Abstract
ClpB-cytoplasmic (ClpB-cyt)/Hsp100 is an important chaperone protein in rice. Cellular expression of OsClpB-cyt transcript is governed by heat stress, metal stress, and developmental cues. Transgenic rice plants produced with 2 kb OsClpB-cyt promoter driving Gus reporter gene showed heat- and metal-regulated Gus expression in vegetative tissues and constitutive Gus expression in calli, flowering tissues, and embryonal half of seeds. Rice seedlings regenerated with OsClpB-cyt promoter fragment with deletion of its canonical heat shock element sequence (HSE−273 to −280) showed not only heat shock inducibility of Gus transcript/protein but also constitutive expression of Gus in vegetative tissues. It thus emerges that the only classical HSE present in OsClpB-cyt promoter is involved in repressing expression of OsClpB-cyt transcript under unstressed control conditions. Yeast one-hybrid assays suggested that OsHsfA2c specifically interacts with OsClpB-cyt promoter. OsHsfA2c also showed binding with OsClpB-cyt and OsHsfB4b showed binding with OsClpB-cyt; notably, interaction of OsHsfB4b was seen for all three OsClpB/Hsp100 protein isoforms (i.e., ClpB-cytoplasmic, ClpB-mitochondrial, and ClpB-chloroplastic). Furthermore, OsHsfB4b showed interaction with OsHsfA2c. This study suggests that OsHsfA2c may play a role as transcriptional activator and that OsHsfB4b is an important part of this heat shock responsive circuitry.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Heat shock proteins (Hsps) represent one of the best-characterized heat shock (HS) induced gene systems. Several different classes of Hsps (i.e., Hsp100, Hsp90, Hsp70, Hsp40, and sHsps) have been characterized in plants (Milioni and Hatzopoulous 1997; Rajan and D’Silva 2009; Sarkar et al. 2009; Singh et al. 2010). At the genetic level, significant modulation of gene expression is noted at the transcriptional level accompanying exposure of cells to high temperature. Promoters of a range of different plant Hsp genes have been analyzed for characterization of developmental and tissue specific expression patterns, both with and without heat stress (Crone et al. 2001; Prandl et al. 1995; Takahashi et al. 1992; Yabe et al. 1994). Analyses of 5′ and 3′ promoter deletions of the GmHsp17.3B in transgenic tobacco showed that the functional unit underlying this regulation process is the heat shock element (HSE) for the HS-dependent transcription of the native gene or with fused reporter genes. The functional HSE element usually contains a minimum of three 5′nGAAn3′ repeated motifs with a maximum insertion of 5 bp between each motif. Sugio et al. (2009) noted that the induced response to HS is lost if the canonical HSE in the AtHsp70A promoter is mutated, but the mutation was seen not to completely abolish the response of the promoter, indicating that there are other sequences that contribute to the activity of the promoter. Apart from HSEs, metal stress responsive elements (STREs) are present in HS promoters. STREs are shown to be important in recruiting metal toxicity specific transcription factors such as MTF-1 in yeast (Grably et al. 2002).
Hsfs possess a common core structure comprising an N-terminal DNA binding domain, an adjacent domain involved in oligomerization, a short peptide motif essential for nuclear import and export, and a C-terminal AHA type activation domain (Kotak et al. 2004). Genome-wide analysis has shown that rice and Arabidopsis contain 26 and 21 Hsf genes, respectively (Mittal et al. 2009; Swindell et al. 2007). Binding specificities of plant Hsfs to different Hsp promoters have not been much analyzed. In vertebrate cells, Hsps are thought to have a role in the positive regulation of Hsf2 (Mathew et al. 2001). According to Bjork and Sistonen (2010), a negative feedback loop operates as evidenced by interactions between human Hsf1 and Hsps (such as Hsp70/Hsp40 and Hsp90) as well as by the fact that Hsps keep Hsf1 in an inactive form under non-stress conditions. Interactions among CI and CII Hsps, Hsp70 and Hsp90 proteins with specific Hsfs have been reported (Baniwal et al. 2004; Lee et al. 1995; Nishizawa-Yokoi et al. 2010; Yamada and Nishimura 2008). The feedback control mechanism in Arabidopsis is considered to involve Hsp70 as a negative regulator of Hsfs (Lee et al. 1995). In tomato, LpHsp17.4-CII functions as putative repressor of LpHsfA2 and probably acts to recruit LpHsfA2 in heat stress granule complex (Port et al. 2004). Cytosolic Hsp90 in Arabidopsis interacts with constitutively expressed HsfA4 to down-regulate its function. Detailed analysis further reflected that Hsp90.2 interacts with AtHsfA7a and AtHsfB1 and not with AtHsfA4c (Yamada et al. 2007; Yamada and Nishimura 2008).
In rice, ClpB/Hsp100 proteins belonging to the group class I Clp ATPase proteins act as chaperone, mediating disaggregation of denatured proteins (Katiyar-Agarwal et al. 2001; Singh and Grover 2010). Rice has three ClpB/Hsp100 isoforms, one ClpB/Hsp100 each in cytoplasm (Os05g44340; OsClpB-cyt), chloroplast (Os03g31300; OsClpB-c), and mitochondria (Os02g08490; OsClpB-m) (Singh et al. 2010). OsClpB/Hsp100-cyt transcript is noted to be heat shock inducible. OsClpB/Hsp100-c and OsClpB-m transcripts are present constitutively and are upregulated in response to heat stress. OsClpB/Hsp100-cyt also shows induction in response to oxidative stress (Singh et al. 2010). Singla et al. (1998) noted high constitutive levels of Hsp100 protein in embryo and seed tissues of rice. Genevestigator analysis shows that OsClpB/Hsp100-cyt was constitutively expressed in seeds, embryos, and endosperms (Singh et al. 2010). However, detailed studies on regulation of gene expression of ClpB/Hsp100 genes are lacking. We report an analysis of transcriptional regulation of OsClpB-cyt gene expression in this study. Data on the effects of different cis-acting sequences (HSEs) and trans-acting factors (Hsfs) in heat-regulated expression of OsClpB-cyt gene are presented. Evidence suggesting a role of OsHsfs in the OsClpB-cyt expression as interacting partners is presented.
Materials and methods
Plasmid construction for plant transformation
Different OsClpB-cyt promoter fragments (2kbProOsClpB-cyt, ΔPro-HSE−273 to −280, ΔUTR-HSE-like−97 to −107 and 834bpPro OsClpB-cyt ) were PCR amplified and cloned in pCAMBIA1381Z vector (Online Resource 1). All pCAMBIA1381Z-derived constructs were introduced in Agrobacterium tumefaciens strain EHA105 and stably transformed in rice [Oryza sativa L. cultivar Pusa basmati 1 (PB1), an indica type] derived calli. Transformation protocol was employed as previously described in our laboratory (Katiyar-Agarwal et al. 2003). Genomic DNA from transgenic plants was isolated as described by Kobayashi et al. (1998). PCR using primers specific for the first 550 bp of Gus gene was performed to confirm the integration of transfer DNA (T-DNA) in the transgenic plants. Transient assays in onion cells were performed as described earlier (Nigam et al. 2008). 2kbProOsClpB-cyt::Gus construct was used in this experiment. Gus staining was done as described (Jefferson et al. 1987). Photographs were taken in bright field using a fluorescence microscope (Leica, Germany).
Site directed mutagenesis of the HSE sequences
Nucleotide deletions in the putative HSE region of the OsClpB-cyt promoter were performed. The deletions were introduced by PCR amplification of plasmid DNA, using Pfu DNA polymerase and oligonucleotides containing the desired sequence. Three-step PCR protocol described by Picard et al. (1994) was employed in which megaprimers containing the deletions are obtained after a first round of amplification for use in subsequent reactions, which amplify the promoter containing the desired deletion (Online Resource 2). For removal of HSE sequence in promoter and HSE-like sequence in 5′UTR, primers were made to amplify a region of 748 bases upstream of the translation start codon. The actual length of this region came down to 741 bases after the removal of the HSE −273 to −280 and 738 bases in the case of deletion of the HSE-like −97 to −107 sequence. PCR products were run on 6% urea-acrylamide gel and stained with ethidium bromide. The cloned inserts were sequenced manually using Thermosequenase cycle sequencing kit (USB, USA). PCR products were subsequently cloned in pCAMBIA1381Z.
RNA isolation and gel blot analysis
For Northern analysis, total RNA was isolated as described by Chomczynski and Sacchi (1987). Fifteen micrograms of total RNA was electrophoresed on a 1% agarose gel made in 1× morpholinopropanesulphonic acid and 1.2% formaldehyde. The membrane was hybridized with PCR-amplified fragment of Gus. The amplified fragment was radiolabeled by random primer extension labeling kit (NEN, USA).
Gus analysis
Histochemical Gus staining was performed with 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid (X-gluc) as substrate (Jefferson et al. 1987). Control vegetative tissues and seeds were directly stained overnight at 37°C. For HS treatment, tissues were first placed at 42°C for 30 min and then stained overnight at 37°C. For each construct, three independent assays were performed. The fluorimetric Gus assay was performed according to the method of Jefferson et al. (1987). Nearly 100 mg of ground rice tissue was homogenized in 200 μl of Gus extraction buffer. The protein content of extracts was measured using the Bradford method (Bradford1976). A Gus assay was performed by incubation of 5 μl of 100 times diluted extract with 200 μl of 2 mM 4-methylumbelliferyl glucuronide at 37°C for 16 h, and the reaction was terminated with 0.2 M Na2CO3.
DNA-protein interaction in yeast
Yeast strain A2279 (mata his3 leu2 trp1 ura3; Moriya et al. 2001) was used for visualizing the interaction of different Hsfs with upstream region of OsClpB-cyt. The OsClpB-cyt promoter was cloned in the vector placZi in the sites EcoRI and XhoI when cloned upstream of CYC1 minimal promoter and in EcoRI and BamHI when β-galactosidase was under direct control of the OsClpB-cyt promoter. In this experiment, the length of the promoter employed was 698 bp, in order to omit NcoI site, which was used for linearizing the plasmid. These vectors were linearized and integrated into the genome at the URA3 locus and transformants were selected on medium lacking uracil. The transformed cells were subsequently used for transformation with a plasmid containing the various OsHsfs under the control of ADH1 promoter provided that leucine and subsequent transformants were selected on a medium lacking leucine. β-Galactosidase assay was done as per the standard protocol using O-nitrophenyl β-d-galactopyranoside (ONPG) as substrate (Miller 1972).
Yeast two-hybrid assay
Yeast two-hybrid assays were carried out using pAD [activation domain (AD) fusion, prey] and pBD [binding domain (BD) fusion, bait] vectors (Stratagene, USA). The various rice Hsf genes were PCR amplified using gene-specific primer sets (Online Resource 1) and cloned in the pBD-GAL4 vector in EcoRI and SmaI sites. The OsClpB genes were cloned in pAD-Gal4 vector. The yeast strain used was YRG2 (MATα ura3-52 his3-200 ade2-101 lys2-801 trp1-901 leu2-3 112 gal4-542 gal80-538 LYS2::UASGAL1-TATAGAL1-HIS3 URA3::UASGAL4 17mers (x3)-TATACYC1-lacZ). For testing OsHsf/OsClpB-cyt protein–protein interactions, the OsHsf (bait) and ClpB (prey) pairs were co-transformed into yeast cells and transformants were selected on a medium lacking the amino acids leucine and tryptophan. Different dilutions of the transformed yeast cells were dotted on medium lacking histidine to check the HIS3 activity. β-Galactosidase activity was measured by quantitative liquid culture method using ONPG as substrate and by filter lift assay (Yeast protocols handbook, Clontech). Each experiment was separately repeated three times.
BiFC assays and subcellular localization
PCR-amplified OsClpB-cyt, OsHsfA2c, and OsHsfB4b genes were cloned into BiFC vectors pUC-SPYCE and pUC-SPYNE (Walter et al. 2004). For transient expression in onion epidermal cells, fusion proteins with N- or C-terminal parts of yellow fluorescent protein in pUC-SPYCE and pUC-SPYNE vectors were introduced into onion epidermal cells by particle bombardment as described previously (Singh et al. 2010). After incubation for 16 h, the cells were visualized by confocal laser scanning microscope (Leica TCS SP5). Green fluorescent protein (GFP) and yellow fluorescent protein (YFP) were excited with an argon laser at 488 and 514 nm, respectively. For subcellular localization of OsClp-B-cyt, the OsClpB-cyt gene was cloned in frame with GFP gene in pCAMBIA1302 vector. The cells were stained with DAPI (4′, 6′-diamidino-2-phenylindole) to identify the nucleus.
Results
Expression mediated by native version OsClpB-cyt promoter
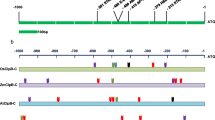
The nucleotide sequence of 2-kb region upstream of translation start codon of the OsClpB-cyt gene was downloaded from the NCBI database. An in silico search indicated the presence of a single HSE sequence (gaatattc; this sequence with configuration of nnnnnnGAAnnTTC is a variant of perfect HSE, which has the configuration of nTTCnnGAAnnTTC) at −273 to −280 position from the translation initiation site (HSE−273 to −280; Fig. 1). A single STRE sequence was present in this region (on the strand opposite to where HSE is marked). An AP-1 binding element (−411 to −416) and a C/EBP binding element (−1,424 to −1,428) were noted in the 2 kb region. In addition, an HSE-like element (gaagcaattc) was marked in the 5′UTR region of the OsClpB-cyt gene at −97 to −107 position (named as HSE−97 to −107). All the above sequences are marked taking “A” of ATG translation initiation codon as +1 (Fig.1). Canonical TATA-box like sequence is not marked in the vicinity of the transcription start site in the OsClpB-cyt promoter, indicating that the OsClpB-cyt promoter is a TATA-less promoter.
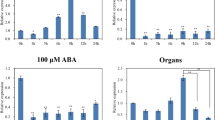
The 2-kb promoter fragment from rice genomic DNA was amplified by PCR. This promoter was cloned in pBCSK vector and subsequently sequenced (Online Resource 3). The 2-kb promoter fragment was cloned in pCAMBIA1381Z binary vector (construct referred to as 2kbProOsClpB-cyt::Gus; Fig. 2a). Transient transformation of 2kbProOsClpB-cyt::Gus in onion epidermal cells was carried out using a biolistic gun, and Gus expression was examined after 16 h. Several cells were seen to express Gus protein even though the cells were not subjected to HS treatment (shown by arrows in Fig. 2b). 2kbProOsClpB-cyt::Gus construct was employed for callus mediated, stable rice transformation. After 48 h of co-cultivation of 2kbPro OsClpB-cyt ::Gus construct harboring A. tumefaciens cells with PB1 rice calli, a set of calli pieces was analyzed for Gus expression. Gus expression was noted in calli even without HS. However, the heat stressed calli showed a distinctly enhanced accumulation of Gus as compared to the unstressed (control) calli pieces (Fig. 2c). T0 plants were analyzed for integration of T-DNA by PCR reaction using primers specific for Gus. All the tested plants obtained after selection on hygromycin containing medium were PCR positive for T-DNA integration. Leaf segments from mature plants exposed to 42°C for 1 h showed heat inducible expression of Gus in histochemical reaction (Fig. 2d). RNA isolated from heat stressed (42°C, 1 h) and unstressed (control) leaf segments were resolved on agarose gel, and Northern probing was done using radiolabeled Gus probe. The Gus transcript was noted to be noticeably heat stress inducible in six independent seedlings analyzed (T1-T6; Fig. 2e). T1 seedlings were subsequently heat stressed (42°C, 1 h). GUS accumulation was observed to be regulated by high temperature in these seedlings (Fig. 2f). 2kbProOsClpB-cyt::Gus plants were analyzed for Gus expression in response to different metals (Fig. 2g). Gus expression was noted to be induced in response to all the metals tested except Zn (Fig. 2g). The histochemical level of Gus expression was relatively low in the case of Cu treatment. In seedlings treated with Cd and As, Gus expression was validated by Northern blotting using radiolabeled Gus gene as probe. Expression of Gus in response to Cd application was observed to be almost of the same magnitude as that in heat stress (Fig. 2h). Expression of Gus was analyzed in the reproductive organs of unstressed (control) 2kbProOsClpB-cyt::Gus plants by histochemical staining. Gus expression was prominent in the anther tissues (Fig. 2i-i). Expression of Gus was seen in the style and ovary tissues (Fig. 2i-ii). Gus expression was significantly higher in the embryonal half of the seeds (Fig. 2i-iii).
Analysis of 2 kb OsClpB-cyt promoter. a Schematic representation of the T-DNA region of 2kbProOsClpB-cyt::Gus construct. b Histochemical expression of Gus in onion epidermal cells shot with 2kbProOsClpB-cyt::Gus construct. Cells showing transient expression of Gus are shown by arrows. c Expression of Gus in heat stressed (42°C, 1 h) calli transformed with 2kbProOsClpB-cyt::Gus construct. d Leaf segments from T1 rice seedlings expressing Gus protein following heat treatment (42°C, 1 h). e Northern analysis showing heat induction of Gus transcript in leaf segments of 2kbProOsClpB-cyt::Gus plants. Probing was done with radiolabeled Gus probe. f Histochemical analysis of Gus expression in 2kbProOsClpB-cyt::Gus plants following heat stress (42°C, 1 h). g Expression of Gus in 2kbProOsClpB-cyt::Gus plants in response to various metals. Treatments were given for 6 h at 28°C in beakers containing 20 μM solutions of arsenic (As), cadmium (Cd), cobalt (Co), copper (Cu), and zinc (Zn). Heat stress was given in a water bath maintained at 42°C for 1 h. Gus expression was analyzed histochemically. h Northern blot showing expression of Gus transcript in metal and heat shocked tissues. Fifteen micrograms of RNA was loaded in each lane. Lower panel depicts rRNA bands as loading control. i Gus expression in the reproductive organs of 2kbProOsClpB-cyt::Gus rice plants. i Whole mount of anther. ii Whole mount of gynoecium tissues. iii Whole mount of seeds. In all the histochemical assays, seven lines were employed; data from one representative transgenic line are shown
PCR amplified 834 bp long OsClpB-cyt promoter fragment containing 724 bp portion upstream to the transcription initiation site and 110 bp downstream to the transcription initiation site (lacking 26 bp sequence from the 3′end of 5′UTR) was cloned next to Gus gene in a separate experiment (construct referred to as 834bpPro OsClpB-cyt ::GUS; Online Resource 4). Rice calli transformed with 834bpPro OsClpB-cyt ::GUS construct showed low levels of Gus histochemical staining in unstressed conditions. There was an increase in the amount of Gus staining after the transformed 834bpProOsClpB-cyt::GUS calli were subjected to HS. In transgenic rice raised with 834bpProOsClpB-cyt::Gus construct, Gus transcript and protein were clearly heat stress inducible.
Mutations in the OsClpB-cyt promoter modify the gene expression
Using PCR, HSE −273 to −280 nucleotide sequence was deleted from the promoter region. Additionally, HSE-like −97 to −107 element noted in 5′UTR of the OsClpB-cyt gene was deleted in a separate experiment. The constructs for deleted HSE −273 to −280 (Fig. 3a) and HSE-like −97 to −107 (Fig. 3d) were named as ΔPro-HSE−273 to −280::Gus and ΔUTR-HSE-like−97 to −107::Gus, respectively. The length of the promoter fragment was 741 bp after removal of the HSE-273 to −280 and 738 bp in the case of deletion of the HSE-like −97 to −107 elements. Transgenic rice plants were raised with ΔPro-HSE−273 to −280::Gus and ΔUTR-HSE-like−97 to −107::Gus constructs were positive by PCR for the presence of the transgene. T0 plants were examined for the effect of deleted HSE sequences on Gus expression. RNA isolated from heat stressed leaf segments was probed using Gus gene. In the case of ΔPro-HSE-273to-280::Gus plants, Gus transcript was constitutively expressed (i.e., unstressed conditions). The levels of this transcript increased in response to HS in all the six independent plants analyzed (T1–T6; Fig. 3b). A similar trend was noted with respect to the expression of Gus protein in fluorimetric analysis, carried out using seven independent plants (P1–P7; Fig. 3c). ΔUTR-HSE-like−97 to −107::Gus plants showed expression in a way similar to 2kbProOsClpB-cyt::Gus plants. There was no expression of either Gus transcript or protein under unstressed (control) conditions. Gus expression was HS inducible both at the transcript (analyzed in T1–T4 independent seedlings) and protein levels (analyzed in T1–T5 independent seedlings) in ΔUTR-HSE-like−97 to −107::Gus plants (Fig. 3e, f).
Analysis of mutant forms of OsClpB-cyt promoter. a Schematic representation of ΔPro-HSE−273 to −280::Gus construct. b Northern analysis of ProΔHSE−273 to −280::Gus plants using radiolabeled Gus as probe. T1–T6 represent independent transgenic plants. c Fluorimetry of ProΔHSE−273 to −280::Gus plants using 20 μg of protein. Ratios of Gus activity in unstressed (control) and HS samples were plotted. For both RNA (b) and protein (c) isolation, seedlings given heat stress at 42°C for 1 h were used and unstressed seedlings were taken as control. P1–P7 represent independent transgenic plants. d Schematic representation of ΔUTR-HSE-like−97 to −107::Gus construct. e Northern analysis of ΔUTR-HSE-like−97 to −107::Gus plants, using radiolabeled Gus as probe. T1–T4 represent independent transgenic plants. f Fluorimetric Gus expression in ΔUTR-HSE-like−97 to −107::Gus plants using 20 μg of protein. Ratios of Gus activity in control and HS samples were plotted. For both RNA (e) and protein (f) isolation, seedlings given heat stress at 42°C for 1 h were used. Unstressed seedlings were taken as control. T1–T5 represent independent transgenic plants
In vivo binding of OsClpB-cyt promoter with heat shock factors
For examining binding of OsClpB-cyt promoter with OsHsfA2a, OsHsfA7, OsHsfA9, OsHsfA2c, OsHsfA2d, OsHsfB4b, OsHsfB4c, OsHsfC1b, and OsHsf26, a system was constructed to express LacZ under the control of OsClpB-cyt promoter in yeast cells. OsClpB-cyt promoter (in this experiment the size of the promoter used was 698 bp taking “A” of ATG translation initiation codon as +1) cloned in the vector pLacZi (Clontech) was integrated into the genome of A2279 cells. The minimal promoter of yeast cytochrome1 (CYC1) is present upstream of lacZ in placZi vector. Plasmid constructs were designed to express the genes for the above OsHsfs driven by ADH1 promoter as fusion proteins to GAL4-AD (Fig. 4a). These plasmid constructs were transformed into strains containing OsClpB-cyt promoter + CYC1. High β-galactosidase activity was seen in the strain expressing OsClpB-cyt promoter + CYC1 and OsHsfA2c (Fig. 4b). The levels of β-galactosidase activity were not significant when strains carrying OsClpB-cyt promoter + CYC1 with other OsHsfs were analyzed.
Yeast one-hybrid assay for analyzing binding of OsClpB-cyt promoter with rice heat shock factor proteins. a Schematic representation of the constructs. b Binding assay of OsClp-B-cyt promoter with OsHsfs. Activity of β-galactosidase in yeast cells transformed with different constructs is shown. c Schematic representation of different versions of the construct containing OsClpB-cyt promoter cloned in the vector pLacZi (CYC1, OsClpB-cyt promoter + CYC1, OsClpB-cyt promoter-CYC1 and OsClpB-cyt promoter ΔHSE −273 to −280 + CYC1). d Binding assay of the above OsClp-B-cyt promoter constructs with OsHsfA2c. OsHsfA2c gene was transformed in yeast cells with the above constructs. Liquid β-galactosidase assay is shown in the left panel. Plate assay is shown in the right panel. For plate assays, equal number of cells were taken, spotted, and allowed to grow for 48 h. Thereafter, plates were overlaid with 0.1 M NaPO4 buffer containing 1% agar and 25 mg/ml X-gal. Plates were kept at 28°C for 24 h
Yeast cells were next transformed with different versions of the construct containing OsClpB-cyt promoter cloned in the vector pLacZi. These included (1) construct CYC1 (lacking OsClpB-cyt promoter), (2) construct OsClpB-cyt promoter + CYC1 (as in Fig. 4b), (3) construct OsClpB-cyt promoter-CYC1 (lacking CYC1 region), and (4) construct ΔPro-HSE−273 to −280 + CYC1 (which had deletion of HSE region in OsClpB-cyt promoter; Fig. 4c). Plasmid with OsHsfA2c was transformed in each of the above yeast cells. The presence of different OsClpB-cyt promoter versions as well as the OsHsfA2c gene was confirmed by PCR, using genomic DNA (with primers specific for OsClpB-cyt promoter) and plasmid DNA (with primers specific for OsHsfA2c gene) preparations from transformed yeast cells. High β-galactosidase activity was seen in strains expressing both OsClpB-cyt promoter (with CYC1) and OsHsfA2c. There was negligible β-galactosidase activity when CYC1 construct was transformed along with OsHsfA2c (liquid assay shown in left panel of Fig. 4d, plate assay shown in right panel of Fig. 4d). β-Galactosidase expression was also below detection limits when OsHsfA2c was not included in the strain as well as when CYC1 minimal promoter was lacking from the construct. The removal of the HSE caused a drastic decrease in LacZ expression as compared to the native promoter. As there was negligible LacZ expression when OsHsfs were not introduced in yeast, possible binding of the native yeast Hsf in mediating LacZ expression in this experiment is ruled out.
Binding of OsClpB-cyt with OsHsfs
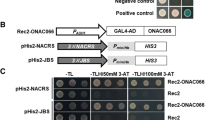
Yeast two-hybrid assays were carried out to analyze bindings among OsClpB-cyt and OsHsfA7, OsHsfA2a, OsHsfB4b, OsHsfB4c, and OsHsf26. OsHsf genes were cloned in pBD [binding domain (BD) fusion, bait vector], and OsClpB-cyt was cloned in pAD [activation domain (AD) fusion, prey] vector (Fig. 5a). Results based on-His + 3AT growth (data not shown) and ONPG assay (Fig. 5b) showed that OsHsfB4b interacts with OsClpB-cyt. For this analysis, pSE1111-ScSNF1 (ScSNF1; AMP-activated serine/threonine protein kinase) and pSE1112-ScSNF4 (ScSNF4; activating gamma subunit of the AMP-activated Snf1p kinase complex) transformed into YRG2 strain to yield YRG2-pSE1111-ScSNF1 + pSE1112-ScSNF4 cells were used as positive control (Fields and Song 1989). Furthermore, binding of OsClpB-m and OsClpB-c with OsHsfB4b was examined. OsClpB-m, OsClpB-c, and OsClpB-cyt genes cloned in pAD vector and OsHsfB4b gene cloned in pBD vector were analyzed. As seen for OsClpB-cyt, OsClpB-m and OsClpB-c proteins showed positive interactions with OsHsfB4b (Fig. 5c), suggesting that OsHsfB4b interacts with all the three ClpB proteins.
Yeast two-hybrid assay for analyzing binding between selected OsHsfs and OsClpB proteins. a Schematic representation of the constructs used. b Yeast two hybrid assay showing β-galactosidase activity in YRG2 yeast cells transformed with different constructs. c Growth assay for –His reporter gene. Serial dilutions of YRG2 cells co-transformed with pAD-OsClpB-cyt/chloroplastic/mitochondrial and pBD-OsHsfB4b constructs were spotted on –Leu Trp, –His Leu Trp, and –His Leu Trp containing 1 mM 3AT. PC: positive control (pSE1111-ScSNF1 and pSE1112-ScSNF4 transformed YRG2 strain to yield YRG2-pSE1111-ScSNF1 + pSE1112-ScSNF4 cells). NC Negative control (pAD + pBD vector transformed YRG2 cells). Three independent sets of yeast two hybrid assays were undertaken; data from one representative experiment are shown
A BiFC assay showed that OsClpB-cyt forms a complex with OsHsfB4b and OsHsfA2c (Fig. 6a, b). In both cases, YFP fluorescence was predominantly detected in the nucleus indicating that the complexes were formed due to an interaction between OsHsfB4b:OsClpB-cyt as well as between the OsHsfA2c:OsClpB-cyt, which are nucleus-localized (Fig. 6a, b). Binding between OsHsfA2c and OsHsfB4b was also noted in a separate BiFC assay (Fig. 6c). Yeast two-hybrid assay showed that OsClpB-cyt forms an oligomeric form in vivo (data not shown). The formation of an oligomeric form of OsClpB-cyt was noted in the BiFC experiment (Fig. 6d). This analysis revealed that OsClpB-cyt was predominantly localized in cytoplasm (Fig. 6d). Cytoplasmic localization of OsClpB-cyt was also noted in an experiment involving fusing this protein with GFP and transiently transforming the construct in onion epidermal cells (Online Resource 5).
BiFC assays for analyzing OsClpB-cyt:OsHsfB4b, OsClpB-cyt:OsHsfA2c, OsHsfA2c:OsHsfB4b, and OsClpB-cyt:OsClpB-cyt interactions, using onion epidermal cells. a Cells co-transformed with OsClpB-cyt-YFPN and OsHsfB4b-YFPC fusion constructs. b Cells co-transformed with OsHsfA2c-YFPN and OsClpB-cyt-YFPC fusion constructs. c Cells co-transformed OsHsfA2c-YFPN and OsHsfB4b-YFPC fusion constructs. d Cells co-transformed with OsClpB-cyt-YFPN and OsClpB-cyt-YFPC fusion constructs. i YFP signal, ii bright field images, iii DAPI (4′,6′-diamidino-2-phenylindole)-stained images and iv merged images of i, ii, and iii
Discussion
OsClpB-cyt/Hsp100 transcript in rice seedlings is not expressed at detectable levels under unstressed control conditions but imposition of heat stress results in its rapid and predominant expression (Agarwal et al. 2003; Singh et al. 2010). A 2-kb OsClpB-cyt promoter contains an HSE, a STRE, an AP-1 binding element as well as a C/EBP binding element (Fig. 1). Stably transformed rice seedlings with 2kbProOsClpB-cyt::Gus construct showed expression of Gus in a heat-regulated manner (Fig. 2). Stably transformed rice seedlings with 834bpProOsClpB-cyt::Gus construct also showed heat inducible Gus transcript and protein. 5′UTR of OsClpB-cyt gene contains TCTCAA (leader motif 1) sequence (Online Resource 6), which is considered to be a translational enhancer element in Fed1A and PsaDb genes (Caspar and Quail 1992; Yamamoto et al. 1995). Both 834 bp and 2 kb OsClpB-cyt promoters contain this translational enhancer sequence. From this study, the OsClpB-cyt promoter appears to be a classical HS promoter. The regulation of stress-related genes with constitutive promoters (i.e., CaMV35S promoter, actin promoter, and ubiquitin promoter) in the plant biotechnology industry is considered undesirable as it may lead to unwanted and hence wasteful metabolisms, causing ultimately a reduction in the growth and yield of transgenic plants (Ito et al. 2006). Wu et al. (2009) employed OsHsp100 promoter for driving OsWRKY11 gene for imparting desiccation tolerance to rice seedlings.
Heat and metal stresses show an overlapping signal reception. The AtHsp90-1 promoter induced gene expression at high levels after HS and As treatments (Haralampidis et al. 2002). 2kbProOsClpB-cyt::Gus rice seedlings in this study showed expression of Gus in response to treatment with Co, Cd, and As. In yeast, Grably et al. (2002) noted that the appropriate transcription of Hsp104 is usually obtained through cooperation between the Msn2/4/STRE and the Hsf/ HSE systems and that each factor could activate the promoter alone, backing up the other. Fine mapping of the elements underlying the response to heat and metals in the OsClpB-cyt promoter remains to be carried out.
In reproductive tissues, a constitutive presence of transcripts/proteins for Hsp100 has been noted (Hong and Vierling 2001; Young et al. 2001). A publicly available microarray database shows that the OsClpB-cyt transcript is expressed in embryo, endosperm, seed, and panicle tissues (https://www.genevestigator.ethz.ch/; Singh et al. 2010). T1 seeds of 2kbProOsClpB-cyt::Gus rice plants showed significant Gus expression histochemically, under unstressed control conditions (Fig. 2). In seeds, the Gus expression was seen predominantly towards the embryonal part. Earlier work showed that the expression of the OsHsp100 protein is mostly localized to the embryonal half of seeds (Singla et al. 1998). Rice is most susceptible to heat injury during flowering, as pollen viability is particularly sensitive to heat stress; even 1–2 h of high temperature at anthesis results in high spikelet sterility (Jagadish et al. 2010). Above-optimal temperatures reduce yield in tomato largely because of the high heat stress sensitivity of the developing pollen grains. Pressman et al. (2007) have shown that a heat tolerant cultivar of tomato expressed higher constitutive levels of Hsp100 in anthers as compared to a sensitive cultivar. From the data on high expression of Hsp100 in anther tissues and high OsClpB-cyt promoter activity noted in this study, it may be inferred that Hsp100 is an important component in pollen physiology of rice plants at high temperature.
In transgenic rice plants raised with a deletion in HSE−273 to −280 sequence (ΔPro-HSE−273 to −280::Gus plants), HS inducibility of this promoter was not affected as Gus expression in ΔPro-HSE−273 to −280::Gus plants was increased above the control values upon HS (Fig. 3). However, unlike in 2kbProOsClpB-cyt::Gus seedlings where Gus transcript/protein expression was at undetectable levels under unstressed control conditions (Fig. 2), expression of Gus was noted under control conditions in ΔPro-HSE−273 to −280::Gus seedlings (Fig. 3). It thus emerges that the HSE−273 to −280 sequence has a role in repressing expression of the downstream transcript under control, uninduced conditions. The relationship of HSEs to HS expression appears to be of a complex nature. Trinklein et al. (2004) showed that Hsf1 binding to HSE in Drosophila by itself does not confer heat inducibility. This group supports the idea that transcription initiation might often require the assembly of several different factors in the promoter. HSE−273 to −280 deletion may be influencing the transcription apparatus such that the constitutive control has been affected while the induced control has remained unaffected. There was no change in the expression profiling of Gus in ΔUTR-HSE-like−97 to −107::Gus plants: The heat stress inducibility of Gus as well as the non-inducibility under unstressed conditions was maintained (Fig. 3). It is thus likely that the HSE-like−97 to −107 is not an important region for regulation of the OsClpB-cyt transcription. A primer extension experiment further showed that HSE-like −97 to −107 falls within the 5′ untranslated region of OsClpB-cyt gene (data not shown).
OsHsfA2a, OsHsfA2c, OsHsfA7, OsHsfA9, OsHsfA2d, OsHsfB4b, OsHsfB4c, OsHsfC1b, and OsHsf26 showed differential transcript expression kinetics in microarray and real-time PCR assays: OsHsfA2a, OsHsfA2c, OsHsfA2d, and OsHsfB4b transcripts are present below detection levels in unstressed conditions but increased immensely upon HS. OsHsfA7, OsHsfA9, and OsHsfC1b transcripts are noted at low levels in unstressed conditions but increased upon HS and OsHsfB4c and OsHsf26 lack HS expression (Mittal et al. 2009). Significant increase in downstream LacZ expression was noted from OsClpB-cyt promoter in yeast cells on transformation with OsHsfA2c only in this assay (Fig. 4), suggesting that OsHsfA2c is specifically involved in regulation of the OsClpB-cyt promoter. However, it is possible that some other OsHsfs apart from the nine tested, which are not induced during heat stress, may also bind to the OsClpB-cyt promoter in planta. Apart from OsHsfA2c binding to OsClpB-cyt promoter, we noted that OsHsfA2c binds with OsClpB-cyt based on BiFC assays (Fig. 6b). OsHsfA2c thus appears to be an important player in the rice heat shock response. Earlier work has documented that OsHsfA2c is one of the most rapidly induced Hsf genes in response to heat stress, that OsHsfA2c possesses transactivation activity, that OsHsfA2c shows trimer formation activity from its monomeric forms, and that OsHsfA2c binds to perfect-type HSE (Mittal et al. 2009; Mittal et al. 2011). This study further noted that OsHsfB4b binds with OsClpB-cyt based on yeast two-hybrid and BiFC assays. Positive interaction was marked for all three isoforms of OsClpB isoforms, namely, OsClpB-cyt, OsClpB-c, and OsClpB-m with OsHsfB4b (Fig. 5). BiFC assays confirmed binding of OsHsfB4b with OsClpB-cyt as well as OsHsfA2c (Fig. 6a, c). Our earlier work has shown that OsHsfB4b is induced in response to heat stress, OsHsfB4b lacks transactivation activity and shows trimer formation activity and maximum binding activity to perfect-type HSE in electrophoretic mobility shift assays (Mittal et al. 2009; Mittal et al. 2011). It may be inferred that HS-induced OsHsfA2c binds to OsClpB-cyt promoter, possibly controlling synthesis of the OsClpB-cyt transcript. We suggest that the binding of OsClpB-cyt with OsHsfA2c may work in a negative feedback loop manner as reported for Hsp90 (Yamada et al. 2007). Interactions between HSPs and HSFs have been documented previously. AtHSP70 was seen to modulate the activity of AtHsf1 by interacting directly with the DNA binding domain and activation domains of AtHsf1 (Kim and Schoffl 2002). Hsp27 in mammalian cells has been shown to be present in the nucleus where it interacts with the transcription factor SP1 and stimulates SP1-mediated gene expression (Friedman et al. 2009). More recently, Hsp70 and Hsp90 have been shown to regulate the activities of HsfA1, HsfA2, and HsfB1 in tomato (Hahn et al. 2011). Notably, OsClpB-cyt is localized in cytoplasm while OsHsfA2c:OsClpB-cyt complex is localized in nuclei. The significance of OsClpB-cyt:OsHsfB4b complex may lie in controlling levels of transcript formation, translational activity, and in stability or functional role of OsClpB-cyt. As OsHsfB4b also showed binding to OsHsfA2c, there is a possibility that a supra complex of OsHsfB4b, OsHsfA2c, and OsClpB-cyt is formed. Additionally, OsHsfB4b also interacts with OsHsfA7, OsHsfA2a, OsHsfB4c, and OsHsf26 proteins (Mittal et al. 2011). Thereby, OsHsfB4b appears to be a critical protein in governing OsHsf interactions. Since OsHsfB4b lacks transactivation potential, it may have a regulatory role, which may be prevent the binding of the class A Hsfs to heat shock promoters by sequestering them inactive. Further work needs to be undertaken to elucidate the amino acid residues important in these interactions. As OsClpB-cyt gene is one of the early heat stress genes in rice, and since the protein has the capability to interact with OsHsfA2c in nucleus, OsClpB-cyt may act as a transcriptional cofactor for OsHsfA2c. However, additional experiments will be needed to determine whether the interaction between the two proteins is direct or mediated by some other protein(s). The present study suggests that heat stress transcription factors and heat shock proteins orchestrate an Hsf/Hsp circuitry, which may involve several additional proteins.
References
Agarwal M, Sahi C, Katiyar-Agarwal S, Agarwal S, Young T, Gallie DR, Sharma VM, Ganesan K, Grover A (2003) Molecular characterization of rice hsp101: complementation of yeast hsp104 mutation by disaggregation of protein granules and differential expression in indica and japonica rice types. Plant Mol Biol 51:543–553
Baniwal SK, Bharti K, Chan KY, Fauth M, Ganguli A, Kotak S, Mishra SK, Nover L, Port M, Scharf KD, Tripp J, Weber C, Zielinski D, von Koskull-Döring P (2004) Heat stress response in plants: a complex game with chaperones and more than twenty heat stress transcription factors. J Biosci 29:471–487
Bjork JK, Sistonen L (2010) Regulation of the members of the mammalian heat shock factor family. FEBS J 277:4126–4139
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Caspar T, Quail PH (1992) Promoter and leader regions involved in the expression of the Arabidopsis ferredoxinA gene. Plant J 3:161–174
Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidiniumthiocyanate-phenol-chloroform extraction. Anal Biochem 162:156–159
Crone D, Rueda J, Martin K, Hamilton D, Mascarenhas J (2001) The differential expression of a heat shock promoter in floral and reproductive tissues. Plant Cell Environ 24:869–874
Fields S, Song OK (1989) A novel genetic system to detect protein-protein interactions. Nature 340:245–246
Friedman MJ, Li S, Li X-J (2009) Activation of gene transcription by heat shock protein 27 may contribute to its neuronal protection. J Biol Chem 284:27944–27951
Grably MR, Stanhill A, Tell O, Engelberg D (2002) HSF and Msn2/4p can exclusively or cooperatively activate the yeast HSP104 gene. Mol Microbiol 44:21–35
Hahn A, Bublak D, Schleiff E, Scharf K-D (2011) Crosstalk between Hsp90 and Hsp70 chaperones and heat stress transcription factors in tomato. Plant Cell 23:741–755
Haralampidis K, Milioni D, Rigas S, Hatzopoulos P (2002) Combinatorial interaction of cis elements specifies the expression of the Arabidopsis AtHsp90-1 gene. Plant Physiol 129:1138–1149
Hong SW, Vierling E (2001) Hsp101 is necessary for heat tolerance but dispensable for development and germination in the absence of stress. Plant J 27:25–35
Ito Y, Katsura K, Maruyama K, Taji T, Kobayashi M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2006) Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol 47:141–153
Jagadish SVK, Muthurajan R, Oane R, Wheeler TR, Heuer S, Bennett J, Craufurd PQ (2010) Physiological and proteomic approaches to address heat tolerance during anthesis in rice (Oryza sativa L.). J Exp Bot 61:143–156
Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6:3901–3907
Katiyar-Agarwal S, Agarwal M, Gallie DR, Grover A (2001) Search for cellular functions of plant Hsp100/Clp family proteins. Crit Rev Plant Sci 20:277–295
Katiyar-Agarwal S, Agarwal M, Grover A (2003) Heat-tolerant basmati rice engineered by over-expression of hsp101. Plant Mol Biol 51:677–686
Kim B-H, Schoffl F (2002) Interactions between Arabidopsis heat shock transcription factor 1 and 70 kDa heat shock proteins. J Expt Bot 53:371–375
Kobayashi N, Horikoshi T, Katsuyama H, Handa T, Takayanagi K (1998) A simple and efficient DNA extraction method for plants, especially woody plants. Plant Tissue Culture Biotech 4:76–80
Kotak S, Port M, Ganguli A, Bicker F, von Koskull-Doring P (2004) Characterisation of C-terminal domains of Arabidopsis heat stress transcription factors (Hsfs) and identification of a new signature combination of plant class A Hsfs with AHA and NES motifs essential for activator function and intracellular localization. Plant J 39:98–112
Lee JH, Hübel A, Schöffl F (1995) Derepression of the activity of genetically engineered heat shock factor causes constitutive synthesis of heat shock proteins and increased thermotolerance in transgenic Arabidopsis. Plant J 4:603–612
Mathew A, Mathur SK, Jolly C, Fox SG, Kim S, Morimoto RI (2001) Stress-specific activation and repression of heat shock factors 1 and 2. Mol Cell Biol 21:7163–7171
Milioni D, Hatzopoulos P (1997) Genomic organization of hsp90 gene family in Arabidopsis. Plant Mol Biol 35:955–961
Miller JH (1972) Experiments in molecular biology. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Mittal D, Chakrabarti S, Sarkar A, Singh A, Grover A (2009) Heat shock factor gene family in rice: genomic organization and transcript expression profiling in response to high temperature, low temperature and oxidative stresses. Plant Physiol Biochem 47:785–795
Mittal D, Enoki Y, Lavania D, Singh A, Sakurai H, Grover A (2011) Binding affinities and interactions among different heat shock element types and heat shock factors in rice (Oryza sativa L.). FEBS J 278:3076–3085
Moriya H, Shimizu-Yoshida Y, Omori A, Iwashita S, Katoh M, Sakai A (2001) Yak1p, a DYRK family kinase, translocates to the nucleus and phosphorylates yeast Pop2p in response to a glucose signal. Genes Dev 15:1217–1228
Nigam N, Singh A, Sahi C, Chandramouli A, Grover A (2008) SUMO-conjugating enzyme (Sce) and FK506-binding protein (FKBP) encoding rice (Oryza sativa L.) genes: genome-wide analysis, expression studies and evidence for their involvement in abiotic stress response. Mol Genet Gen 279:317–383
Nishizawa-Yokoi A, Tainaka H, Yoshida E, Tamoi M, Yabuta Y, Shigeoka S (2010) The 26S Proteasome function and Hsp90 activity involved in the regulation of HsfA2 expression in response to oxidative stress. Plant Cell Physiol 51:486–496
Picard V, Ersdal-Badju E, Lu A, Bock SC (1994) A rapid and efficient one-tube PCR-based mutagenesis technique using Pfu DNA polymerase. Nucleic Acids Res 22:2587–2591
Port M, Tripp J, Zielinski D, Weber C, Heerklotz D, Winkelhaus S, Bubla KD, Scharf KD (2004) Role of Hsp17.4-CII as coregulator and cytoplasmic retention factor of tomato heat stress transcription factor HsfA2. Plant Physiol 135:1457–1470
Prandl R, Kloske E, Schoffl F (1995) Developmental regulation and tissue-specific differences of heat shock gene expression in transgenic tobacco and Arabidopsis plants. Plant Mol Biol 28:73–82
Pressman E, Shaked R, Firon N (2007) Tomato (Lycopersicon esculentum) response to heat stress: focus on pollen grains. Plant Stress 1:216–227
Rajan VBV, D'Silva P (2009) Arabidopsis thaliana J-class heat shock proteins: cellular stress sensors. Funct Intg Genome 9:433–446
Sarkar NK, Kim YK, Grover A (2009) Rice sHsp genes: genomic organization and expression profiling under stress and development. BMC Genomics 10:393
Singh A, Grover A (2010) Plant Hsp100/ClpB-like proteins: poorly-analyzed cousins of yeast ClpB machine. Plant Mol Biol 74:395–404
Singh A, Singh U, Mittal D, Grover A (2010) Genome-wide analysis of rice ClpB/HSP100, ClpC and ClpD genes. BMC Genomics 11:95
Singla SL, Pareek A, Kush AK, Grover A (1998) Distribution patterns of 104 kDa stress-associated protein in rice. Plant Mol Biol 37:911–919
Sugio A, Dreos R, Aparicio F, Maule AJ (2009) The cytosolic protein response as a subcomponent of the wider heat shock response in Arabidopsis. Plant Cell 21:642–654
Swindell WR, Heubner M, Weber AP (2007) Transcriptional profiling of Arabidopsis heat shock proteins and transcription factors reveals extensive overlap between heat and non-heat stress response pathways. BMC Genomics 8:125
Takahashi T, Naito S, Komeda Y (1992) Isolation and analysis of the expression of two genes for the 81-kilodalton heat-shock proteins from Arabidopsis. Plant Physiol 99:383–390
Trinklein ND, Murray JI, Hartman SJ, Botstein D, Myers RM (2004) The role of heat shock transcription factor 1 in the genome-wide regulation of the mammalian heat shock response. Mol Biol Cell 15:1254–1261
Walter M, Chaban C, Schutze K, Batistic O, Weckermann K, Nake C, Blazevic D, Grefen C, Schumacher K, Oecking C, Harter K, Kudla J (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40:428–438
Wu X, Shiroto Y, Kishitani S, Ito Y, Toriyama K (2009) Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. Plant Cell Rep 28:21–30
Yabe N, Takahashi T, Komeda Y (1994) Analysis of tissue-specific expression of Arabidopsis thaliana HSP90-family gene HSP81. Plant Cell Physiol 35:1207–1219
Yamada K, Nishimura M (2008) Cytosolic heat shock protein 90 regulates heat shock transcription factor in Arabidopsis thaliana. Plant Signal Behav 3:660–662
Yamada K, Fukao Y, Hayashi M, Fukazawa M, Suzuki I, Nishimura M (2007) Cytosolic HSP90 regulates the heat shock response that is responsible for heat acclimation in Arabidopsis thaliana. J Biol Chem 282:37794–37804
Yamamoto YY, Tsuji H, Obokata J (1995) 5′-leader of a photosystem I gene in Nicotiana sylvestris, psaDb, contains a translational enhancer. J Biol Chem 270:12466–12470
Young TE, Ling J, Geisler-Lee CJ, Tanguay RL, Caldwell C, Gallie DR (2001) Developmental and thermal regulation of the maize heat shock protein, HSP101. Plant Physiol 127:777–791
Acknowledgments
This work was supported by the Centre for Plant Molecular Biology, Department of Biotechnology and University Grants Commission, Government of India. We thank the Yeast Genetic Resource Centre, Japan, for giving us the strain A2279. BiFC vectors pUC-SPYCE and pUC-SPYNE were kindly provided by F. Schoffl and C. Oecking, University of Tubingen, Germany. We thank CIF at University of Delhi South Campus for confocal microscopy work. AS, DM, DL, MA, and RCM thank the Council of Scientific and Industrial Research, Government of India for their fellowship awards.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online Resource 1
Primers used in the current study (PDF 69 kb)
Online Resource 2
(PDF 96 kb)
Online Resource 3
(PDF 43 kb)
Online Resource 4
(PDF 225 kb)
Online Resource 5
(PDF 334 kb)
Online Resource 6
(PDF 142 kb)
Rights and permissions
About this article
Cite this article
Singh, A., Mittal, D., Lavania, D. et al. OsHsfA2c and OsHsfB4b are involved in the transcriptional regulation of cytoplasmic OsClpB (Hsp100) gene in rice (Oryza sativa L.). Cell Stress and Chaperones 17, 243–254 (2012). https://doi.org/10.1007/s12192-011-0303-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12192-011-0303-5