant. Int J Med Microbiol 299:479–488
Mullins MA, Register KB, Bayles DO, Dyer DW, Kuehn JS, Phillips GJ (2011) Genome sequence of Haemophilus parasuis strain 29755. Stand Genomic Sci 5:61–68
Oliveira S, Blackall PJ, Pijoan C (2003) Characterization of the diversity of Haemophilus parasuis field isolates by use of serotyping and genotyping. Am J Vet Res 64:435–442
Olvera A, Pina S, Perez-Simo M, Aragon V, Segales J, Bensaid A (2011) Immunogenicity and protection against Haemophilus parasuis infection after vaccination with recombinant virulence associated trimeric autotransporters (vtaa). Vaccine 29:2797–2802
Parsons LM, Yeh DC, Orban J (2004) Solution structure of the highly acidic protein hi1450 from Haemophilus influenzae, a putative double-stranded DNA mimic. Proteins 54:375–383
Parsons LM, Liu F, Orban J (2005) Hu-alpha binds to the putative double-stranded DNA mimic hi1450 from Haemophilus influenzae. Protein Sci 14:1684–1687
Riley MG, Russell EG, Callinan RB (1977) Haemophilus parasuis infection in swine. J Am Vet Med Assoc 171:649–651
Schmitt L, Tampe R (2002) Structure and mechanism of abc transporters. Curr Opin Struct Biol 12:754–760
Shi R, Proteau A, Wagner J, Cui Q, Purisima EO, Matte A, Cygler M (2009) Trapping open and closed forms of fite: a group iii periplasmic binding protein. Proteins 75:598–609
Siddiqui MA, Fujiwara S, Takagi M, Imanaka T (1998) In vitro heat effect on heterooligomeric subunit assembly of thermostable indolepyruvate ferredoxin oxidoreductase. FEBS Lett 434:372–376
Sturgill D, Malone JH, Sun X, Smith HE, Rabinow L, Samson ML, Oliver B (2013) Design of RNA splicing analysis null models for post hoc filtering of drosophila head RNA-seq data with the splicing analysis kit (spanki). BMC Bioinforma 14:320
Takahashi K, Naga S, Yagihashi T, Ikehata T, Nakano Y, Senna K, Maruyama T, Murofushi J (2001) A cross-protection experiment in pigs vaccinated with Haemophilus parasuis serovars 2 and 5 bacterins, and evaluation of a bivalent vaccine under laboratory and field conditions. J Vet Med Sci 63:487–491
Tang C, Zhang B, Yue H, Yang F, Shao G, Hai Q, Chen X, Guo D (2010) Characteristics of the molecular diversity of the outer membrane protein a gene of Haemophilus parasuis. Can J Vet Res 74:233–236
Vergauwen BEJ, Dansercoer A, Devreese B, Savvides SN (2010) Glutathione import in Haemophilus influenzae rd is primed by the periplasmic heme-binding protein hbpa. Proc Natl Acad Sci U S A 107:13270–13275
Yang YZG, Winkler ME (1996) Identification of the pdxk gene that encodes pyridoxine (vitamin B6) kinase in Escherichia coli k-12. FEMS Microbiol Lett 141:89–95
Yue M, Yang F, Yang J, Bei W, Cai X, Chen L, Dong J, Zhou R, Jin M, Jin Q, Chen H (2009) Complete genome sequence of Haemophilus parasuis SH0165. J Bacteriol 191:1359–1360
Zeng J, Spiro S (2013) Finely tuned regulation of the aromatic amine degradation pathway in Escherichia coli. J Bacteriol 195:5141–5150
Zhai Z, Cheng L, Tang F, Lu Y, Shao J, Liu G, Bao Y, Chen M, Shang K, Fan H, Yao H, Lu C, Zhang W (2012) Immunoproteomic identification of 11 novel immunoreactive proteins of Riemerella anatipestifer serotype 2. FEMS Immunol Med Microbiol 65:84–95
Zhou M, Guo Y, Zhao J, Hu Q, Hu Y, Zhang A, Chen H, Jin M (2009) Identification and characterization of novel immunogenic outer membrane proteins of Haemophilus parasuis serovar 5. Vaccine 27:5271–5277
Zhou X, Xu X, Zhao Y, Chen P, Zhang X, Chen H, Cai X (2010) Distribution of antimicrobial resistance among different serovars of Haemophilus parasuis isolates. Vet Microbiol 141:168–173
Acknowledgments
This work was supported by grants from the Program for New Century Excellent Talents in the University of Ministry of Education of China (no. NCET-110671) and the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yu, Y., Wu, G., Zhai, Z. et al. Fifteen novel immunoreactive proteins of Chinese virulent Haemophilus parasuis serotype 5 verified by an immunoproteomic assay. Folia Microbiol 60, 81–87 (2015). https://doi.org/10.1007/s12223-014-0343-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12223-014-0343-1