Abstract
SLIT2 has been classified as a major tumour suppressor gene due to its frequent inactivation in different cancer types. However, alterations of SLIT2 expression and relation to patient outcomes in diffuse large B cell lymphoma (DLBCL) remain undefined. The aim of this study was to investigate the expression and the methylation status of SLIT2 gene as well as its relation to patient outcomes in DLBCL. Immunohistochemical (IHC) staining was carried out to detect the expression of SLIT2 in a series of 108 DLBCL cases. Re-analysis of previously published dataset (GSE10846) that measured gene expression in DLBCL patients who had received CHOP or R-CHOP therapy was performed to identify associations between SLIT2 and patients survival. Laser capture microdissection was performed to isolate GC B cells and DLBCL primary tumor cells. Bisulfite treatment and methylation-specific PCR (MSP) analysis were done to assess SLIT2 promotor methylation status. We report that the expression of SLIT2 protein was reduced in a subset of DLBCL cases and this was significantly correlated with advanced clinical stage (p = 0.041) and was an independent predictor of worse overall survival (OS) (p = 0.012). Re-analysis of published gene expression data showed that reduced SLIT2 mRNA expression was significantly correlated with worse OS in R-CHOP-treated ABC DLBCL patients (p = <0.01). Hypermethylation of the SLIT2 promotor was significantly correlated with low SLIT2 expression (p = 0.009). Our results provide a novel evidence of reduced expression of SLIT2 that is associated with promoter hypermethylation and adverse outcomes in patients with DLBCL.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Diffuse large B cell lymphoma (DLBCL) is a heterogeneous disease with distinct molecular features and clinical behavior [1]. It is the most common of the non-Hodgkin lymphomas [2]. About 30–40% of the non-Hodgkin lymphomas are DLBCL. Based on gene expression profiling, two main classes of DLBCL can be identified, these are the germinal center B cell (GCB) and the activated B cell like subtypes (ABC) with a small percentage (10–15%) being un-classifiable [3, 4].
DLBCL is usually an aggressive cancer, with a median survival of less than 12 months in untreated patients. Since the 1970s, the standard treatment has been the CHOP regimen consisting of adriamycin, prednisone, vincristine, and cyclophosphamide [5]. Use of this regimen is associated with around 40% progression-free survival and 50% long-term survival [6]. There has been a considerable improvement in the prognosis of DLBCL within the last few years, mainly due to the inclusion of rituximab, an anti-CD20 monoclonal antibody [7]. Notwithstanding these notable achievements, a sizable percentage of patients have refractory disease or will relapse reducing their chances of survival [8].
The pathogenesis of DLBCL is complex involving interactions between genomic and epigenomic changes [9, 10]. SLIT2 has been classified as a major tumour suppressor gene due to its frequent inactivation in different cancer types, including deletion or hyper-methylation of its promoter [11, 12]. SLIT2 plays a role in the suppression of tumour growth [13, 14] via the regulation of the β-catenin and PI3-K signaling pathways leading to enhancement of β-catenin and E-cadherin mediated cell-cell adhesion [15].
In hematolymphoid malignancies, only in leukemias and mantle cell lymphoma, few reports are available for the role of SLIT2 and its main receptor ROBO1 [16,17,18]. These reports showed frequent SLIT2 methylation in leukemia cell lines, also in acute lymphocytic leukemia (ALL) and chronic lymphocytic leukemia (CLL) primary samples [17]. Moreover, the expression of SLIT2 was restored after treating ALL lines with the demethylating agent (5-aza-2′-deoxycytidine) [17]. In Mantle cell lymphoma, ROBO1 displayed a high level of methylation, which correlated well with its low mRNA expression [16, 18], also correlated with aggressive clinciopathological features and shorter survival [18]. Here, we have studied the expression and the methylation status of SLIT2 gene as well as its relation to patient outcome in DLBCL.
Materials and Methods
Tissue Specimens
This is a retrospective cohort study in which Formalin-fixed paraffin-embedded tissues were used from 108 DLBCL patients diagnosed between 1997 and 2012 at the University Hospital, Birmingham. Diagnosis was confirmed by histological review. Detailed clinical and follow-up data was available for all patients. Follow up ranged from 1 to 180 months with a mean of 45 months. Based on the Hans algorithm, DLBCL were grouped by IHC into either GCB or ABC-DLBCL types [19]. The baseline pathologic and clinical characteristics of the patients are listed in Table 1. Different normal/reactive lymphoid tissues (tonsils and lymph nodes) obtained through tonsillectomy or excision biopsy, fixed in formalin and embedded with paraffin, were used for both positive and negative control as well as for GC B cells microdissection for the methylation analysis (see below).
Immunohistochemistry
Paraffin-embedded tissues from both DLBCL samples and normal lymphoid tissues were sectioned at a thickness of 4 μm and placed on X-tra Adhesive micro slides. The expression of SLIT2 protein was detected by IHC using Leica BOND-MAXTM autostainer (Leica Biosystems) according to the protocol instruction. In brief, the tissue sections were deparaffinized and then rehydrated through graded alcohols. Sections were immersed into preheated EDTA (PH 8.0) for 20 min. Endogenous peroxidase activity inhibited in 0.3% hydrogen peroxide, and non-specific binding sites blocked with normal goat serum at room temperature. Primary mouse monoclonal anti-SLIT2 [Clone EPR2771; Abcam Ltd. 134,166 USA; diluted at 1:800) was used for 1 h. A cold PBS was used in place of the primary antibody for the negative control staining.
Microdissection of GC B Cells and DLBCL Cells
DLBCL cases were selected with variable SLIT2 staining intensity as well as GC B-cells from normal/reactive lymphoid tissues expressing strong SLIT2 staining. A total of 6 unstained sections per each case were cut at 8 μm thickness and stained with Haematoxylin with available matched SLIT2 immunostained sections for each case. Sections were placed onto membrane-covered slides for microdissection (PEN membrane slide, Carl Zeiss Ltd. Germany). Using a PALM Microbeam microdissection (Carl Zeiss Ltd., Germany), laser capture microdissection was performed to capture GC B cells and DLBCL tumor cells from areas of which matched to the SLIT2 stained sections. Microdissected tissues were put into PALM adhesive cap (Carl Zeiss Ltd.) tubes for further methylation analysis.
Analysis of SLIT2 Expression
The entire tumour section was scanned at low power to survey overall staining intensity in DLBCL tumour cells. SLIT2 protein was found to be expressed in both tumour cell cytoplasm and the membrane. The intensity was judged by the comparing the intensity between tumour and GCs of normal lymphoid tissue (tonsillar/lymph nodal tissue as an external positive control or residual GCs in the affected lymph node as an internal positive control).
The expression intensity was scored as follows: 0 = negative, 1 = weak, 2 = moderate, comparable to normal germinal centers and 3 = strong, presence of cells with staining obviously stronger than in normal GCs. Staining percentage was scored 4 (if greater than 75% of tumour cells were positive), 3 (50–75%), 2 (25–50%), or 1(< 25%). The staining was graded as “low expression” if staining intensity multiplied by percentage was less than or equal to 4 [20, 21].
Re-Analysis of Gene Expression Data
A previously published dataset (GSE10846) that had measured gene expression in DLBCL patients who received CHOP or R-CHOP therapy [20] was downloaded from GEO (https://www.ncbi.nlm.nih.gov/geo/). The Bioconductor project’s affy package was used to conduct a probe level quantile normalization [21] and robust multi-array analysis [22] on the raw CEL files. The R survival package was used to perform survival analysis (http://www.r-project.org/), including log-rank test with samples split by median SLIT2 expression.
Bisulfite Treatment and Methylation-Specific PCR (MSP) Analysis
DNA was extracted from the microdissected samples using QIAamp DNA FFPE Tissue Kit (Catalogue no. 56404). Microdissected cells were placed in proteinase K digestion solution, then incubated at 90 °C. Residual contaminants were washed away. DNA was then eluted in Buffer ATE. Purified DNA was checked on Nanodrop spectrophotometer. DNA was stored at -20 °C.
Bisulfite modification of genomic DNA was carried out as described previously [23, 24]. 50 ng of bisulfited DNA for each sample was used for methylation-specific PCR (MSP) analysis. Multiple methylated primer sets targeting SLIT2 promoter were designed and tested. The best pair (m5/m7) with clear methylation band, inversely correlated with expression, and not amplifying any not-bisulfited DNA, was chosen. MSP primer set used in the study was: SLIT2m5: 5’-GATCGGTTTAGGTTGCGG C; SLIT2m7: 5’-AACAACTAAACATAACGCGCG. MSP was performed for 38 cycles using AmpliTaq Gold® DNA Polymerase (Thermo Fisher Scientific) [23,24,25].
Statistical Methods
Data were analyzed using IBM SPSS advanced statistics (Statistical Package for Social Sciences), version 23 (SPSS Inc., Chicago, IL). Numerical data was described as median and interquartile range or range or mean and standard deviation as appropriate, while qualitative data were described as number and percentage. Chi-square (Fisher’s exact) test was used to examine the relation between qualitative variables as appropriate. Survival analysis was done using the Kaplan-Meier method. Comparison between two survival curves was done using the log rank test. Multivariate analysis was done by Cox regression model to test for independent prognostic effects of statistically significant variables on univariate level with calculating hazard ratio and its 95% confidence interval. A p value less than 0.05 was considered statistically significant. All tests were two tailed. Overall survival (OS) was calculated from date of diagnosis till date of death or last follows up. Event free survival (EFS) was calculated from date of treatment till date of relapse, death or last known follow up.
Results
Pattern of SLIT2 Protein Expression in Normal Lymphoid Tissue and DLBCL
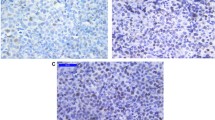
Immunohistochemical analysis of SLIT2 was performed for both normal lymphoid tissue (tonsil/lymph nodes) and primary DLBCL. In normal lymphoid tissue, high SLIT2 expression was restricted to the germinal centers with only few inter-follicular lymphocytes showing low SLIT2 expression (Fig. 1a& b). SLIT2 was located in both the cytoplasm and membrane of DLBCL tumour cells. Different intensities of SLIT2 protein expression were observed among different cases; 41/108 cases (38%) showed high expression (SLIT2 intensity multiplied by the percentage > 4), 66/108 (61.1%) had low expression (SLIT2 intensity multiplied by the percentage ≤ 4). Only one case (1/108, 0.9%) was negative (Low expression ≤4). Figure 1c& d show representative DLBCL cases with different levels of SLIT2 protein expression. Of the 108 cases, 49 (45.4%) were of GCB type, while 58 (53.7%) cases were of ABC-DLBCL type. One case was unclassifiable due to inconclusive IHC results. 55.1% (27 cases) of GCB DLBCL and 65.5% (38 cases) of the ABC-DLBCL showed low SLIT2 expression. There was no significant correlation between subtype and SLIT2 protein expression (p = 0.27).
SLIT2 protein expression in normal lymphoid tissue and DLBCL primary tumours (a) High SLIT2 protein expression in the reactive GCs, also in normal squamous epithelium of the tonsil, with low expression in few scattered interfolliclular cells (Mag. 10x). (b) Higher magnification of one of the GCs expressing high SLIT2 protein. (c) Example of high SLIT2 expression (score 3) in DLBCL, sparing the residual normal/reactive lymphocytes. (d) A representative case of DLBCL with low SLIT2 expression (score 1)
Association between SLIT2 Protein Expression and Clinicopathological Features
In our study, a significant correlation between low SLIT2 intensity and advanced clinical stage was found (stages 3&4 vs 1&2; p = 0.041). There was a tendency for cases with low SLIT2 expression to be associated with the poor prognostic group as defined by the Revised International Prognostic Index (R-IPI) (p = 0.099). No significant relationships between SLIT2 protein expression and other parameters were found [Table 2].
Impact of SLIT2 Protein Expression on the Survival of DLBCL Patients
SLIT2 protein expression was then assessed for its impact on patients outcomes. A univariate Kaplan-Meier (K-M) analysis revealed a significant reduction in overall survival (OS) for DLBCL patients with low SLIT2 protein, with a median of 28 months, while the median survival was not reached in patients with strong SLIT2 expression (p = 0.044) (Fig. 2a). In a multivariate analysis, SLIT2 was an independent significant predictor of OS (p = 0.012). Although the median EFS was only 12.6 months for patients with low tumour expression of SLIT2, compared with 88.9 months for those with strong SLIT2 expression, though this difference was not significant (p = 0.108), (Fig. 2b). We found no correlation between SLIT2 protein expression and OS or EFS in patients with either GC- (p = 0.26 and p = 0.35, respectively) or ABC-DLBCL (p = 0.26 and p = 0.31, respectively).
Kaplan-Meier survival analysis for DLBCL cases expressing low/high SLIT2 protein. (A) Low SLIT2 protein expression correlates with poor OS in DLBCL patients (p = 0.044). (B) Patients with low SLIT2 protein or negative expression show difference in Event-free survival with a median of only 12.6 months as compared to 88.9 median survival of patients experienced high SLIT2 protein, however that was not statistically significant (p = 0.108)
SLIT2 Gene Expression Predicts Survival in DLBCL (ABC Subtype) Patients Treated with R-CHOP
To further explore the potential effects of SLIT2 expression on outcome we took advantage of the dataset previously published by Lenz et al. (2008) (GSE10846) that had measured gene expression in the tumour tissues of 181 DLBCL patients treated with either CHOP or R-CHOP and which had used microarray gene expression to define subtype (Lenz et al. 2008). We found that low SLIT2 mRNA expression was significantly correlated with worse OS in R-CHOP-treated ABC DLBCL patients (p = <0.01) [Fig. 3]. SLIT2 expression was not significantly correlated with OS or EFS in patients with the GC subtype.
Methylation of the SLIT2 Promotor in DLBCL
We measured SLIT2 promoter methylation in 26 DLBCL cases with different SLIT2 protein expression as well as microdissected GC B cells with high SLIT2 expression, 14/26 (54%) DLBCL had low SLIT2 protein expression and 12/26 (46%) had strong expression.
Higher SLIT2 methylation (strong/moderate) was found in cases with reduced SLIT2 protein expression, and that was statistically significant (Fisher’s exact test, p = 0.009). In samples with low/negative SLIT2 expression, 13/14 (92.9%) had a strong/moderate-methylation status. On the other hand, among samples with higher SLIT2 expression, 7/12 (58.3%) had weak/un-methylation status, while 5/12 (41.7%) had strong/moderate-methylation status (Fig. 4a). In contrast, the microdissected GC B cells with strong SLIT2 protein expression showed unmethylated SLIT2 gene (Fig. 4b). In this small series there was no statistically significant correlation between SLIT2 methylation and DLBCL subtypes (7/18 ABC-DLBCL and 4/8 GCB DLBCL had weak/un-methylation status; p = 0.65).
Methylation-specific PCR (MSP) analysis of the SLIT2 promotor in microdissected GC B cells and DLBCL samples. (A) DLBCL cases with variable SLIT2 protein intensities as detected by IHC. Samples highlighted in red express a low/negative SLIT2 protein. Reduced SLIT2 protein expression correlates with promotor methylation status (strong/moderate) (p = 0.009). (B) GC B cells expressing high SLIT2 protein show un-methylated status Methylated (OCI-LY3) and unmethylated (HEK293) cell lines were used as positive controls. M: methylated; U: unmethylated
Discussion
The SLIT family of genes is large extracellular matrix-secreted and membrane-associated glycoproteins. SLIT2 is known to function through its two main receptors Robo1 and Robo4. Robo1 is mainly expressed in cancer cells [26].
The present study aimed to identify if SLIT2 might be a potential tumour suppressor also in DLBCL. After careful search of existing literatures, no such study has yet been reported in DLBCL. Our results show that a subset of DLBCL is characterized by reduced SLIT2 expression correlated significantly with promotor hypermethylation. Furthermore, reduced SLIT2 expression was also associated with poor prognosis, suggesting that SLIT2 might serve as a prognostic biomarker in DLBCL patients.
In our study, reduced expression of SLIT2 protein was correlated significantly with advanced clinical stage (p = 0.041). Moreover, a significant reduction in OS of DLBCL patients was correlated with low expression of SLIT2 protein (p = 0.044). This has been supported by multivariate analysis, where poor survival was associated with lower SLIT2 expression (p = 0.012).
Consistent with our data, several studies indicate that inactivation of the SLIT/ROBO1 pathway is important for cancer development and that its low expression is associated with a poor prognosis [13, 27]. Thus, Tseng et al. (2015), found that low SLIT2 expression is correlated with late stage and with poor OS and DFS in oesophageal cancer patients [28], effects which might be explained by their observation that cancer cell motility is reduced when SLIT2 is stably overexpressed. These observations were confirmed by Kim et al. (2008), who found that SLIT2 expression was decreased or abolished in human oesophageal squamous cell carcinomas (SCCs) compared to normal tissues as detected by in situ hybridization [29]. Low expression of SLIT2 also increased cell migration in lung cancer and invasive cervical cancer [30, 31]. The underlying mechanism is that inactivation of the SLIT2-ROBO1 pathway enhances cell migration through accumulation of activated Cdc42 (a member of the Rho-family of GTPases), resulting in loss of the interaction between E-cadherin and B-catenin and the inactivation of EGFR degradation [15, 28, 32,33,34].
SLIT2 protein can also inhibit the migration of endothelial cells which might explain the promotion of angiogenesis following SLIT2 loss [35]. Apoptosis was also shown to be increased and proliferation decreased in SLIT2-transfected tumours, suggesting another mechanism for SLIT2-mediated tumour suppression [29]. Interestingly, Chang et al. reported a correlation between low SLIT2 expression in the stromal fibroblasts with lymph node metastasis in breast cancer, indicating an important role for the expression of SLIT2 in the tumour microenvironment on the progression of tumours [36].
In our study, re-analysis of the published dataset that had measured mRNA expression among DLBCL patients treated with either CHOP or R-CHOP [20], revealed that downregulation of SLIT2 mRNA was significantly correlated with reduced overall survival of R-CHOP-treated ABC DLBCL patients. We did not find such an association in our IHC analysis possibly because of the smaller numbers of FFPE samples studied or because of the reported deficiencies in the correct delineation of gene expression sub-entities by IHC.
It was evident that SLIT2 gene methylation was significantly correlated with its low protein expression in DLBCL, whereas normal GC B cells strongly expressed SLIT2 and lacked methylation of the promoter. Our results suggest that promoter methylation could explain the reduced expression of SLIT2 in DLBCL. In keeping with our findings, previous studies have reported that promoter hypermethylation is responsible for the down-regulation of SLIT2/ROBO1 signaling in various solid tumours, including breast, head and neck squamous cell carcinoma (HNSCC), cervical, oesophageal, hepatocellular carcinomas and gliomas, where it is also associated with poor prognosis [27, 28, 37,38,39]. In hematolymphoid malignancies, Dunwell et al. (2009) found that SLIT2 was methylated in all ten leukemia cell lines analyzed, whilst DNA from peripheral blood and bone marrow from healthy control samples showed no SLIT2 methylation [17]. The expression of SLIT2 was restored after treating ALL lines with 5-aza-2′-deoxycytidine [17]. These findings suggest that reactivating the SLIT2 function by reversing epigenetic inactivation could represent a novel therapeutic opportunity to attenuate human cancer.
In conclusion, the results of the present study indicate for the first time that reduced expression of SLIT2 gene is associated with poorer patient outcomes in DLBCL. Furthermore, we have shown that SLIT2 protein down-regulation correlates with promoter hypermethylation. The functional impact of reduced SLIT2 inhibition in DLBCL remains to be established.
References
Hunt KE, Reichard KK (2008) Diffuse large B-cell lymphoma. Arch Pathol Lab Med 132(1):118–124
Teras LR, DeSantis CE, Cerhan JR, Morton LM, Jemal A, Flowers CR (2016) US lymphoid malignancy statistics by World Health Organization subtypes. CA Cancer J Clin 66(6):443–459. https://doi.org/10.3322/caac.21357
Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, Powell JI (2000) Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403(6769):503. https://doi.org/10.1038/35000501
Sujobert P, Salles G, Bachy E (2016) Molecular classification of diffuse large B-cell lymphoma: what is clinically relevant? Hematol Oncol Clin N 30(6):1163–1177. https://doi.org/10.1016/j.hoc.2016.07.001
McKelvey EM, Gottlieb JA, Wilson HE, Haut A, Talley RW, Stephens R, Lane M, Gamble JF, Jones SE, Grozea PN, Gutterman J (1976) Hydroxyldaunomycin (Adriamycin) combination chemotherapy in malignant lymphoma. Cancer 38(4):1484–1493. https://doi.org/10.1002/1097-0142(197610)38:4%3C1484::aid-cncr2820380407%3E3.0.co;2-i
Fisher RI, Gaynor ER, Dahlberg S, Oken MM, Grogan TM, Mize EM, Glick JH, Coltman CA Jr, Miller TP (1993) Comparison of a standard regimen (CHOP) with three intensive chemotherapy regimens for advanced non-Hodgkin's lymphoma. New England Journal of Medicine 328(14):1002-6. https://doi.org/10.1056/nejm199304083281404
Coiffier B, Lepage E, Briere J, Herbrecht R, Tilly H, Bouabdallah R, Morel P, Van Den Neste E, Salles G, Gaulard P, Reyes F (2002) CHOP chemotherapy plus rituximab compared with CHOP alone in elderly patients with diffuse large-B-cell lymphoma. N Engl J Med 346(4):235–242. https://doi.org/10.1056/nejmoa011795
Coiffier B, Thieblemont C, Van Den Neste E, Lepeu G, Plantier I, Castaigne S, Lefort S, Marit G, Macro M, Sebban C, Belhadj K (2010) Long-term outcome of patients in the LNH-98.5 trial, the first randomized study comparing rituximab-CHOP to standard CHOP chemotherapy in DLBCL patients: a study by the Groupe d'Etudes des Lymphomes de l'Adulte. Blood 1:blood-2010. https://doi.org/10.1182/blood-2010-03-276246
Cerhan JR, Berndt SI, Vijai J, Ghesquieres H, McKay J, Wang SS, Wang Z, Yeager M, Conde L, de Bakker PI et al (2014) Genome-wide association study identifies multiple susceptibility loci for diffuse large B cell lymphoma. Nat. Genet 46:1233–1238. https://doi.org/10.1158/1538-7445.am2014-lb-272
Skibola CF, Curry JD, Nieters A (2007) Genetic susceptibility to lymphoma. Haematologica 92(7):960–969
Gröne J, Doebler O, Loddenkemper C, Hotz B, Buhr HJ, Bhargava S (2006) Robo1/Robo4: differential expression of angiogenic markers in colorectal cancer. Oncol Rep 15(6):1437–1443. https://doi.org/10.3892/or.15.6.1437
Xian J, Clark KJ, Fordham R, Pannell R, Rabbitts TH, Rabbitts PH (2001) Inadequate lung development and bronchial hyperplasia in mice with a targeted deletion in the Dutt1/Robo1 gene. Proc Natl Acad Sci 98(26):15062–15066
Dallol A, Da Silva NF, Viacava P, Minna JD, Bieche I, Maher ER, Latif F (2002) SLIT2, a human homologue of the Drosophila Slit2 gene, has tumor suppressor activity and is frequently inactivated in lung and breast cancers. Cancer Res 62(20):5874–5880
Qiu H, Zhu J, Yu J, Pu H, Dong R (2011) SLIT2 is epigenetically silenced in ovarian cancers and suppresses growth when activated. Asian Pac J Cancer Prev 12(3):791–795
Prasad A, Paruchuri V, Preet A, Latif F, Ganju RK (2008) Slit-2 induces a tumor-suppressive effect by regulating β-catenin in breast cancer cells. J Biol Chem 283(39):26624–26633. https://doi.org/10.1074/jbc.m800679200
Appe AJ, Aggerholm A, Hansen MC, Ebbesen LH, Hokland P, Bentzen HH, Nyvold CG (2017) Differential expression levels and methylation status of ROBO1 in mantle cell lymphoma and chronic lymphocytic leukaemia. Int J Lab Hematol 39(3):e70–e73. https://doi.org/10.1111/ijlh.12615
Dunwell TL, Dickinson RE, Stankovic T, Dallol A, Weston V, Austen B, Catchpoole D, Maher ER, Latif F (2009) Frequent epigenetic inactivation of the SLIT2 gene in chronic and acute lymphocytic leukemia. Epigenetics 4(4):265–269. https://doi.org/10.4161/epi.9137
Enjuanes A, Fernàndez V, Hernández L, Navarro A, Beà S, Pinyol M, López-Guillermo A, Rosenwald A, Ott G, Campo E, Jares P (2011) Identification of methylated genes associated with aggressive clinicopathological features in mantle cell lymphoma. PLoS One 6(5):e19736. https://doi.org/10.1371/journal.pone.0019736
Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G, Müller-Hermelink HK, Campo E, Braziel RM, Jaffe ES, Pan Z (2004) Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood 103(1):275–282. https://doi.org/10.1182/blood-2003-05-1545
Lenz G, Wright G, Dave SS, Xiao W, Powell J, Zhao H, Xu W, Tan B, Goldschmidt N, Iqbal J, Vose J (2008) Stromal gene signatures in large-B-cell lymphomas. N Engl J Med 359(22):2313–2323. https://doi.org/10.1056/nejmoa0802885
Bolstad BM, Irizarry RA, Åstrand M, Speed TP (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19(2):185–193. https://doi.org/10.1093/bioinformatics/19.2.185
Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP (2003) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Research 31(4):e15. https://doi.org/10.1093/nar/gng015
Tao Q, Swinnen LJ, Yang J, Srivastava G, Robertson KD, Ambinder RF (1999) Methylation status of the Epstein-Barr virus major latent promoter C in iatrogenic B cell lymphoproliferative disease: application of PCR-based analysis. Am J Pathol 155(2):619–625. https://doi.org/10.1016/s0002-9440(10)65157-7
Tao Q, Huang H, Geiman TM, Lim CY, Fu L, Qiu GH, Robertson KD (2002) Defective de novo methylation of viral and cellular DNA sequences in ICF syndrome cells. Hum Mol Genet 11(18):2091–2102. https://doi.org/10.1093/hmg/11.18.2091
Murray PG, Fan Y, Davies G, Ying J, Geng H, Ng KM, Li H, Gao Z, Wei W, Bose S, Anderton J (2010) Epigenetic silencing of a proapoptotic cell adhesion molecule, the immunoglobulin superfamily member IGSF4, by promoter CpG methylation protects Hodgkin lymphoma cells from apoptosis. Am J Pathol 177(3):1480–1490 https://doi.org/10.2353/ajpath.2010.100052
Andrews W, Liapi A, Plachez C, Camurri L, Zhang J, Mori S, Murakami F, Parnavelas JG, Sundaresan V, Richards LJ (2006) Robo1 regulates the development of major axon tracts and interneuron migration in the forebrain. Development 133(11):2243–2252 https://doi.org/10.1242/dev.02379
Qin F, Zhang H, Ma L, Liu X, Dai K, Li W, Gu F, Fu L, Ma Y (2015) Low expression of Slit2 and Robo1 is associated with poor prognosis and brain-specific metastasis of breast cancer patients. Sci Rep 5:14430. https://doi.org/10.1038/srep14430
Tseng RC, Chang JM, Chen JH, Huang WR, Tang YA, Kuo IY, Yan JJ, Lai WW, Wang YC (2015) Deregulation of SLIT2-mediated Cdc42 activity is associated with esophageal cancer metastasis and poor prognosis. J Thorac Oncol 10(1):189–198. https://doi.org/10.1097/jto.0000000000000369
Kim HK, Zhang H, Li H, Wu TT, Swisher S, He D, Wu L, Xu J, Elmets CA, Athar M, Xu XC (2008) Slit2 inhibits growth and metastasis of fibrosarcoma and squamous cell carcinoma. Neoplasia 10(12):1411–1420. https://doi.org/10.1593/neo.08804
Mitra S, Mazumder-Indra D, Mondal RK, Basu PS, Roy A, Roychoudhury S, Panda CK (2012) Inactivation of SLIT2-ROBO1/2 pathway in premalignant lesions of uterine cervix: clinical and prognostic significances. PLoS One 7(6):e38342. https://doi.org/10.1371/journal.pone.0038342
Tseng RC, Lee SH, Hsu HS, Chen BH, Tsai WC, Tzao C, Wang YC (2010) SLIT2 attenuation during lung cancer progression deregulates β-catenin and E-cadherin and associates with poor prognosis. Cancer Res 12:0008–5472. https://doi.org/10.1158/0008-5472.can-09-2084
Dickinson RE, Duncan WC (2010) The SLIT-ROBO pathway: a regulator of cell function with implications for the reproductive system. Reproduction 139:697–704. https://doi.org/10.1530/rep-10-0017
Wu WJ, Tu S, Cerione RA (2003) Activated Cdc42 sequesters c-Cbl and prevents EGF receptor degradation. Cell 114:715–725. https://doi.org/10.1016/s0092-8674(03)00688-3
Wong K, Ren XR, Huang YZ et al (2001) Signal transduction in neuronal migration: roles of GTPase activating proteins and the small GTPase Cdc42 in the Slit-Robo pathway. Cell 107:209–221. https://doi.org/10.1016/s0092-8674(01)00530-x
Park KW, Morrison CM, Sorensen LK, Jones CA, Rao Y, Chien CB, Wu JY, Urness LD, Li DY (2003) Robo4 is a vascular-specific receptor that inhibits endothelial migration. Dev Biol 261(1):251–267. https://doi.org/10.1016/s0012-1606(03)00258-6
Chang PH, Hwang-Verslues WW, Chang YC, Chen CC, Hsiao M, Jeng YM, Chang KJ, Lee EY, Shew JY, Lee WH (2012) Activation of Robo1 signaling of breast cancer cells by Slit2 from stromal fibroblast restrains tumorigenesis via blocking PI3K/Akt/β-catenin pathway. Cancer Res 23:canres-0877. https://doi.org/10.1158/0008-5472.can-12-0877
Legg JA, Herbert JM, Clissold P, Bicknell R (2008) Slits and roundabouts in cancer, tumour angiogenesis and endothelial cell migration. Angiogenesis 11(1):13–21. https://doi.org/10.1007/s10456-008-9100-x
Maiti GP, Ghosh A, Mondal P, Ghosh S, Chakraborty J, Roy A, Roychowdhury S, Panda CK (2015) Frequent inactivation of SLIT2 and ROBO1 signaling in head and neck lesions: clinical and prognostic implications. Oral surgery, oral medicine, oral pathology and oral radiology1 119(2):202–212. https://doi.org/10.1016/j.oooo.2014.09.029
Dallol A, Forgacs E, Martinez A, Sekido Y, Walker R, Kishida T, Rabbitts P, Maher ER, Minna JD, Latif F (2002) Tumour specific promoter region methylation of the human homologue of the Drosophila roundabout gene DUTT1 (ROBO1) in human cancers. Oncogene 21(19):3020. https://doi.org/10.1038/sj.onc.1205421
Acknowledgements
This work was supported by Ministry of Higher Education fund support (#2012-13), Egypt and by Hong Kong HMRF grant (#16151042).
Funding
This work was supported by Ministry of higher education fund support (#2012–13), Egypt and by Hong Kong HMRF grant (#16151042). The funding sources had no influence on the study design, analysis, and interpretation of the data.
Author information
Authors and Affiliations
Contributions
Ghada Mohamed undertook pathologic review of the cases, interpreted and analyzed the IHC results, performed laboratory work and co-wrote the manuscript.; Soha Talima undertook clinical data analysis and co-wrote the manuscript; Lili Li performed laboratory work and co-wrote the manuscript; Wenbin Wei undertook bioinformatic and statistical analyses, interpreted the bioinformatic data and co-wrote the manuscript; Zbigniew Rudzki Undertook pathologic review of the cases; Rasha Allam undertook statistical analysis and interpreted the data; William Simmons undertook cases’ data collection; Qian Tao supervised laboratory work and co-wrote the manuscript; Paul G Murray designed and oversaw the research and co-wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no conflict of interest.
Ethical Approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the UK Health Research Ethics Committee (North West-Haydock; reference 15/NW/0079).
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Mohamed, G., Talima, S., Li, L. et al. Low Expression and Promoter Hypermethylation of the Tumour Suppressor SLIT2, are Associated with Adverse Patient Outcomes in Diffuse Large B Cell Lymphoma. Pathol. Oncol. Res. 25, 1223–1231 (2019). https://doi.org/10.1007/s12253-019-00600-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12253-019-00600-9