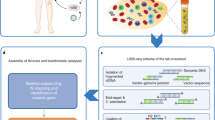
Abstract
Background
Host genome integration of HBV sequence is considered to be significant in HBV antigen expression and the development of hepatocellular carcinoma (HCC).
Method
We developed a probe-based capture strategy to enrich integrated HBV DNA for deep-sequencing analysis of integration sites in paired patient samples derived from tumor, liver tissue adjacent to tumor, saliva and plasma, as a platform for exploring the correlation, significance and utility of detecting integrations in these sample types.
Results
Most significantly, alpha fetoprotein levels significantly correlated to the amounts of integrations detected in tumor. Viral-host chimeric DNA fragments were successfully detected at high sequencing coverage in plasma rather than saliva samples from HCC patients, and each fragment of this type was only seen once in plasma from chronic hepatitis B patients. Almost all plasma chimeric fragments were derived from integrations in tumor rather than in adjacent liver tissues. Over 50% of them may produce viral-host chimeric transcripts according to deep RNA sequencing in paired tissue samples. Particularly, in patients with low HBV DNA level (< 250 UI/ml), the seemingly normal HBsAg titers may be explained by larger amounts of integrations detected. Meanwhile, we developed a strategy to predict integrants by pairing breakpoints for each integration event. Among four resolved viral patterns, the majority of Pattern I events (81.2%) retained the complete opening reading frame for HBV surface proteins.
Conclusion
We achieve the efficient enrichment of plasma cell-free chimeric DNA from integration site, and demonstrate that chimeric DNA profiling in plasma is a promising noninvasive approach to monitor HBV integration in liver cancer development and to determine the ability of integrated sequences to express viral proteins that can be targeted, e.g. by immunotherapies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
Hepatitis B virus (HBV) integration has been known to occur in hepatocellular carcinoma (HCC) as well as chronic liver infection [1, 2]. Double-stranded linear DNA (dslDNA) seems to be preferred DNA substrate for integration [2]. Recently, the resequencing of HCC genomes has identified hundreds of integration sites in the human genome implicating many cellular genes [3,4,5]. HBV integrations were more likely to occur in chromosome sites of genomic instability such as long interspersed nuclear elements (LINEs), short interspersed nuclear elements (SINEs), microsatellites and telomeres [6, 7], and further studies also showed integration sites tend to occur within boundaries of the altered copy numbers of a gene [8, 9]. Viral integrations may occur early during HBV infection and will constantly happen during the long infection history, and their patterns have already been applied to detect cell clonal expansion in both tumor and non-tumor tissues [10, 11]. Chimeric DNA fragments from integration sites released by tumor cells account for a proportion of plasma cell-free DNA (cfDNA), could serve as a biomarker, and reflect a part of the genetic changes in tumor genomes [12].
Theoretically, the analysis of the plasma cfDNA pool is not only more accessible but also more representative of the entire liver than single liver biopsies. In addition, it can theoretically be repeated at each patient visit. The scarcity of chimeric virus-host DNA in cfDNA requires deep sequencing depth similar to circulating tumor DNA detection, which currently limits the wide-scale implementation in a clinical scenario [13]. Hence, we aimed at enriching viral-host fragments using capture probes prior to deep sequencing to reduce the sequencing volume and increase the sensitivity to detect HBV integration events. Although it has been shown that chimeric DNAs reflect tumor recurrence after surgery [14], no efforts have been made to examine if integrants originating from tumor lesions may be detected once HCC occurred or to identify integrations in plasma from chronic hepatitis B patients.
Recently, it has been shown that integrated HBV DNA, containing an intact open reading frame (ORF) of envelope proteins, can serve as an additional template for producing hepatitis B surface antigen (HBsAg) and influences antiviral treatment responses and disease prognosis [15]. Furthermore, a study has demonstrated, that liver cells harboring integrated HBV DNA sequences can express peptides that can be recognized by HBsAg-specific T cells [16]. Nevertheless, almost all previous studies adopting short read sequencing have only taken each breakpoint as single integration event and were unable to predict the viral integrants, which would require that the two ends have been identified. Thus, we intended to evaluate a novel prediction strategy in identifying those integrants with HBsAg production ability. It may facilitate novel immunotherapies targeting viral proteins as neoantigens, and monitor liver disease progression.
In this study, we designed viral DNA probes covering the whole HBV genome so that we could enrich plasma cfDNAs for HBV-containing sequences for deep sequencing. The small size of the HBV genome enabled us to achieve a deep sequencing coverage at a small sequencing volume, with a significantly increased ability to detect viral integrants. Using this technique, we analyzed tumor and adjacent non-tumor liver samples alongside cfDNA derived from either plasma or saliva sampled from the same patient. We found this to be an efficient strategy to characterize viral integration events stemming from tumors with a potential use to characterize liver cancers in patients with HBV infection in a noninvasive fashion. In addition, we use the short read sequencing data to predict the orientation and length of integration by pairing the two ends of each integration event.
Methods
Patients and samples
The present study was conducted in You’an Hospital (Beijing, China). A total of 42 patients were enrolled in the stages of sample collection (Fig. 1a). Among these patients, 27 patients had HCC, 5 patients had bile duct carcinoma (BDC, all HBV positive), and 10 patients had chronic hepatitis B. Blood samples from HCC and BDC patients were collected before surgery, and the corresponding liver tissues were obtained afterwards. A total of four samples, which included two tumor sites and two adjacent non-tumors, were used for the analysis. Chronic hepatitis B patients only provided blood samples for analysis. The diagnosis was made according to the guidelines for the prevention and treatment of chronic hepatitis B: a 2015 update [17]. The BCLC staging criteria were used to classify HCC patients. The laboratory findings are summarized in Tables S1 and Table S2. The study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki and was approved by the Ethics Committee of You’an Hospital. An informed consent was obtained from all patients. In Stage II of sample collection (Fig. 1a), 16 HBV-related cancer patients (11 HCC and 5 bile duct carcinoma, BDC, Tables S2-S3). Besides, four patients without either HBV infection or HBV DNA detected, including two HCV-related HCC patients, one HCC and one BDC, served as negative controls, and status for other markers, see Table S1 (I008, N001, N002, and N003). DNA sample of HepG2.2.15 cell line was provided by Beijing Tricision BioTherapeutics Inc, and cell line authentication was examined by Guardian Technology Co. Ltd. using short tandem repeat loci. We performed three capture experiments (replicate 1–3) using HepG2.2.15 DNA samples following the below procedure, and obtained 1G, 1.5G, and 2G raw sequencing data, respectively (Table S3). Sample collection and DNA extraction, see Supplementary Methods for details.
Study design. a Sample collection in five stages. HCC, hepatocellular carcinoma; BDC, bile duct carcinoma; negative, three patients with liver cancer but without HBV infection. b DNA amounts for sequencing library construction in tissue and plasma (left). Integrations lead to two host breakpoints and two viral breakpoints in the human genome and HBV genome, respectively. Two host breakpoints are located at upstream and downstream of the integrated viral fragment. Most of integration sites have deletions in human genome, leading two 35- < 4500 base pairs in distance between two host breakpoints (Pattern I). In some cases (Pattern II), sequences of both breakpoints are consistent. c Experiment work flow for the capture assay. d Sequencing volumes for captured fragments (left), and junction read mapping to the reference region of integration sites. Human fragments in the virus-host junction reads can be mapped to either the upstream or downstream of the breakpoints. Theoretically, each integration event should be supported by these four types of junction read pairs with adequate read depth at both upstream and downstream breakpoints. In pair-end sequencing, at most, one read in a read pair would represent the junction read covering the integration boundary. The other read would either be a host fragment (read pairs with a dashed line in blue) or a viral fragment (read pairs with a dashed line in light green), and the read alignment shows the mapping of these two groups of read pairs in the corresponding color
Viral capture experiment: design and bioinformatic analysis
Probe design and experiment, Sequencing experiments and Integration calling, and Breakpoint annotation and visualization, see Supplementary Methods for details.
DNA microarray experiment for structure variation analysis
The genome-wide genotyping of tumor tissues was performed using the HumanCoreExome-24 BeadChip (Illumina Inc.), which was scanned by the iScan Reader (Illumina Inc). The LogR ratio (LRR) and B allele frequency (BAF) of each genotyped locus were extracted by Illumina GenomeStudio 2011. Variations in copy numbers by each sample were determined by pennCNV [18].
Statistical analysis
Statistical analysis, including Chi-square test, t test, linear regression, and Pearson correlation, was performed using R packages (https://www.r-project.org/).
Results
Landscape of HBV integration in cancer and adjacent non-cancer liver tissues
To enrich for viral integrants in cfDNA isolated from plasma, we designed DNA probes specific for HBV DNA sequences (Method). In probe validation in HepG2.2.15 cells known to harbor integrated copies of HBV (Stage I, Fig. 1a), we identified sequencing five integration sites in three replicates (Table S3). The virus-cell DNA junctions in the integrant fragments consisted of both the viral genome ends (viral breakpoints, Fig. 1b) and cellular genome ends (host breakpoints, Fig. 1b). We then applied the method to 80 biopsy samples collected from 20 liver cancer patients (study design in Fig. 1a), and identified 424 integration events (Fig. 2a). The number of detected integration events detected per sample varied from 2 to 82 (average: 26) among the 16 HBsAg-positive patients. Nevertheless, there was no significant difference in integration events between the tumor sites and paired adjacent non-tumor sites of the same individuals (t test, p > 0.05, Figures S1-S2). In four HBsAg-negative patients as negative control, three (N001, N002, and N003) had none integrations detected. I008 with HCV infection, negative for all viral markers and HBV DNA, had three integration at extreme low coverage (< 10 sequencing depth) identified. Additionally, the detection of viral integrations in five patients with bile duct carcinoma may indicate contribution of hepatocytes in their tumor origins.
a All integration events connecting the viral and human genomes. Each light blue line indicates one integration event, with one end showing the breakpoint in the HBV genome and the other in human chromosomes [23]. Each integration event was only observed in one patient. Bubbles with diverse diameters, between chromosomes and central connections, illustrate the sequencing read depth in multiple samples from each patient. T two samples from tumor tissues; N adjacent non-HCC tissues. The hotspot for viral breakpoints locates at approximately nt 1600–1900. b Integration events with the same sequence features at the disrupted human genome regions and integration burden in all patients. The top panel shows the diagnosis of liver cancer, the age of the patient and the Barcelona clinic liver cancer (BCLC) stages. The middle panel provides the number of integration events per MB according the total events observed in all four solid tissue samples obtained from each individual. At the bottom panel, patients were listed by frequencies (left part) of the 26 types of integrations observed in > 2 patients (right part). c Comparison of sequencing read depths of integrations in tumors and adjacent liver tissues [23]. The values for the sequencing depths were log transformed. Each dot indicates the sequencing read depth of an integration in tumor (y axis) and non-HCC liver (x axis). A higher read depth in two sites of each sample was used for plotting. Diverse colors indicate the different patients. d The amounts of integrations detected in all tumor samples significantly associate with the amounts detected in all paired adjacent liver tissues (adj_liver). The amount value is normalized by sequencing volume in corresponding samples (per GB). AFP levels relate to amounts of detected integrations in tumor samples (e), rather than in paired adjacent liver samples (f)
Genomic annotation of host breakpoints specific to tumor or non-tumor tissues revealed no significant differences in cellular genome locations of the integration sites in these two types of tissues, which can be either between genic and intergenic regions (Chi-square test, p = 0.9; Table S4), or between repeated and non-repeated regions (Chi-square test, p = 0.09). The most commonly directly interrupted gene was FN1, which was detected in 8 out of 17 patients; while the most common directly interrupted repeat sequence was ALR/Alpha, which was also found in 8 patients (Fig. 2b). Furthermore, integrations in the telomeres of chromosomes, characterized by the repeat sequence of (TTAGGG) n, were also very common (23.5%, 4/17). We also observed 29.4% (5/17) patients had integration sites in the promoter region of TERT, which was consistent with a previous finding (23.7%, 18/76) [3]. Besides, GO analysis for genes directly interrupted by integrations or locating within 20 K around integration sites did not identify significantly enriched GO items (Figure S2), indicating the random nature of viral integrations.
Most patients were infected by genotype type C (88%, 15/17) (Table S3), and about 71% (12/17) had single tumor lesions detected and had multiple lesions or diffuse liver tumor. We compared the amounts of integrations detected in different groups of patients, including genders, e antibody status, presence of multiple lesions, antiviral therapies, and BCLC stages A and B. We also examined the correlation between the number of detected integration events and alpha fetoprotein (AFP), HBV DNA level, or HBsAg titers (Figure S1). We observed a correlation between the quantity of integrations, the AFP levels (Pearson correlation coefficient = 0.641, p value = 0.01, Figure S1). Although the amount of integrations detected in adjacent liver tissues did correlate with those found in tumor samples (Pearson correlation coefficient = 0.683, p-value = 0.0025, Fig. 2d), AFP levels were better correlated with integrations for tumor tissue (Pearson correlation coefficient = 0.767, p value = 0.00085, Fig. 2e) compared to adjacent liver tissues (R = 0.498, p value = 0.059, Fig. 2f).
Our capture strategy achieved high sequencing coverage (number of junction reads) of integration breakpoints, with 70% (296/424) of integrations having sequencing coverage over 100 in at least 1 of 4 aliquots analyzed in the same individual. The sequencing coverage of a breakpoint could indicate the abundance of the integrant, reflecting the size of the clone carrying the corresponding integration. Integration events in tumor samples had sequencing coverage around tenfold higher than that in adjacent non-tumor samples (Figs. 2c and 3b), indicating significant expansion of tumor clones. In conclusion, our assay can be applied to efficiently capture and characterize integration events.
Detection of integration events in saliva and plasma obtained from HCC patients. a The integration event in saliva. Only one non-redundant junction read is obtained in comparison to high sequencing coverage in paired plasma sample. b The cfDNA was successfully extracted in seven of eight patients, and viral integration events were detected in all seven plasma samples. More integration events were identified in paired liver tissues. The limited integration events were shared between tumors and paired non-HCC liver samples (green). In particular, the detected integrations in plasma well reflected the counterparts uncovered in tumor tissues (orange and purple), and the integration events specific to paired non-HCC liver tissues (blue) were not observed in the corresponding plasma samples. c The read depths of integration events in tumors, non-tumor liver tissues and plasma samples. For those detected in all three tissues (T.N.P.). d HBsAg titers associated with the amount of integrations in patients with low HBV DNA level
Capturing the HBV integrations in body fluids
To examine the suitability of saliva, a recently adopted liquid biopsy solution for other cancers [19], we collected the saliva samples along with plasma samples for seven liver cancer patients (Stage III, Fig. 1a; Table S1). In total, 32 integration events were successfully detected in 5 patients (Table S5). Only one patient had integration detected in saliva (Table S5). Although she had 5 integrants with high abundance (250–881 junction read pairs) in plasma (Table S5), only 1 integrant (279 junction read pairs in plasma) was seen in saliva supported by 1 non-redundant junction (Fig. 3a). Therefore, only 1 of 32 (3%) integrations were detected in the saliva samples suggesting that saliva is not as informative or accurate as plasma samples and thus not a suitable sample for detection of viral integration in liver cancer, possibly due to the distance between the liver and the oral cavity—a parameter previously reported as important in the detection of tumor DNA-based biomarkers in head and neck squamous cell carcinomas [20].
To trace the origin of integrants in plasma cfDNA, we collected paired tumor and adjacent normal tissues from HCC patients (Stage IV, Fig. 1a; Clinical information, Table S1). The cfDNA in all seven HBV-positive HCC patients was analyzed (one plasma sample failed in the cfDNA extraction) (Table S2). Overall, the plasma integration events predominantly reflected the tumor tissues. First of all, we detected 29 integration events from 7 plasma cfDNAs (MaxCF > 0, Table S6) and all of them (29/29, 100%) could be detected in the corresponding liver tumor samples (Fig. 3b, Table S6). Notably, junction abundance for integration in plasma samples correlated with that in tumor samples (R2 = 0.64, p = 6.2 × 10–29, Table S6; Fig. 3c), but not in the paired non-tumor liver tissues (R2 = 0.32, p = 6.2 × 10–12). This also supported cell-free junctional DNA was shed by the tumor tissue compared to the non-tumor tissue.
Deep RNA sequencing for the same tumor and adjacent liver aliquots were performed for four patients (c001, c002, c003, c005 in Table S6). Totally, chimeric RNA transcripts were observed for 17 integration sites, among which 13 were also seen in plasma cfDNA and the rest 4 were only observed in the transcriptome data. As a result, among 24 integrants in cfDNAs from these 4 patients, 54% (13/24) had transcription activity. Besides, the amounts of chimeric transcripts from integration sites showed a better association with DNA junction abundance in tumor tissues (R2 = 0.27, p = 1.5 × 10–8, Table S6), than that in adjacent liver tissues (R2 = 0.01, p = 0.15). It suggested integration events observed in cfDNAs were more likely to have transcription activity in tumor than in adjacent liver tissues, and they would contribute to viral antigen production if they kept intact ORFs of these proteins.
A lot of factors may hinder the attempt to dissect the correlation between HBsAg titers and integrations, one of the major factors is the contribution to viral antigen production by HBV cccDNA. To minimize its influence, we examined 11 patients having integrations detected along with low HBV level (< 250 IU/ml), including I001, I003, I007, I008, I012, I014, I016, s003, s004, c001, and c002 (Table S1 and S2) in Stage II–IV of this study (Fig. 1). In this group of patients, HBsAg titers reached the level observed in those patients with relative high HBV DNA level, and over 50% (6/11) had a titer of more than 5000 IU per ml. Their HBsAg levels were significantly positively correlated to the amount of integrations detected in their samples (Pearson correlation coefficient = 0.718, p-value = 0.0128, Fig. 3d), suggesting their serological HBsAg may be attributed to viral integrations.
Taken together, we find plasma integration profile is valuable for the detection of tumor-derived integration events, and RNA sequencing data not only confirmed that some of these integrations are transcriptionally active but information about tumor expression could be derived from the plasma. Then, we also sought HBV integrations in the plasma cfDNAs of ten chronic hepatitis B patients without liver cancer (Stage V, Fig. 1a; Clinical information, Table S2). No events met the integration criteria in all these patients (“Methods” section). We did observe a lot of single-junction reads indicating the existence of integration events, and breakpoint distribution of them was consistent with that of breakpoints found in tumor and liver tissues (Figure S3). However, their authenticity should be supported by analyzing paired tissue samples from liver biopsy. The scarcity of DNA fragments derived from chronically infected liver was likely due to a combination of limited turnover of infected cells and clonality. It would be interesting to evaluate the cfDNA from patients where there is an active HBsAg-specific T-cell response where selective killing could increase the overall number of HBV integrations detected.
HBV integrant prediction: sequence boundary of a single integration in human genome
To accurately infer the HBV integration, sequence and orientation will be valuable for assessing the viral protein/peptide potential of integrations. Each integration should have two viral breakpoints and two host breakpoints (Fig. 3a). As a premise, we assumed that two independent integration events in host genome should be far away from each other (Method). Therefore, we applied a rule that two cellular breakpoints that occurred within 20 K base pairs (bp) were a single integration event (Fig. 4). Among the total 424 integrations observed in paired tumor and adjacent non-tumor tissues, we were able to map 218 of these accurately at each end of the integrated sequence. The genomic distance between breakpoints ranged mostly from 0 to 50 bp (87%, 189/218; Fig. 4a, b Figure S4). The probability of breakpoints belonging to two independent integrations located within such a small region under current integration rate was extremely small (p50bp = 5 × 10–7, the calculation see Supplementary Method). The frequency of large deletions (> 1000 bp, 5%, 11/218; Fig. 4c) and redundant human DNA fragments (13%, 30/218; Fig. 4d) were relatively low. Notably, 7 of the 11 integrations in the intronic region of FN1 gene had repeated sequences at the breakpoints.
Patterns of integration events according to the read alignment features of host breakpoints in the human genome. At the integration sites of the human genome, there can be lost sequences with diverse length (d, the position of the downstream breakpoint minus the upstream one). There can also be no loss of host sequences in the integration sites, and the d should be 1 bp (a). Deletions with a diverse length can also be observed at the integration site of the human genome. For instance, a 11 bp deletion (b) and a 2012 bp deletion (c). For host pattern II, the longest redundant sequence is 35 bp (d). For these integrations, the top panel shows the transcripts of genes and repeat sequences near the corresponding regions in the human genome according to the online UCSC genome browser (hg19). The shading of repeat elements reflects the amount of sequence variations associated with the repeat element. The darker it is, the fewer such variations are observed
Obviously, genomic structure variation may influence the pairing analysis of the remaining 206 integrations (Fig. 5a). To test this hypothesis, the whole genome genotyping of tumor tissues from four HCC patients was performed. Among the 19 unpaired breakpoints identified in these patients, 9 breakpoints were located at the telomere or centromere regions, while 10 host breakpoints were located at the boundary of large structure variations (SV in Table S6 and Figures S5A-S5H). Particularly, two sites in chromosome 9 were separated by 1.7 M bp, and each was located at one end of a same-length deletion region in the human genome (Fig. 5b). Thus, the alterations in tumor genomes and the inaccurate mapping of junction reads in repetitive sequences were the two major reasons for the inability to pair some host breakpoints.
a Chromosome locations for all integration events. Events with identified paired boundaries (PB) are presented in green, while single boundaries (SB) are presented in grey. b The influence of structure variations on the boundary or breakpoint pairing analysis. An example was provided to show that two breakpoints at chromosome 9, which were separated by 1.7 M bp, were located exactly at the two boundaries of a same-length genomic deletion
HBV integrant prediction: four patterns of integrated viral fragments
After pairing the host breakpoints for individual integration, we obtained the corresponding sequences covering viral boundary of this integration event. Then, accurate mapping of the integrated viral sequence could be achieved (Fig. 6). Among 218 integration events with known host breakpoints, 215 integrated sequences could be characterized including the orientation of the viral sequence. Four distinct viral sequence patterns were observed (Fig. 6a, b). Their viral breakpoints were relatively consistent, showing similar distributions between the integration patterns. The majority of integrated sequences consisted of nt 1600–1900 of the viral genome (64.2%, 138/215) (Fig. 6c, d). This region included the cohesive ends of DR1 and DR2, which also are features of dslDNA ends. Almost all Pattern I integrations had viral ends consistent with the ends of the dslDNA and the viral segments in this group were shorter than the full-length HBV genome, ranging from 952 to 3214 bp (Fig. 6e). Interestingly, viral breakpoints in viral pattern II were located more commonly between nt 1–1000 than those in viral pattern I (p = 3.1 × 10–7, t test), and most of them had a length under 1000 bp (Fig. 6f). The viral segments in viral pattern II (21.4%, 46/215) were shorter than pattern I, ranging from 32 to 1584 bp. Viral integrants in viral pattern III (10.2%, 22/215) and IV (4.2%, 9/215) appeared to be formed by ligation of the ends of least two viral fragments in a 3′-to-3′ or 5′-to-5′ manner. In addition, most individual samples contained all four patterns (Figure S6). We acknowledged here that the method infers patterns by assembling sequence data from multiple 150 bp reads. Clearly, a direct verification would require a sequence of long DNA segments without fragmentation.
Patterns of integration events according to the features of paired breakpoints in the HBV genome (a). The mapping of viral fragments in virus-human junctions to the HBV genome is shown. The reads mapped to the plus strand are in red, and those to the minus strand are in dark red. The read depth shows the sequencing coverage of each base along the viral genome, and the read alignment demonstrates the mapping of all reads to the genome [23]. The arrows illustrate the extending direction according to the tail of the peak at the breakpoints. The integrants were predicted according to the directions at two boundaries of the integration. All four combinations of two directions at breakpoints, and their schematic diagram are illustrated in (b). The red fans summarize the features of the junction read mapping at the breakpoints of both 5′ and 3′ ends. The curved edge indicates the inconsistent ending of the reads, and the vertical edge indicates the consistent boundary. The yellow parts of the circle indicate the estimated integrants. Viral Patterns III and IV seem to have multiple viral fragments firstly ligated in different ways before integrating into the host genome. c The breakpoint distribution across the HBV genome is shown. The distributions of the four viral patterns (I–IV) and the unpaired breakpoints (NA) in different colors, as well as the hotspot for viral breakpoints located around the DR1-DR2 region, are shown [23]. d All inferred integrants for viral pattern I and II are shown using the dslDNA format of the HBV genome as a reference sequence. e The length distribution of viral integrates in pattern I and pattern II
Furthermore, we observed chimeric RNA transcripts from all four viral patterns of integration sites in tissue samples with both DNA capture experiments and deep RNA sequencing. In all, 76 integrations were identified in the DNA capture experiments (Table S6). Among them, viral patterns of breakpoints were determined in 42 integrants, and each pattern had 19% integrations with transcription activity (Table S6). Therefore, there were no obvious differences in transcription activity among these four patterns of integration events.
Discussion
DNA capture increases the sensitivity to detect integration and reduces the interference from non-integrated HBV DNA in samples. In our experiments, we have shown that the concentration of adequate probes is a critical parameter to optimize capture which has not been discussed so far. To this end, we determined that applying 200 ng of probes for each sample to ensure that all viral fragments would be captured. Theoretically, a 200-ng probe scan can capture at least 1011 target molecules. The DNA extracted from each liver tissue consisted of 105 cells that resulted in 600 ng of double-stranded DNA. Unlike circulating tumor DNA which is confounded by the DNA released from blood cells, detection of virus-host DNA junction may be influenced by both integrated and non-integrated HBV DNA. Each HBV-infected cell can contain up to 1000 copies of replicative intermediates, resulting in approximately 108 copies of non-integrated HBV DNA, which can only consume a maximal of 1/1000 of input probes. Thus, there were sufficient probes to capture all HBV integration events.
Although this assay was not designed to perform a quantitative analysis on integrations, the same number of viral probes and a relatively equal amount of input DNA enabled the comparison of the relative abundance of individual as well as total integration events among all analyzed samples. The same, unique HBV integration site carried by many liver tumor cells was uncovered by a higher number of sequencing reads, supporting the idea that the relative abundance of HBV integrations can serve as a genetic marker for clonal selection and the expansion of affected hepatocytes [11]. Notably, the highest sequencing read depth for the cellular sequence breakpoints from liver tissues reached 11,579 in the present study. Hence, we assume that the potential for identification of the majority of integration events in a given sample was adequately addressed in the present study. However, we cannot rule out the possibility that more tissue aliquots, more probes or ultra-deep sequencing may identify additional integrations at low frequencies or increase the read depth for breakpoints.
The detection of new integration events or changes in integration events over time may help to monitor disease progression in the liver and may have the potential to indicate metastases or secondary tumors both of which are hard to detect in particular in a cirrhotic liver. Theoretically, cfDNA in blood HBV-containing HBV-host DNA junctions should mirror integration events in the liver, since it must have been released from the liver as HBV only infects hepatocytes and only integrates into hepatocytes. The present data revealed that the HBV integration events detected in plasma cfDNA predominantly originated from liver cancer cells. In the cancer and paired non-HCC liver tissues from the patients at Stage II (Fig. 1a), we observed that the integration events had much higher sequencing read numbers in non-tumor liver tissues, indicating that some non-tumor clones with integrations might have already significantly expanded before an HCC grew out (Fig. 2c). Nevertheless, it was regrettable that corresponding plasma samples were not available to compare the abundance of HBV DNA fragments in the circulation with that in liver tissue. However, investigation of the patients enrolled in Stage IV revealed that HBV integration events identified in HCC but not in those detected in non-tumor liver tissues had correspondent counter-part fragments detected in plasma (Fig. 3b). Circulating cfDNA in plasma is most likely released from dying or circulating tumor cells and less likely from injured adjacent tissue. Thus, one would expect that less DNA is released from non-HCC liver tissue compared to liver tumor tissue.
According to the paired viral breakpoints we detected, the sequences of the integrated viral segments for viral pattern I and II were assembled (Fig. 6d). It was considered that a majority of viral pattern I events (81.2%, 112/138) preserved the ORF of the large surface protein, and 14 of the remaining 26 events had an intact ORF of the middle surface protein. All these integrations would of course also encode for the small envelop protein S and have the potential to secrete HBsAg. These observations support the suggestion that integrated HBV DNA provides significant additional capacity for HBsAg production besides HBV cccDNA, and represents a challenge to reduce HBsAg production [15, 21]. Particularly, our patients with low HBV DNA levels (< 250 IU/ml) had HBsAg levels significantly positively correlated to the amount of integrations detected (Fig. 3d), suggesting the contribution of viral integrations to serological HBsAg. Recently, Tan et al. demonstrated the first attempt to target integrated viral proteins as neoantigens in immune therapies for liver cancer [16], and our study explored the feasibility of a noninvasive method to identify the candidate integrations for this kind of therapy, which will be a promising companion diagnostic test in the future.
Conclusion
The present data provide evidence that the integration patterns varied among individual patients. Diverse patterns and different percentages of different integration sites in the same individual imply distinct HBV antigen expression patterns, which are expected to impact therapeutic responses to HBV treatment or efforts targeting tumor cells expressing viral proteins. The HBV integration detected in the plasma cfDNA pool may potentially become a new plasma biomarker that could complement present biomarkers to monitor HBV related to liver disease stage, including liver tumor occurrence. However, future prospective studies with a larger sample size are required to validate our findings.
Data availability
The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive in BIG Data Center [22], Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under accession number CRA000925 that is publicly accessible at https://bigd.big.ac.cn/gsa.
Abbreviations
- HBV:
-
Hepatitis B virus
- HCC:
-
Hepatocellular carcinoma
- ORF:
-
Open reading frame
- HBsAg:
-
Hepatitis B surface antigen
- dslDNA:
-
Double-stranded linear DNA
- LINEs:
-
Long interspersed nuclear elements
- SINEs:
-
Short interspersed nuclear elements
- cccDNA:
-
Closed circular DNA
- cfDNA:
-
Cell-free DNA
- BDC:
-
Bile duct carcinoma
- SV:
-
Structure variations
- AFP:
-
Alpha fetoprotein
References
Koshy R, Koch S, Von Loringhoven AF, et al. Integration of hepatitis B virus DNA: evidence for integration in the single-stranded gap. Cell 1983;34:215–223
Yang W, Summers J. Integration of hepadnavirus DNA in infected liver: evidence for a linear precursor. J Virol 1999;73:9710–9717
Sung W-K, Zheng H, Li S, et al. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat Genet 2012;44:765–769
Fujimoto A, Totoki Y, Abe T, et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet 2012;44:760–764
Zhao L-H, Liu X, Yan H-X, et al. Genomic and oncogenic preference of HBV integration in hepatocellular carcinoma. Nat Commun 2016;7:12992
Tu T, Budzinska MA, Shackel NA, et al. HBV DNA integration: molecular mechanisms and clinical implications. Viruses 2017;9(4):75
Heikenwalder M, Protzer U. LINE(1)s of evidence in HBV-driven liver cancer. Cell Host Microbe 2014;15:249–250
Meyer M, Wiedorn KH, Hofschneider PH, et al. A chromosome 17: 7 translocation is associated with a hepatitis B virus DNA integration in human hepatocellular carcinoma DNA. Hepatology 1992;15:665–671
Tokino T, Matsubara K. Chromosomal sites for hepatitis B virus integration in human hepatocellular carcinoma. J Virol 1991;65:6761–6764
Dandri M, Locarnini S. New insight in the pathobiology of hepatitis B virus infection. Gut 2012;61(Suppl 1):i6–i17
Mason WS, Gill US, Litwin S, et al. HBV DNA integration and clonal hepatocyte expansion in chronic hepatitis B patients considered immune tolerant. Gastroenterology 2016;151:986–998 e4
Zhao Y, Xue F, Sun J, et al. Genome-wide methylation profiling of the different stages of hepatitis B virus-related hepatocellular carcinoma development in plasma cell-free DNA reveals potential biomarkers for early detection and high-risk monitoring of hepatocellular carcinoma. Clin Epigenetics 2014;6:30
Guo S, Diep D, Plongthongkum N, et al. Identification of methylation haplotype blocks aids in deconvolution of heterogeneous tissue samples and tumor tissue-of-origin mapping from plasma DNA. Nat Genet 2017;49:635–642
Wang YC, Li CL, Ho MC, et al. Cell-free junctional DNA fragment from hepatitis B virus integration in HCC for monitoring postresection recurrence and clonality. J Clin Oncol 2019;37(15_suppl):4090–4090
Wooddell CI, Yuen MF, Chan HL, et al. RNAi-based treatment of chronically infected patients and chimpanzees reveals that integrated hepatitis B virus DNA is a source of HBsAg. Sci Transl Med 2017;9(409):eaan0241
Tan AT, Yang N, Lee Krishnamoorthy T, et al. Use of expression profiles of HBV-DNA integrated into genomes of hepatocellular carcinoma cells to select T cells for immunotherapy. Gastroenterology 2019;156:1862–1876 e9
Hou J, Wang G, Wang F, et al. Guideline of prevention and treatment for chronic hepatitis B (2015 update). J Clin Transl Hepatol 2017;5:297–318
Wang K, Li M, Hadley D, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res 2007;17:1665–1744
Siravegna G, Marsoni S, Siena S, et al. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol 2017;14:531–548
Wang Y, Springer S, Mulvey CL, et al. Detection of somatic mutations and HPV in the saliva and plasma of patients with head and neck squamous cell carcinomas. Sci Transl Med 2015;7:293ra104
Hu B, Wang R, Fu J, et al. Integration of hepatitis B virus S gene impacts on hepatitis B surface antigen levels in patients with antiviral therapy. J Gastroenterol Hepatol 2018;33:1389–1396
Wang Y, Song F, Zhu J, et al. GSA: genome sequence archive. Genomics Proteomics Bioinform 2017;15:14–18
Zhang D, Chen W, Zhang K, et al. Viral integration profiles in the plasma cell-free DNA from patients with HBV infection well represent tumor clone compositions during HCC development. J Hepatol 2018;68:S121–S122
Acknowledgements
This manuscript has been edited and proofread by Medjaden Bioscience Limited.
Funding
This project was partially supported by the Innovation Promotion Association CAS (2016098), the National Natural Science Foundation of China (81201700), the Major State Basic Research Development Program (2014CB542006), the Key Research Program of the Chinese Academy of Sciences (KJZD-EW-L14), National Science and Technology Support Program (2012BAI11B05), Beijing Natural Science Foundation (7192158), the Fundamental Research Funds for the Central Universities (3332018032) and the Capital's Funds for Health Improvement and Research (2018-1-1151). The sponsor or funding organization had no role in the design or conduction of this research.
Author information
Authors and Affiliations
Contributions
The concept and study design was provided by D.Z., K.Z, W.C., C.Z. and U.P.; patient enrollment, the sample and clinic data collection were performed by P.D., Z.W. X.Y. and H.D.; the sequencing experiments were performed by C.T., and D.Z.; the data analysis and interpretation were performed by W.C., D.Z., K.Z. P.D., Y.H., S.G, H.Z., U.P. and G.F.; the manuscript preparation was performed by: D.Z., K.Z., G.F. and U.P.; overall responsibility was given to D.Z., W.C., K.Z, P.D., U.P. and C.Z
Corresponding authors
Ethics declarations
Conflict of interest
Dr. Dake Zhang has a patent pending for the probe-based HBV DNA capture in plasma as a liquid biopsy to monitor HCC development. The authors declare no other potential conflicts of interests.
Ethics approval
The study was approved by the Institutional Review Board at Beijing institute of Genomics (IRB number: 2016H005). The study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki and was approved by the Ethics Committee of You’an Hospital.
Informed consent
An informed consent was obtained from all patients.
Consent for publication
All authors reviewed and approved the final version of the manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chen, W., Zhang, K., Dong, P. et al. Noninvasive chimeric DNA profiling identifies tumor-originated HBV integrants contributing to viral antigen expression in liver cancer. Hepatol Int 14, 326–337 (2020). https://doi.org/10.1007/s12072-020-10016-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12072-020-10016-2