Abstract
The aim of the study was to understand the role of SLIT2–ROBO1/2–CDC42 signalling pathways in development of breast cancer (BC). Primary BC samples (n=150), comprising of almost equal proportion of four subtypes were tested for molecular alterations of SLIT2, ROBO1, ROBO2 and CDC42, the key regulator genes of this pathway. Deletion and methylation frequencies of the candidate genes were seen in the following order: deletion, SLIT2 (38.6%) >ROBO1 (30%) >ROBO2 (7.3%); methylation, SLIT2 (63.3%) >ROBO1 (26.6%) >ROBO2 (9.3%). Majority (80%, 120/150) of the tumours showed alterations (deletion/methylation) in at least one of the candidate genes. Overall, alterations of the candidate genes were as follows: SLIT2, 75.3% (101/150); ROBO1, 45.3% (68/150); ROBO2, 15.3% (23/150). Significantly, higher alteration of SLIT2 locus was observed in triple negative breast cancer (TNBC) over HER2 subtype (P=0.0014). Similar trend is also seen in overall alterations of SLIT2 and/or ROBO1, in TNBC than HER2 subtype (P=0.0012); of SLIT2 and/or ROBO2 in TNBC than luminal A (P=0.014) and HER2 subtype (P=0.048). Immunohistochemical analysis of SLIT2, ROBO1/2 showed reduced expression, concordant with their molecular alterations. Also, high expression of total CDC42 (49/52; 94.2%) and reduced expression of phospho Serine-71 CDC42 (41/52; 78.8%) was observed. Coalterations of SLIT2 and/or ROBO1, SLIT2 and/or ROBO2 had significant association with reduced expression of phospho Serine-71 CDC42 (P=0.0012–0.0038). Alterations of SLIT2 and/or ROBO1, reduced expression of phospho Serine-71 CDC42 predicted poor survival of BC patients. Results indicate the importance of SLIT2–ROBO1–CDC42 signalling pathway in predicting tumour progression.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer (BC) is the most common cause of cancer among women in both developed and developing countries. It has been reported that one million new cases of breast cancer occur each year worldwide (Mc Pherson et al. 2000). Determinants of poor prognosis of BC include young age (<40 years), early menstruation, late menopause, family history, multiparity, nulliparity, use of contraceptive pills, obesity etc. (Mukherjee et al. 2012). Study of molecular pathogenesis of breast cancer is essential for early diagnosis and development of individual-based treatment.
Cytogenetic analysis and chromosome mapping revealed deletions in several chromosomal regions like 3p12 and 4p15 to be associated with development of BC (Shivapurkar et al. 1999; Dallol et al. 2001, 2002; Alvarez et al. 2013). The candidate tumour suppressor genes ROBO1, located at 3p12.3 and SLIT2, located at 4p15.2 were shown to be altered in BC by different studies (Shivapurkar et al. 1999; Dallol et al. 2002). SLIT2 is the cognate ligand for different receptors including ROBO1/ROBO2 and could control different pathways (Dickinson and Duncan 2010). Interaction of SLIT2 with ROBO1 leads to SrGAP1 recruitment to ROBO1, resulting conversion of active CDC42–GTP to inactive CDC42–GDP complex (Wong et al. 2001). SLIT2–ROBO1 signalling mediating intrinsic dephosphorylation of CDC42–GTP has been reported to prevent tumour progression in glioma (Yiin et al. 2009; Xu et al. 2010) and in medulloblastoma (Werbowetski-Ogilvie et al. 2006) with no prior report on the role of this pathway in BC progression.
Different studies showed frequent deletion (57–63%) and methylation (48–80%) of SLIT2 locus in BC compared to ROBO1 (deletion, 15–27%; methylation, 5–19%) (Shivapurkar et al. 1999; Dallol et al. 2001; Martinez et al. 2001; Dallol et al. 2002; Alvarez et al. 2013). Alterations of ROBO2 (located 1.3 Mb telomeric to ROBO1 at chromosome 3p12.3) is yet to be studied in BC, as its alterations have already been studied in head and neck squamous cell carcinoma (HNSCC) and carcinoma of the cervix (CACX) (Ghosh et al. 2009; Mitra et al. 2012). No somatic mutation in SLIT2 and ROBO1/2 genes was reported in BC (Dallol et al. 2001, 2002). In immunohistochemical (IHC) analysis, weak or absence of SLIT2 expression was seen in BC compared to ROBO1 expression in 45–59% samples (Wang et al. 2011). However, to the best of our knowledge, alterations of SLIT2 and ROBO1 were not analysed in the same set of samples to understand their role in development of BC.
IHC data showed upregulation of Rho GTPase Cdc42 protein (82.35% of studied BC population), without emphasizing on active CDC42–GTP expression in BC (Halon et al. 2013). It has also been evident that, besides SLIT2–ROBO1 mediated inactivation of CDC42, Serine-71 phosphorylation could inhibit the binding of downstream effector proteins like PAK1, WASP etc. (Taegun et al. 2000; Schwarz et al. 2012), resulting in deregulation of this pathway. The expression pattern of phosphorylated CDC42 at Serine 71 (pSer71 CDC42) has not been studied in BC. Therefore, it is imperative to analyse the alterations (deletion/ methylation/mutation/expression) of SLIT2 and ROBO1/ ROBO2 along with expressions of CDC42 and pSer71 CDC42 in same set of BC samples to understand the association of SLIT2–ROBO1–CDC42 pathway in the development of BC.
Thus, our study has been focussed on the following aspects in primary BC of Indian patients of different clinical stages: (i) analysis of alterations (deletion/methylation/mutation) of SLIT2, ROBO1 and ROBO2 genes, (ii) expression analysis of SLIT2, ROBO1 and ROBO2, CDC42, pSer71 CDC42 proteins by IHC, (iii) clinicopathological correlation of alterations of these genes with progression of BC. Our data showed frequent inactivation of SLIT2 and ROBO1/2 genes by deletion and/or methylation followed by their reduced expression along with pSerCDC42 in BC, indicating importance of this pathway in development of this tumour.
Materials and methods
Collection of clinical specimens
DNA was isolated from freshly operated 150 primary pretherapeutic BC samples and their adjacent normal tissues for molecular analysis. One part of each sample was fixed in formalin and paraffin embedded for IHC. Remaining part of the samples were stored at −80 ∘C until further use. Informed consent and approval were obtained from patient for sample collection from the Research Ethics Committee of the institute. All tumours were staged according to the International Union against Cancer (UICC) tumour-node-metastasis (TNM) classification (table ?? in electronic supplementary material at http://www.ias.ac.in/jgenet/). Detailed clinicopathological history of the patients is provided in table 1.
Microdissection and DNA extraction
Prior to DNA extraction, the contaminant normal cells in the tumour specimen were removed by manual microdissection from cryosections using surgical knives under a dissecting microscope (Leica MZ 16, Bannockburn, USA). Tumours containing at least 70–80% tumour cells after microdissection were taken for DNA isolation. For each sample, DNA was isolated from tumour, adjacent normal tissue following a previously established protocol, i.e. proteinase K digestion followed by phenol–choloroform extraction (Dasgupta et al. 2002).
Deletion analysis
Deletion analysis of SLIT2 and ROBO1/2 loci for the tumours (n = 150) was carried out by microsatellite/exonic markers (table 2 in electronic supplementary material) Mitra et al. (2012). Briefly, polymerase chain reaction (PCR) was carried out by using a [ γp32] ATP-lablled forward primer. PCR products were electrophoresed on 7% denaturing polyacrylamide gel and autoradiographed on X-ray film Dasgupta et al. (2002). The scoring of loss of heterozygosity (LOH), hemizygous deletion (HED), microsatellite size alterations of one allele (MAI), homozygous deletion (HD) were done on autoradiogram as described by Mitra et al. (2012).
Promoter methylation analysis
Promoter methylation analysis of SLIT2 and ROBO1/2 using methylation sensitive restriction analysis (MSRA) was carried out in the entire 150 primary BC and adjacent normal tissue pairs using methylation sensitive restriction enzyme HpaII and its methylation insensitive isoschizomer MspI following a previously established protocol (Singh et al. 2005). The β-3A adaptin gene (K1) was used as digestive control and RAR β2 (K2) was used as the control for DNA integrity (Loginov et al. 2004). Methylation status of the candidate genes were also checked in a BC cell line, MCF7, by MSRA. The methylation primers of SLIT2 and ROBO1/2 genes, and the control genes are described in table 1 in electronic supplementary material.
Methylation analysis was further validated in 15 randomly selected primary BC sample pairs by methylation-specific-PCR (MSP) (Herman et al. 1996) after bisulphite modification of DNA using primers listed in table 1 in electronic supplementary material. Genomic DNA (5 μg) was subjected to bisulphite modification followed by PCR amplification of the modified DNA using primers for nonmethylation (U) or methylation (M) specific alleles. Details of MSRA/MSP protocols have been described in electronic supplementary data.
Validation of methylation status of candidate genes in MCF7 cells
MCF7 cell line was cultured for five days at 5 μM and 10 μM concentrations of 5-Aza-2 ′-deoxycytidine (5-aza-dC) (Sinha et al. 2011). The cells were harvested followed by RNA isolation, cDNA preparation and real time quantitation of the candidate genes. Briefly, total RNA was isolated from the 5-aza-dC treated/untreated MCF-7 samples using TRIzol reagent according to manufacturer’s protocol (Invitrogen, USA). The real time quantitation of RNA expression of SLIT2, ROBO1/ROBO2 was done by Power SYBR-green PCR assay (Applied Biosystems, USA) using ddCt method (Livak and Schmittgen 2001; Schmittgen and Livak 2008), with β2-microglobulin gene as internal control. Details of this method have been described in electronic supplementary data.
Mutation analysis
Mutation analysis for the SLIT2–ROBO1 interacting regions (exons 9–14 of SLIT2; exons 2–4 of ROBO1) was carried out by SSCP in all 150 sample pairs followed by PCR amplification and direct sequencing of the target region (Maiti et al. 2015) in 10 random pairs of BC samples. Sequencing of both strands of each PCR product was done with ABI PRISMTM BD Terminator Cycle Sequencing kit (PE Applied Biosystems, Foster City, USA) and electrophoresed in 3100-Avant Genetic Analyzer (PE Applied Biosystems). Sequence of the samples (tumour and normal) were analysed and compared with reference human genome sequence GRCh37 (Ensemble) to identify mutations in the regions, if any.
IHC
Molecular subtyping
Randomly collected BC samples from hospital section of Chittaranjan National Cancer Institute (CNCI) were subtyped for oestrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor 2 (HER2) expressions by IHC (Reiner et al. 1990; Perrone et al. 2006). Briefly, about 5 μm paraffin sections of primary BC samples were dewaxed, rehydrated and reacted overnight with primary antibodies: mouse monoclonal IgG2a (sc-787) for ER α; rabbit polyclonal IgG (sc-7208) for PR and mouse monoclonal IgG2a (F-11, sc-7301) for HER2, Santa Curz Biotehcnology, USA. Horseradish peroxidase (HRP)-conjugated secondary antibodies (goat antimouse IgG (sc-2005); goat antirabbit IgG (sc-2004)) were added at 1:500 dilutions. The slides were developed using 3-3 ′ diaminobenzidine (DAB) as the chromogen and counterstained with hematoxylin. Scoring was done as per the recommended guidelines of American Society of Clinical Oncology (ASCO).
In the present study, a total of 150 BC samples were selected which comprised of comparable frequencies (24.6–25.3%) of the four different subtypes of BC samples for better comparative analysis of molecular alterations among the subtypes (figure 1 in electronic supplementary material).
Expression of candidate genes
Protein expressions of SLIT2, ROBO1 and ROBO2 were studied by IHC in 68 primary BC (from the same pool of BC samples) and in adjacent normal breast tissues using primary antibodies (goat polyclonal IgG sc-16611, sc-16615, sc-1661 for SLIT2, ROBO1 and ROBO2, respectively; mouse polyclonal sc-8401 for CDC42; rabbit polyclonal sc-135641 for phosphor Serine-71 CDC42) and HRP conjugated secondary antibodies (rabbit antigoat sc-2768; goat antimouse IgG (sc-2005); goat anti-rabbit IgG (sc-2004)) from Santa Cruz Biotehcnology, USA, following a previously established protocol (Mitra et al. 2012). Of the 68 samples, protein expression of total CDC42 and phospho Serine-71 CDC42 could be further carried out in 52 BC samples. Scoring for SLIT2, ROBO1, ROBO2, CDC42 and pSer71 CDC42 expression was carried out following the method of Perrone et al. (2006).
Statistical analysis
Fisher’s exact test was used to determine different clinico-pathological association with alterations of the tumours. All statistical tests were two-sided and considered significant at probability value, P≤0.05. Survival curves were obtained according to Kaplan–Meier. Cox proportional hazards regression model predicted patient’s survival. Overall survival (OS) was measured from the date of surgery to the date of most recent follow-up or death (up to five years). The detailed follow-up records are available for 83 BC patients. All the statistical analyses were performed using statistical programs Epi Info 6.04, SPSS 10.0 (SPSS, Chicago, USA).
Results
Deletion analysis
In BC, SLIT2, ROBO1 and ROBO2 showed deletion and microsatellite size alteration of one allele (MA1) (figure 1a). However, differential frequencies of deletions of these genes have been seen in the following order: SLIT2 (38.6%) >ROBO1 (30%) >ROBO2: 7.3% (table 1 in electronic supplementary material). The frequencies of MA1 in SLIT2 and ROBO1 were infrequent in BC (0.6% for SLIT2, 1.3% for ROBO1) (table ?? in electronic supplementary material). A nonsignificant trend in deletion frequencies of SLIT2 and ROBO1/2 genes among different subtypes were observed, indicating deletions in these genes could be independent events in development of BC (figure 1b; table 1 in electronic supplementary material).
(a) Deletion and microsatellite size alterations of different marker loci of SLIT2, ROBO1 and ROBO2 (i) LOH, loss of heterozygosity; (ii) HE, hemizygous deletion; (iii) MA1, microsatellite size alteration of one allele. (b) Deletion pattern of individual candidate genes, SLIT2–ROBO1 ligand–receptor pair, SLIT2– ROBO2 ligand–receptor pairs in different subtypes. Bars represent percentage ±SD.
Methylation analysis
Variable frequencies of promoter methylation of SLIT2, ROBO1 and ROBO2 were seen in the BC samples (figure 2a; table 1 in electronic supplementary material). High frequency of methylation was seen in SLIT2 (63.3%) followed by ROBO1 (26.6%) and ROBO2 (9.3%) in the BC samples (table 1 in electronic supplementary material; figure 2a). Comethylation frequencies of SLIT2 and ROBO1 varied from 5.4–26.3% whereas comethylation of SLIT2 and ROBO2 was observed only in luminal B and TNBC (7.8–13.1%) (figure 2a). MSP analysis in 15 randomly selected tumours from the same pool of BC samples also showed concordant results (figure 2 and table 4 in electronic supplementary material).
(a) Methylation pattern of SLIT2, ROBO1, ROBO2, SLIT2–ROBO1 ligand–receptor pair, SLIT2–ROBO2 ligand–receptor pair. (b) Confirmation of methylation status of the candidate genes in MCF7 cell line by 5-aza-dC mediated reactivation of RNA expression, as observed in qRT-PCR. (c) Overall alteration pattern of individual candidate genes, SLIT2–ROBO1 ligand–receptor pair, SLIT2–ROBO2 ligand–receptor pairs. Bars represent percentage ±SD (2a, 2c) and fold change ±SD (2b).
Significantly higher frequency of methylation of SLIT2 was observed in TNBC compared to luminal A (P=0.041) and HER2 subtype (P=0.0032; figure 2a) of BC. A statistically significant association was seen methylation frequency between ROBO1 and ROBO2 genes in BC (table 3 in electronic supplementary material). In addition, the promoter methylation was analysed in MCF7 cell line by MSRA. MCF7, SLIT2 and ROBO1 genes were found to be methylated (figure 3a in electronic supplementary material).
Validation of promoter methylation of the genes
In confirmation of the promoter methylation of SLIT2 and ROBO1 in MCF7, it was evident that gradual increase in RNA expression of these genes were seen with increase in concentration of 5-aza-dC. At 10 μm 5-aza-dC treatment in MCF7 cells, the candidate genes showed increased expression in the following order SLIT2: 3.74 fold >ROBO1: 9.8 fold >ROBO2: 1.47 fold (figure 2b; figure 3b in electronic supplementary material).
Mutation analysis
No altered band was observed in the SLIT2–ROBO1 interacting domains of all the 150 sample pairs. The SSCP analysis followed by sequencing of 10 pairs of BC samples showed no somatic mutation in the SLIT2–ROBO1 interacting domain (exons 9–14 of SLIT2; exons 2–4 of ROBO1). Thus, we restricted further screening of mutation by sequencing, considering that mutation of this region might be a rare event in development of BC.
Overall alteration of the candidate genes
Majority (80%; 120/150) of the tumours showed genetic/epigenetic alteration in at least one of the SLIT2 and/or ROBO1/ROBO2 genes, indicating importance of these genes in development of BC. Overall alterations (deletion and/or methylation) of the candidate genes were seen in the following order: SLIT2: 75.3% (101/150), ROBO1: 45.3% (68/150), ROBO2: 15.3% (23/150) (figure 2c). Frequency of overall alterations of SLIT2 and/or ROBO1 (80.6%) was high compared to that of SLIT2 and/or ROBO2 (46%) (figure 2c). Significantly, higher overall alteration of SLIT2 gene was observed in TNBC compared to HER2 subtype (P=0.0014) (figure 2c; table 2). Similar trend is also seen in overall alterations of SLIT2 and/or ROBO1, in TNBC than HER2 subtype (P value: 0.0012) (figure 2c; table 2). Whereas, alterations of SLIT2 and/or ROBO2 was observed to be significantly higher in TNBC over luminal A (P value: 0.014) and HER2 subtype (P value: 0.0048) of BC patients (figure 2c). Overall alterations of the candidate genes did not show any significant difference with disease progression (figure 4 in electronic supplementary material).
Protein expression analysis
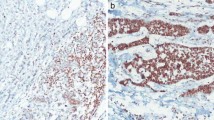
IHC analysis of SLIT2 and ROBO1/ROBO2 proteins showed expression of these proteins in the membrane and cytoplasm of luminal and myoepithelial cells of normal breast duct (figure 3, a(i), b(i), c(i)), while expression of these proteins were mainly localized in the cytoplasm of primary tumours (figure 3, a(ii, iii), b(ii, iii), c(ii, iii)). Reduced or absence of expression of SLIT2, ROBO1 and ROBO2 were observed in 82.4, 55.7 and 40% of BC samples (n=68) respectively (table 3). Significant concordance was observed in the alterations (deletions and/or methylation) of SLIT2 and ROBO1 with their expression (P=0.026 and P=0.00012 for SLIT2 and ROBO1 respectively) (table 3).
Expression of total CDC42 and pSer71 CDC42 was localized in the membrane and cytoplasm of luminal and myoepithelial cells of normal breast duct (figure 4, a(i), b(i)) and in the cytoplasm of primary BC (figure 4, a(ii), b(ii)). Moderate to high expression of total CDC42 was observed in 94.2% (49/52) of BC tumour samples, while reduced expression of pSer71 CDC42 was observed in 78.8% (41/52) of the samples (table 3). Also, a significant association was observed with coalterations of SLIT2 and/or ROBO1, SLIT2 and/or ROBO2 with reduced expression of pSer71 CDC42 (P value: 0.0012–0.0038) (n=52) (table 3). It was observed that 88.4% (46/52) of the samples showed reduced expression of at least one of the candidate proteins (SLIT2, ROBO1, ROBO2 and pSer71-CDC42) (table 3).
Clinicopathological correlations and survival analysis among BC samples
The deletion and methylation of SLIT2, ROBO1 and ROBO2 did not show any significant association with age at onset, grade and lymph node involvement, except significant association of SLIT2 methylation with early stage (P value: 0.013) and lymph node involvement (P value: 0.0093) (table 2). Expression pattern of SLIT2, ROBO1/2 showed no significant association with stage, grade, lymph node or age at onset among BC samples (table 2).
The Kaplan–Meier (K–M) survival analysis revealed poor prognosis of BC patients showing alterations of SLIT2 (P value: 0.0267) and/or ROBO1 (P value: 0.006) (figure 5), indicating their prognostic significances. This also indicated that abrogation of these ligand–receptor interactions predicted poor prognosis in BC patients. Survival analysis also showed that BC patients with alterations of SLIT2, ROBO1 and/or ROBO2 and reduced expression of pSer71 CDC42 had poor disease prognosis compared to alterations positive or alterations negative BC with moderate level of expression of pSer71 CDC42 (P value: 0.0399; figure 6), indicating importance of pSer71 CDC42 in disease prognosis (figure 6). Together, these results suggested a bidirectional regulation mechanism in SLIT2–ROBO1–CDC42 mediated tumourigenesis (figure 7).
Survival analysis of BC patients with/without alterations of SLIT2, ROBO1, and/or ROBO2 with reduced expression of pSer71 CDC42. A, alteration of SLIT2, ROBO1, and/or ROBO2; E, expression of pSer71 CDC42. + , With alterations and/or expressions of pSer71 CDC42. −, No alterations and/or reduced expression.
From our study, it is also evident that for both hormone receptor positive and negative subtypes, alterations of candidate genes predicted poor prognosis for the patient (figure ?? in electronic supplementary material). The multivariate Cox model showed alterations of SLIT2 (P=0.0268; HR: 2.4882), ROBO1 (P=0.0007; HR: 3.2180), coalterations of SLIT2 and ROBO1 (P=0.0357; HR 2.62) as well as SLIT2 and ROBO2 (P=0.0125; HR: 2.68), alterations of ROBO1 and reduced expression of pSer71-CDC42 (P=0.0187; HR: 1.1707), high grade (grade III/IV) predicted poor outcome of the BC patients (table 4).
Discussion
The aim of this study was to understand the importance of SLIT2–ROBO1/ROBO2–CDC42 pathway in the development of BC. For this reason, the alterations (deletion and/or methylation) of SLIT2 and ROBO1/ROBO2 followed by protein expression of these genes along with CDC42 and pSer71-CDC42 were analysed in the primary BC samples. Our data showed low frequency of deletion and high promoter methylation in SLIT2 (38.6 and 60.6% respectively), indicating the importance of epigenetic inactivation of this gene in development of BC. Unlike SLIT2, ROBO1 had comparable frequencies of deletion (30%) and methylation (26.6%) while low frequencies were observed for overall alterations of ROBO2 (7.3% deletion; 8.6% methylation). Overall alteration frequencies of the candidate genes showed maximum alterations in SLIT2, followed by ROBO1 and very low frequency alterations in ROBO2. As confirmed by MSRA and 5-aza-dC mediated demethylation experiments, SLIT2 and ROBO1 were found to be methylated in BC. In conformity with our data, earlier studies also showed frequent deletion (57–63%) and methylation (48–80%) of SLIT2 locus in BC compared to ROBO1 (deletion: 15–27%; methylation: 5–19%) (Shivapurkar et al. 1999; Dallol et al. 2001; Martinez et al. 2001; Dallol et al. 2002; Alvarez et al. 2013). Frequent alterations of SLIT2 and ROBO1 have been reported in other malignancies namely CACX, HNSCC, hepatocellular carcinoma, glioma, prostate carcinoma (Latil et al. 2003; Yiin et al. 2009; Zheng et al. 2009; Mitra et al. 2012; Maiti et al. 2015).
Absence of any somatic mutation in SLIT2–ROBO1 interacting domain was also in conformity with earlier observation of Dallol et al. (2001, 2002) in a cohort study involving BC patients from UK, indicating mutation in this pathway as a rare event in development of BC.
To the best of our knowledge, our study is the first approach to study subtype specific association of the alterations of the candidate genes. From our data, it is evident that methylation frequency of SLIT2 was significantly high in TNBC compared to luminal A (P = 0.041) or HER2 (P=0.0032) subtype of BC (figure 2a). Significantly, higher overall alterations of SLIT2 locus was observed in TNBC over HER2 subtype (P = 0.0014) (figure 2c). Our data showed higher frequencies of overall alterations of SLIT2 and/or ROBO1 in luminal B as well as in TNBC subtype over that of luminal A or HER2 subtype (figure 2c). Similar observation was reported by Guedj et al. (2012), showing higher frequencies of molecular alterations in luminal B as well as in basal-like tumours over other subtypes, owing to the high proliferative index of these subtypes (Guedj et al. 2012).
Alterations of SLIT2 and ROBO1/2, correlated with their reduced protein expression profile from our study (reduced expression of SLIT2, ROBO1 and ROBO2 in 82.4, 55.7 and 40% of BC samples, respectively). Similar to our study, reduced expression of SLIT2, ROBO1/ROBO2 have been reported in several other malignancies like HNSCC, liver, lung, oral, cervical, breast, kidney (Dallol et al. 2001, 2002; Zabrovsky et al. 2002; Ghosh et al. 2009; Yiin et al. 2009; Zheng et al. 2009; Mitra et al. 2012; Maiti et al. 2015) though overexpression of these genes have also been reported in prostrate and breast cancers (Latil et al. 2003; Bieche et al. 2004). From our study, reduced expression of SLIT2 and ROBO1 in majority of primary BC with alterations of SLIT2 (63.2%), ROBO1 (45.5%) indicated the importance of genetic and/or epigenetic alterations in regulating gene expression (table 3). However, a moderate level of expression of ROBO2 was observed in 50% of the cases (n=68). These indicate that ROBO2 alteration might be rare event in predicting BC progression.
Our next aim was to analyse the alterations in expression pattern of active CDC42, the downstream key regulator protein of SLIT2–ROBO1 signalling pathway. Earlier results had established that alterations of SLIT2 correlated with poor tumour prognosis through upregulation of active CDC42 (Bieche et al. 2004; Yiin et al. 2009). Previous reports have shown that apart from SLIT2–ROBO1 pathway mediated deactivation of CDC42-GTP, AkT mediated Serine-71 phosphorylation of CDC42 results in inefficient downstream effector coupling through PAK1, N-WASP etc., leading to cell cycle arrest (Taegun et al. 2000; Schwarz et al. 2012). To understand whether SLIT2–ROBO1 mediated deactivation as well as AkT mediated Serine-71 phosphorylation of CDC42 occur in tandem, we checked the expression pattern of total CDC42 and pSer71 CDC42, in the context of alteration of SLIT2/ROBO1/ROBO2. From our data, high expression of total CDC42 was observed in 94.2% of BC cases (49 of 52) whereas reduced expression of pSer71 CDC42 was observed in 78.8% of the cases, indicating phosphorylation-mediated inactivation is infrequent in BC and thus CDC42 is mostly retained in the active conformation, capable of binding to downstream effectors, namely PAK1, N-WASP etc., leading to tumour migration. Overexpression of CDC42 has also been reported in BC (Fritz et al. 2002; Halon et al. 2013), though the expression status of pSer71 CDC42 has not been reported in BC or any primary tumour.
In understanding the clinical importance of these alterations, our data showed significant association of methylation of SLIT2 with early stage and with lymph node involvement. KM survival analysis showed alterations of SLIT2 and/or ROBO1 genes or alterations of SLIT2, ROBO1/ ROBO12 genes along with reduced expression of pSer71 CDC42 to be associated with poor prognosis in BC patients, indicating that alterations of SLIT2–ROBO1 signalling are a necessary event for development of BC. Alterations of SLIT2 and ROBO1, SLIT2 and ROBO2, alterations of ROBO1 along with reduced expression of pSer71-CDC42 were found to predict poor disease free survival, according to Cox multivariate analysis (table 4). In conformity with our observation, earlier reports have also established that alterations (deletion/methylation/expression) of SLIT2 and ROBO1 to be associated with poor prognosis of patients in BC and other cancers (Chang et al. 2012; Mitra et al. 2012). No earlier data have established the implication of pSer71-CDC42 in disease prognosis and in predicting patient outcome.
Thus, it was evident from our analysis that inactivation of SLIT2, ROBO1/2 augment tumourigenesis through active CDC42 mediated downstream signalling.
References
Alvarez C., Tapia T., Cornejo V., Fernandez W., Muñoz A., Camus M. et al. 2013 Silencing of tumor suppressor genes RASSF1A, SLIT2, and WIF1 by promoter hypermethylation in hereditary breast cancer. Mol. Carcinog. 52, 475–487.
Bieche I., Lerebours F., Tozlu S., Espie M., Marty M. and Lidereau R. 2004 Molecular profiling of inflammatory breast cancer: identification of a poor-prognosis gene expression signature. Clin. Cancer Res. 10, 6789–6795.
Chang P. H., Hwang-Verslues W. W., Chang Y. C., Chen C. C., Hsiao M., Jeng Y. M. et al. 2012 Activation of Robo1 signaling of breast cancer cells by Slit2 from stromal fibroblast restrains tumorigenesis via blocking PI3K/Akt/ β-catenin pathway. Cancer Res. 72, 4652–4661.
Dallol A., Eva Forgacs, Alonso Martinez, Yoshitaka Sekido, Rosemary Walker, Takeshi Kishida et al. 2001 Tumour specific promoter region methylation of the human homologue of the Drosophila Roundabout gene DUTT1 (ROBO1) in human cancers. Oncogene 21, 3020–3028.
Dallol A., Fernandes Da Silva N., Viacava P., Minna J. D., Bieche I., Maher E. R. et al. 2002 SLIT2, a human homologue of the Drosophila Slit2 gene, has tumor suppressor activity and is frequently inactivated in lung and breast cancers. Cancer Res. 62, 5874–5880.
Dasgupta S., Mukherjee N., Roy S., Roy A., Sengupta A., Roychowdhury S. and Panda C. K. 2002 Mapping of the candidate tumor suppressor genes’ loci on human chromosome 3 in head and neck squamous cell carcinoma of an Indian patient population. Oral Oncol. 38, 6–15.
Dickinson R. E. and Duncan W. C. 2010 The SLIT–ROBO pathway: a regulator of cell function with implications for the reproductive system. Reproduction. (doi:10.1530/REP-10--0017).
Fritz G., Brachetti C., Bahlmann F., Schmidt M. and Kaina B. 2002 Rho GTPases in human breast tumours: expression and mutation analyses and correlation with clinical parameters. Br. J. Cancer 87, 635–644.
Ghosh S., Ghosh A., Maiti G. P., Alam N., Roy A., Roychoudhury S. and Panda C. K. 2009 Alterations of ROBO1/DUTT1 and ROBO2 loci in early dysplastic lesions of head and neck: clinical and prognostic implications. Hum. Genet. 125, 189–198.
Guedj M., Marisa L, de Reynies A., Orsetti B., Schiappa R., Bibeau F. et al. 2012 A refined molecular taxonomy of breast cancer. Oncogene 31, 1196–1206.
Halon A., Donizy P., Surowiak P. and Matkowski R. 2013 ERM/Rho protein expression in ductal breast cancer: a 15 year follow-up. Cell Oncol. 36, 181–190.
Herman J. G., Jeremy R., Graff J. R., Myohanen S., Nelkin B. D. et al. 1996 Methylation-specific PCR: A novel PCR assay for methylation status of CpG islands. Proc. Natl. Acad. Sci. USA 93, 9821–9826.
Kwon T., Kwon D. Y., Chun J., Kim J. H. and Kang S. S. 2000 Akt protein kinase inhibits Rac1-GTP binding through phosphorylation at Serine 71 of Rac1. J. Biol. Chem. 275, 7423–7428.
Latil A., Chene L., Cochant-Priollet B., Mangin P., Fournier G., Berthon P. et al. 2003 Quantification of expression of netrins, slits and their receptors in human prostate tumors. Int. J. Cancer 103, 306–315.
Livak K. J. and Schmittgen T. D. 2001 Analysis of relative gene expression data using real time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408.
Loginov V. I., Maliukova A. V., Seregin I. A., Khodyrev D. S., Kazubskaia T. P., Ermilova V. D. et al. 2004 Methylation of the promoter region of the RASSF1A candidate tumor suppressor gene in primary epithelial tumors. Mol. Biol. 38, 654–667.
Maiti G. P., Ghosh A., Mondal P., Ghosh S., Chakraborty J., Roy A. et al. 2015 Frequent inactivation of SLIT2 and ROBO1 signaling in head and neck lesions: clinical and prognostic implications. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 119, 202–212.
Martinez A., Walker R. A., Shaw J. A., Dearing S. J., Maher E. R. and Latif F. 2001 Chromosome 3p allele loss in early invasive breast cancer: detailed mapping and association with clinicopathological features. Mol. Pathol. 54, 300–306.
Mc Pherson K., Steel C. M. and Dixon J. M. 2000 ABC of breast diseases. Breast cancer-epidemiology, risk factors, and genetics. BMJ 321, 624–628.
Mitra S., Mazumder-Indra D., Mondal R. K., Basu P. S., Roy A., Roychoudhury S. and Panda C. K. 2012 Inactivation of SLIT2-ROBO1/2 pathway in premalignant lesions of uterine cervix: clinical and prognostic significances. PLoS One 7, e38342.
Mukherjee N., Bhattacharya N., Alam N., Roy A., Roychoudhury S. and Panda C. K. 2012 Subtype-specific alterations of the Wntsignaling pathway in breast cancer: clinical and prognostic significance. Cancer Sci. 103, 210–220.
Perrone F., Suardi S., Pastore E., Casieri P., Orsenigo M., Caramuta S. et al. 2006 Molecular and cytogenetic subgroups of oropharyngeal squamous cell carcinoma. Clin. Cancer Res. 12, 6643–6651.
Reiner A., Neumeister B., Spona J., Reiner G., Schemper M. and Jakesz R. 1990 Immunocytochemical localization of estrogen and progesterone receptor and prognosis in human primary breast cancer. Cancer Res. 50, 7057–7061.
Schmittgen T. D. and Livak K. J. 2008 Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108.
Schwarz J., Proff J., Hävemeier A., Ladwein M., Rottner K., Barlag B. et al. 2012 Serine-71 phosphorylation of Rac1 modulates downstream signaling. PLoS One 7, e44358.
Shivapurkar N., Sood S., Wistuba I. I., Virmani A. K., Maitra A., Milchgrub S. et al. 1999 Multiple regions of chromosome 4 demonstrating allelic losses in breast carcinomas. Cancer Res. 59, 3576–3580.
Singh R. K., Dasgupta S., Bhattacharya N., Chunder N., Mondal R., Roy A. et al. 2005 Deletion in chromosome 11 and Bcl-1/CyclinD1 alterations are independently associated with the development of uterine cervical carcinoma. J. Cancer Res. Clin. Oncol. 131, 395–406.
Sinha S., Singh R. K., Bhattacharya N., Mukherjee N., Ghosh S., Alam N. et al. 2011 Frequent alterations of LOH11CR2A, PIG8 and CHEK1 genes at chromosomal 11q24.1-24.2 region in breast carcinoma: Clinical and prognostic implications. Mol. Oncol. 5, 454–464.
Wang J., Wang L., Liu F. F., Ma Y. J., Fu L., Li W. L. and Gu F. 2011 Robo1 expression in breast cancer and its relationship to brain metastasis. Zhonghua Zhong Liu Zazhi 33, 447–451.
Werbowetski-Ogilvie T. E., Seyed Sadr M., Jabado N., Angers-Loustau A., Agar N. Y. R., Wu J. et al. 2006 Inhibition of medulloblastoma cell invasion by Slit. Oncogene 25, 5103–5112.
Wong K., Ren X. R., Huang Y. Z., Xie Y., Liu G., Saito H. et al. 2001 Signal transduction in neuronal migration: roles of GTPase activating proteins and the small GTPase Cdc42 in the Slit-Robo pathway. Cell 107, 209–221.
Xu Y., Li W. L., Fu L., Gu F. and Ma Y. J. 2010 Slit2/Robo1 signaling in glioma migration and invasion. Neurosci. Bull. 26, 474–478.
Yiin J. J., Hu B., Jarzynka M. J., Feng H., Liu K. W., Wu J. Y. et al. 2009 Slit2 inhibits glioma cell invasion in the brain by suppression of Cdc42 activity. Neuro. Oncol. 11, 779–789.
Zabarovsky E. R., Lerman M. I. and Minna J. D. 2002 Tumor suppressor genes on chromosome 3p involved in the pathogenesis of lung and other cancers. Oncogene 21, 6915– 6935.
Zheng D., Liu B. B., Liu Y. K., Kang X. N., Sun L., Guo K. et al. 2009 Analysis of the expression of Slit/Robo genes and the methylation status of their promoters in the hepatocellular carcinoma cell lines. Zhonghua Gan Zang Bing Za Zhi 17, 198–202 (in Chinese).
Acknowledgements
We thank the Director, Chittaranjan National Cancer Institute, Kolkata, India. Financial support for this work was provided by DST Fast Track SR/FT/LS-81/2009 grant to Dr R. Bhattacharya, CSIR RA grant 09/030(0071)/2013-EMR-I (RA) to Dr N. Mukherjee and UGC-NET Fellowship grant F.2-3/2000 (SA-I) (sr. no. 2061030813, ref. no.: 20-06/2010(i)EU-IV dated 22.10.2010) to Mrs H. Dasgupta.
Author information
Authors and Affiliations
Corresponding author
Additional information
[Bhattacharya R., Mukherjee N., Dasgupta H., Islam M. S., Alam N., Roy A., Das P., Roychoudhury S. and Panda C. K. 2016 Frequent alterations of SLIT2–ROBO1–CDC42 signalling pathway in breast cancer: clinicopathological correlation. J. Genet. 95, xx–xx]
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
BHATTACHARYA, R., MUKHERJEE, N., DASGUPTA, H. et al. Frequent alterations of SLIT2–ROBO1–CDC42 signalling pathway in breast cancer: clinicopathological correlation. J Genet 95, 551–563 (2016). https://doi.org/10.1007/s12041-016-0678-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12041-016-0678-2