Abstract
Large body of animal work and emerging clinical findings have suggested that early exposure to anesthetics may result in increased risk of learning disabilities and behavioral impairments. Recent studies have begun to investigate anesthesia-induced epigenetic modifications to elucidate their role in behavioral and neurodevelopmental abnormalities. Here we examine sevoflurane-induced transgenerational modifications of subicular neuronal DNA methylation and expression of immediate early genes (IEGs), arc and junB, crucial to synaptic plasticity and normal neuronal development. We show that 6 h sevoflurane exposure in postnatal day 7 rat pups resulted in decreased neuronal 5-methycytosine, indicating reduced DNA methylation. This effect is transgenerationally expressed in offspring born to exposed mothers which is of importance considering that decreased DNA methylation in the brain has been linked with functional decline in learning and memory. We further show that sevoflurane exposure induces upregulation of Arc and JunB mRNA expression, 42.7% and 35.2%, respectively. Transgenerational changes in Arc and JunB mRNA were sexually dimorphic only occurring in males born to exposed females, expressed as upregulation of Arc and JunB mRNA, 71.6% and 74.0%, respectively. We further investigated correlation between altered arc promoter methylation and observed upregulation of Arc mRNA and observed that sevoflurane reduced methylation in the 5-upstream promoter region of females exposed to sevoflurane. Transgenerational hypomethylation and modifications to IEGs crucial to synaptic plasticity, observed following neonatal sevoflurane exposure could contribute to morphological and cognitive deficits known to occur with neonatal sevoflurane exposure.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In 2016, after a comprehensive analysis of published scientific studies, the Food and Drug Administration issued a Drug and Safety Communication stating that children exposed prenatally or postnatally to repeat or prolonged general anesthesia (GA) may be at risk for neurodevelopmental impairments [1]. We believe that this warning is warranted, as recent clinical evidence suggests that cumulative ≥ 2 h exposure to general anesthetics is associated with postoperative cognitive dysfunction, increased expression of serum inflammatory markers, and increased risk of learning disabilities [2,3,4]. The underlying mechanism(s) for impeded neurodevelopment and cognition is incompletely understood.
Exposure to the inhalational anesthetic sevoflurane during critical stages of brain development induces widespread neuronal apoptosis in various mammalian species [5,6,7,8]. Sevoflurane-induced neurotoxicity and synaptic morphological changes are associated with behavioral deficits exhibited as juveniles and adults [9, 10]. Importantly, cognitive deficits have been reported in unexposed offspring of dams exposed in utero, suggesting that exposure to anesthetics may result in a transgenerational epigenetic modification [12]. Epigenetic modulation is a nongenetic form of inheritance thought to be a key regulator of gene-environment interactions, regulating gene expression via “tags” such as DNA methylation and histone modification. Recent work has begun to investigate anesthesia-induced epigenetic modifications of histones and, to a lesser extent, DNA methylation within the hippocampus, to elucidate the involvement of such modifications in behavioral and neurodevelopmental abnormalities [13,14,15,16]. Recently, we showed GA-induced epigenetic modulation of hippocampal histone-3 via hypoacetylation [13]. This epigenetic histone modulation in addition to fragmentation of cAMP-responsive element binding protein (CREB) resulted in the transcriptional downregulation of bdnf and c-fos, immediate early genes (IEGs) crucial to normal neuronal morphological development [13].
Now, we aim to elucidate the role of DNA methylation in subiculum by examining long-lasting neuronal 5-methylcytosine (5-mC) and 5-hydroxymethylcytosine (5-hmC) DNA modifications resulting from early postnatal exposure to sevoflurane. DNA methylation is crucial to normal development and differentiation by acting as a suppressor of gene expression and is found in the promoter region of critical genes [17]. Additionally, a decrease in DNA methylation in the brain coincides with functional decline in learning and memory seen with aging [18]. DNA methylation occurs on the 5-carbon of cytosine to generate 5-mC; once formed, it is extremely stable and possesses a self-perpetuating capacity which allows its transmission to offspring [19, 20].
Focusing on the subiculum, a highly vulnerable brain region where sevoflurane is known to cause substantial neurotoxicity, we aim to elucidate epigenetic modulations of IEGs, arc and junB, critical for the formation of neuronal circuits and proper synaptic transmission [7, 8]. The subiculum plays an important role in processing and integration of information relayed to other cortical areas. It is vital to synaptic transmission and anatomical connectivity and is intertwined with both the entorhinal and cingulate cortices, anterior thalamic nuclei, and hippocampal CA1 region [21,22,23]. Alterations in the subiculum can directly affect spatial navigation, mnemonic processing, and response to stress [22, 24, 25]. Synaptic plasticity changes in the brain, including those that support memory formation, have been functionally linked to the induction of IEGs via neuronal stimulation [26,27,28].
We show that neonatal sevoflurane exposure causes aberrant, transgenerational DNA methylation in subiculum of offspring which might influence transcription of IEGs, arc and junB, that are essential to synaptic plasticity and neuronal physiology. We further examine the relationship between altered global subicular DNA methylation and promotor-specific DNA methylation changes in target IEG, arc.
Materials and Methods
Animals and Anesthesia Administrations
Animals were purchased from Envigo (USA). These experiments were approved by the Animal Use and Care Committee of the University of Colorado Anschutz Medical Campus, the Office of Laboratory Animal Resources (OLAR), Aurora, CO. All experiments were conducted in accordance with Public Health Service’s Policy on Humane Care and Use of Laboratory Animals. Animals were maintained on a 12-h light cycle and were given free access to food and water. Efforts were made to minimize the number of animals used while being able to conduct meaningful sex difference statistical analyses.
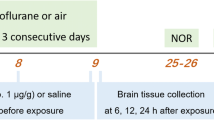
Postnatal day 7 (PND7), Sprague-Dawley rats of equal sex were exposed to sevoflurane at 3% for 2 h followed by 2.4% for 4 h in continuous 30% oxygen/air mixture, with vehicle group exposed to 30% oxygen/air mixture for 6 h. For the duration of exposure to sevoflurane or air, pups were separated from their mother and placed in closed chambers maintained at 34–35 °C. Sevoflurane was administered by calibrated agent-specific vaporizer; gas levels in both sevoflurane and vehicle chambers were monitored in real-time feedback (Datex Capnomac Ultima). After exposure, pups were allowed to recover before being returned to their mothers. Rats were aged to PND28 and sacrificed for whole brain or subiculum collection. A subset of Gen0 females, from sevoflurane and vehicle treatment groups, was separated for future role as breeders. Rats exposed to sevoflurane and vehicle constitute Gen0 experimental and control group, respectively; the first litter born to females of Gen0 was classified as Gen1.
Histological Preparation and Immunohistochemistry
At PND28, rats from Gen0 and Gen1 were transcardially perfused with phosphate buffer solution (PBS at pH 7.4) followed by 4% paraformaldehyde (PFA). Whole brains were extracted and postfixed in 4% PFA for 24 h. Coronal sections (3 mm thick) of forebrain containing the hippocampus were processed and embedded in paraffin. Serial 5-μm sections of subiculum (coordinates approximately Bregma − 4.20 mm through − 5.40 mm, Lambda 3.40 mm through 2.20 mm) were cut using Leitz 1512 Rotary Microtome. Sections were deparrafinized before antigen retrieval with sodium citrate buffer (pH 6.0) was performed. Tissue sections were washed three times in PBS followed by incubation in PBS, containing 1% glycine and 0.01% Triton X-100 for 15 min, to increase membrane permeabilization. Following 5 min of PBS wash, tissue sections were blocked in PBS containing 5% donkey serum and 0.01% Triton-X-100 for 1 h before immediately places in primary antibody solution, consisting of PBS, 0.01% Triton X-100, and mix of primary antibodies 5-mC (mouse monoclonal anti-5mC antibody, 1:200; Gene Tex (#GTX629448), CA, USA), 5-hmC (rabbit anti-5-hmC antibody; Active Motif (#39769), CA, USA), and NeuN (chicken anti-NeuN antibody; ABN91, EMD Millipore Corp, MA, USA) for 72 h at 4 °C. A subset of animals was used as experimental controls to test for nonspecific binding. This subset was incubated in PBS containing 1% donkey serum and 0.01% Triton-X-100 instead of the designated primary antibodies. Unbound primary antibodies were removed via three PBS, 0.01% Triton X-100 washes; samples were then washed 5 min with PBS then incubated with corresponding Alexa Fluor secondary antibodies (1:500, anti-rabbit Alexa Fluor 488 (A21206, Invitrogen), anti-mouse Alexa Fluor 555 (ab150106, Abcam), and anti-chicken Alexa Fluor 647 (AP194SA6; Millipore)) for 2 h at room temperature. Tissue sections were washed three times with PBS, allowed to dry and mounted with DAPI containing mounting medium (Vectashield, Vetor Laboratories, Inc., CA, USA).
Intensity of 5-mC (Fig. 1a, b, d, e) and 5-hmC (Fig. 2a, b, d, e) was determined in subicular neuronal nuclei for Gen0 and Gen1 using merged images of DAPI, NeuN, 5-mC, and 5-hmC staining at 20X magnification (Olympus Fluoview FV1200 Laser Scanning Confocal Microscope; Olympus Life Science, MA, USA). Images were analyzed using Image ProPlus. The dorsal subiculum was visually located, and a predetermined area was defined before individual neuronal nuclei area were determined using DAPI and NeuN as markers. 5-mC and 5-hmC intensities were determined for each neuronal-nuclei within the defined subicular area. An average of 79 neurons per subiculum with 2–6 subicular sections per rat was analyzed. The average 5-mC and 5-hmC for an individual rat is presented as a single point on corresponding graphs (Fig. 1c, f and 2c, f). Prolonged primary antibody incubation was determined not to cause nonspecific binding (Fig. 1g and 2g).
A single-prolonged sevoflurane exposure results in subicular neuron 5-mC DNA hypomethylation. 5-mC DNA staining of subicular neuronal nuclei (a, b, d, e) was quantified using density/intensity of individual cells within a defined subicular area (c, f). Representative images display DAPI (blue), NeuN (white), and 5-mC (red); scale bar represents 100 μm. No effect of sex was observed. An effect of treatment was transgenerationally observed as hypomethylation of rats exposed to sevoflurane (c) and offspring born to exposed mothers (f). Data points of c and f are representative of individual animals’ mean 5-mC intensities. Data is present as mean ± SE; data was analyzed via two-way ANOVA. *p < 0.05. Prolonged incubation did not result in nonspecific binding, as shown by control slide (g) with no primary antibody 5-mC and NeuN
A single-prolonged sevoflurane exposure does not alter subicular neuron 5-hmC DNA methylation status. 5-hmC DNA staining of subicular neuronal-nuclei (a, b, d, e) were quantified using density/intensity of individual cells within a defined area (c, f). Representative images display DAPI (blue), NeuN (white), and 5-hmC (green); scale bar represents 100 μm. No effect of sex or treatment was observed (c, f). Data points of c and f are representative of individual animals’ mean 5-hmC intensities. Data is presented as mean ± SE; data was analyzed via two-way ANOVA. Prolonged incubation did not result in nonspecific binding, as shown by control slide (g) with no primary antibody 5-mC and NeuN
Real-Time RT-PCR
At PND28, Gen0 and Gen 1 rats were transcardially perfused with ice-cold PBS, brains were removed, and subicula were isolated. Subicular tissue from 2 to 3 brains of same sex and treatment group was pooled into one sample, flash frozen in liquid nitrogen, and stored at − 80 °C. RNA was isolated using RNeasy Mini Kit (Qiagen, Germany), and concentrations were determined spectrophotometrically (NanoDrop One; Thermo Scientific, MA, USA). Reverse transcriptase was performed on 500 ng of RNA using iScript cDNA Synthesis Kit (Bio-Rad, CA, USA). Real-time polymerase chain reaction (PCR) primers JunB, Arc, and GAPDH were designed and purchased from Bio-Rad (10025636, CA, USA). Quantitative real-time RT-PCR reaction mixture consisted of 100 ng cDNA, 1× SsoAdvanced Universal SYBR Green Supermix (Bio-Rad, CA, USA), gene specific 1× PrimPCR assay (Primer; Bio-Rad, CA, USA), and nuclease-free water. The following PCR protocol was performed using CFX Connect Real-Time System (Bio-Rad CA, USA): 95 °C for 2 min followed by 40 cycles of 95 °C for 5 s and 60 °C for 30 s, ending with a melting cure performed 65–95 °C (0.5 °C increments) at 5 s per step. PCR was performed in triplicate for target of interest (JunB and Arc) and reference target (GAPDH); threshold cycle numbers were averaged. The expression of target gene was normalized to GAPDH expression and relative value calculated using the 2(−ΔCT) method [29]. For statistical analysis, mRNA levels were expressed as fold change to male vehicle group that was assigned the value 1.
Arc Promotor CpG Methylation Analysis
At PND28, Gen0 and Gen 1 rats were transcardially perfused with ice-cold PBS, brains were removed, and subicula were isolated. Subicular tissue from 2 to 3 brains of same sex and treatment group was pooled into one sample, flash frozen in liquid nitrogen, and stored at − 80 °C. gDNA was isolated using DNeasy Blood and Tissue Kit (Qiagen, Germany); concentrations were determined spectrophotometrically (NanoDrop One; Thermo Scientific, MA, USA). Following gDNA extraction, samples were pyrosequencing by EpigenDx (Worcester, MA, USA) using their validated arc methylation assay [30, 31]. CpG methylation at sites − 29 through − 1 and 10–19, in reference to mRNA start codon (Chr7:115910602–115910070) of arc promoter region, was quantified by pyrosequencing of bisulfite-treated genomic DNA (gDNA).
The following methods were performed by EpigenDx. Five hundred nanograms of the extracted gDNA was bisulfite treated using EZ DNA Methylation kit (Zymo Research, Inc., CA, USA). Using 1 μL of bisulfite treated DNA and 0.2 μM of each primer PCR was performed, followed by HPLC purification of PCR products via the use of one biotin-labeled primer and Sepharose beads. After purification, PCR products still bound to Streptavidin Sepharose HP (GE Healthcare Life Sciences, MA, USA) were washed, denatured with a 0.2-μM NaOH solution, and rewashed using the Pyrosequencing Vacuum Prep Tool (Pyrosequencing, Qiagen, Germany), as per the manufacturer’s protocol. Purified single-stranded PCR products were then annealed with 0.5 μM of sequencing primer followed by pyrosequencing on the PSQ96 HS System (Pyrosequencing, Qiagen, Germany).
The methylation status of each CpG site was determined individually as an artificial C/T SNP using QCP software (Pyrosequencing, Qiagen, Germany). The methylation level at each CpG site was calculated as the percentage of the methylated alleles divided by the sum of all methylated and unmethylated alleles. Each experiment included non-CpG cytosines as internal controls to detect incomplete bisulfite conversion of the input DNA. In addition, a series of unmethylated and methylated DNA are included as controls in each PCR. Furthermore, PCR bias testing was performed by mixing unmethylated control DNA with in vitro methylated DNA at different ratios (0%, 5%, 10%, 25%, 50%, 75%, and 100%), followed by bisulfite modification, PCR, and pyrosequencing analysis. Five separate pyrosequencing assays were conducted by EpigenDx for methylation analysis of arc promoter region; ADS3867-FS1, ADS3867-FS2, ADS3867-FS3, ADS3867-FS4, ADS6522-FS2, and ADS6522-FS3. In reference to arc’s ATG, mRNA start codon site, the pyrosequencing assays examined the methylation status of CpGs located from − 277 to + 256.
Statistical Analysis
All data are presented as mean ± SE and analyzed by GraphPad Prism 7. Two-way ANOVA multiple comparison with Tukey post hoc and 95% confidence interval test for comparison of sex and treatment differences was used unless noted otherwise. Brightness and contrast of example images in Fig. 1 and 2 were adjusted for optimal view; for analysis, no adjustments were made to obtained images. Average methylation of CpG regions was analyzed using multiple unpaired t test, using an option of fewer assumptions in calculating individual p values for higher reliability.
Results
Sevoflurane Decreases Subicular Neuronal DNA Methylation
We set out to examine the previously uninvestigated potential of sevoflurane to alter DNA methylation in the subiculum. Modification of global DNA methylation was examined via IHC staining for 5-mC (Fig. 1) and 5-hmC (Fig. 2). Gen0 results revealed no significant effect of sex on 5-mC (Fig. 1a–c) and 5-hmC (Fig. 2a–c) signal intensity within the nucleus of subicular neurons. However, our 5-mC results demonstrate an effect of treatment (Fig. 1a–c), as sevoflurane-exposed rats (Gen0) exhibit a significant reduction of 5-mC (*p = 0.0124; Fig. 1c) in comparison to the vehicle group. To ensure that active demethylation was not occurring at the age examined (PND 28), 5-hmC was also examined (Fig. 2a–c). 5-hmC is hypothesized to be a potential key intermediate in an active DNA demethylation process [32]. Sevoflurane-exposed Gen0 demonstrate no difference in 5-hmC when compared to vehicle (p = 0.5190; Fig. 2c), suggesting that sevoflurane exposure at early postnatal age caused a stable global subicular-neuronal decrease in 5-mC DNA methylation in rat subiculum.
Since DNA methylation is known to be stable and has the potential to be inherited, we hypothesized that DNA hypomethylation observed in Gen0 could be transgenerational. To test this hypothesis, Gen0 females were bred and offspring were aged to PND28, termed Gen1. Our Gen1 results reveal no significant effect of sex on 5-mC (Fig. 1d–f) and 5-hmC (Fig. 2d–f) signal intensities within the nucleus of subicular neurons. However, similar to Gen0, our Gen1 5-mC results demonstrate an effect of treatment (*p = 0.0266; Fig. 1f), indicating that sevoflurane-exposed rats exhibit transgenerational reduction of 5-mC. To investigate if active demethylation was occurring in Gen1, 5-hmC levels were quantified and we concluded that there was no difference between rats born to mothers exposed to sevoflurane or vehicle treatment (p = 0.2908; Fig. 2f). Our data suggest that neonatal exposure to sevoflurane may cause a transgenerational global hypomethylation, expressed as a decreased level of 5-mC in subicular neurons.
Comparing PND28 rat weight between sex, treatment, and generation, no difference was observed (data not shown).
Sevoflurane Exposure Results in Sex-Specific Transgenerational Upregulation of Target IEGs
Since volatile anesthetics impair synaptic integrity during early stages of brain development, we hypothesized that sevoflurane may alter expression of IEGs that support synaptic plasticity, arc, and neuronal physiology, junB [27, 30, 33].
We observed a main effect of treatment for JunB expression, in which Gen0 sevoflurane–exposed rats exhibit an average increase of 35.2% in JunB mRNA expression (**p = 0.00524; Fig. 3a). There was also a main effect of treatment for Arc expression; Gen0 sevoflurane–exposed rats show an average increase of 42.7% in Arc mRNA expression (Gen0, ***p = 0.0004; Fig. 3c). Additionally, a main effect of sex was observed for Arc mRNA expression, as exhibited by females displaying an overall higher degree of expression compared to males (*p = 0.0242; Fig. 3c).
Sevoflurane exposure induces sex-specific transgenerational upregulation of IEGs, JunB, and Arc mRNA. RT-qPCR was performed on isolated PND28 rat subiculum. Two-way ANOVA with Tukey’s post hoc test demonstrates a sevoflurane-induced effect of treatment shown as upregulation of JunB (a) and Arc (c) mRNA, as indicated by the bracket between vehicle and sevoflurane. Additionally, two-way ANOVA reveals an effect of sex for Gen0 Arc mRNA expression (c) as displayed by increased expression in females over males. Upregulation of JunB and Arc is transgenerationally expressed exclusively in male offspring (Gen1, JunB b, and Arc d). Data points are representative of individual rats. Data is presented as mean ± SE. Sevoflurane versus vehicle *p < 0.05, **p < 0.01. Males versus females #p < 0.05
Based on Gen0 sevoflurane-induced upregulation of JunB and Arc, the potential of transgenerational upregulation of these genes was investigated with offspring born to exposed female rats. Both JunB and Arc Gen1 two-way ANOVA revealed a statistically significant interaction of *p = 0.0183 and *p = 0.0152, respectively. Simple main effects revealed that males (Gen1) born to sevoflurane-exposed mothers had a 74% increase in JunB mRNA expression (**p = 0.0029; Fig. 3b) and 71.6% increase in Arc mRNA expression (**p = 0.003; Fig. 3d), while their female cohort (Gen1) expressed no difference (Fig. 3b, d).
Sevoflurane Exposure Modulates Methylation of arc Promoter
To further our DNA methylation studies, we chose to focus on methylation status of the arc promoter. Arc is a key IEG that directly impacts synaptic plasticity, long-term potentiation (LTP), consolidation of memory, and synaptic weakening and has been implicated in neurological disorders [30, 31, 34]. We performed pyrosequencing to examine the association between global subicular-neuronal DNA hypomethylation and the upregulation of Arc mRNA expression following sevoflurane exposure. CpG sites − 29 through 19, in relation to ATG site, of arc promoter region were examined for altered methylation status by creating a heat map (Fig. 4a). From the arc promoter heat map (Fig. 4a), we observed transgenerationally expressed regions of no methylation for 12CpGs between all sex and treatment groups. Additionally, it was observed that exposure to sevoflurane causes a diverse effect on arc methylation of individual CpGs. Sevoflurane-induced average difference was determined for each CpG in comparison to corresponding vehicle. These values were then expressed as %CpG that displayed an average increase, decrease, or no change in methylation (Fig. 4b–e). In Gen0 males when compared to vehicle males, sevoflurane treatment exhibited no change in methylation of 33.3% CpGs, an increased methylation of 35.9% CpGs, and decreased methylation of 30.7% CpGs (Fig. 4b). Sevoflurane treatments of Gen0 females when compared to their vehicle counterpart exhibited no effect of 30.77% of the CpGs, increased methylation of 23.08% CpGs, and decreased methylation of 46.15% CpGs (Fig. 4c).
Mean arc CpG methylation displayed with heat map and mean difference pie charts. Pyrosequencing was performed on isolated PND28 rat subicula for the methylation status of 39 CpGs. Arc’s promoter sequence − 277 through + 256 in relation to the mRNA start codon (ATG) was divided into three sections: (1) 5-upstream—CpGs prior to transcriptional start site (TSS); (2) 5-untranslated region (5-UTR)—CpGs located after TSS and before mRNA ATG; and (3) exon 1—the CpGs after ATG (a). The scale bar represents the percent methylation of individual CpGs for designated groups; red indicates 0% methylation with purple equal to 9% methylation. Data is displayed as vehicle followed by their corresponding sevoflurane group for males and females. Generation 0 is displayed as the left 4 columns and Gen1 as the right 4 columns. Columns correspond to above brackets and labels. Additionally, data is presented as the mean methylation difference of the sevoflurane treatment to its corresponding vehicle for each sex and generation (b, c, d, e). The mean difference is displayed as sevoflurane-induced % CpG increase (gray), decrease (black), and no change (white) in methylation. n = 5 per each group
Next, we examined sevoflurane-induced transgenerational modifications of arc promoter in Gen1. Gen1 sevoflurane males compared to vehicle males displayed no methylation difference of 23.08% CpGs, increased methylation of 38.46% CpGs, and decreased methylation of 38.46% CpGs (Fig. 4d). Gen1 sevoflurane females compared to vehicle females displayed no methylation difference of 28.21% CpGs, increased methylation of 38.46% CpGs, and decreased methylation of 33.33% CpGs (Fig. 4e).
Arc’s promoter sequence − 277 through + 256 in relation to the ATG (Fig. 5a) was divided into three sections: (1) 5-upstream—CpGs prior to transcriptional start site (TSS); (2) 5-untranslated region (5-UTR)—CpGs located after TSS and before mRNA ATG; and (3) exon 1—the CpGs after ATG (as previously shown in Fig. 4a). We compared the average methylation within these three regions for vehicle and sevoflurane-exposed rats. No change in the methylation of the three regions of the arc promoter was observed in sevoflurane-exposed Gen0 males compared to vehicle (Fig. 5b). As shown in our heat map (Fig. 4a), the prominent sevoflurane-induced DNA methylation decrease occurs in the promoter 5-upstream region of Gen 0 females. When quantified, we confirm that sevoflurane-exposed females have significantly reduced methylation of the 5-upstream region of arc promoter (Fig. 5c; *p = 0.038). This decrease in arc promoter 5-upsteam CpG methylation parallels with observed upregulation of Arc mRNA expression in Gen0 females (Fig. 3c). In relation to the observed sevoflurane-induced methylation difference of Gen1 males and females (Fig. 4d, e), it was revealed that the altered arc promoter methylation does not appear to be focused in one region as shown by no statistical difference in comparison to the vehicles for the three arc promoter region investigated (Fig. 5d, e). It should be noted that arc promotor is overall modestly methylated, which is consistent with previous studies [30, 31].
Postnatal sevoflurane exposure reduces arc 5-upstream CpG promoter methylation in females exposed. Pyrosequencing was performed on isolated PND28 rat subicula. Percent methylation of 39 arc promoter CpGs was determined and grouped based upon the promoter region. Using Rnor_6.0 sequence, location referenced to the ATG (indicated by brackets, a), CpGs were divided into three categories, 5-upstream of the TSS (as indicated by arrow, CpG − 29 through − 21), 5-UTR (CpG − 20 through − 1), and exon 1 that occurs after arc’s ATG sequence (CpG 10 through 19). Data represents the average percent of methylation. Gen0 males (b) display no difference between vehicle and sevoflurane average percent of methylation for any CpG ranges. Gen0 females (c) display a decrease in methylation from sevoflurane in the promoter region 5-upstream of the transcription start site. Gen1 males (d) and females (e), offspring of Gen0 females, display no difference in methylation when averaged across the different CpG ranges. Data points are representative of individual rats. Data is presented as mean ± SE. Data was analyzed via multiple unpaired t test. *p < 0.05
Discussion
In our study, we show that sevoflurane exposure of rats during critical stages of their brain development causes substantial transgenerational epigenetic modifications manifested as global subicular-neuronal hypomethylation and upregulation of IEGs, JunB and Arc mRNA. With the observed subicular-neuronal DNA hypomethylation in Gen0 female, only the arc promoter 5-upstream region mimics the global subicular DNA methylation change.
DNA methylation is critical for synaptic plasticity and memory formation [35, 36]. Aberrant DNA methylation is linked with age-associated functional decline of learning and memory, as well as several disease states such as myelodysplastic syndrome, cancer, and atherosclerosis [18].
We report herein that a single prolonged exposure of PND7 rats to sevoflurane causes significant epigenetic reduction of 5-mC within subicular neurons. Growing number of studies suggests that anesthesia induces a variety of epigenetic modifications, which begs the question how and whether they are related to cognitive deficits reported to be associated with prolonged and cumulative (> 2 h) anesthesia exposures [3, 4, 12, 13, 20]. Transgenerational nature of sevoflurane-induced hypomethylation in subicular neurons suggests that sevoflurane causes epigenetic modification in not only subicular somatic cells but also oocyte germ cells as well. Indeed, a recent study showed that postnatal exposure to sevoflurane alters methylation status of KCC2 gene in male germ cells, thus supporting the hypothesis that sevoflurane is a powerful epigenetic modulator with transgenerational influence [20]. Further studies are needed to investigate the mechanisms contributing to DNA hypomethylation and transmittal to its offspring as relevant to anesthesia-induced long-lasting cognitive impairments.
DNA methylation acts as a suppressor of gene expression. Therefore, hypomethylation is predicted to increase gene transcription [37]. The inherently low methylation status of the arc promotor reported by us and others may explain its robust transcription capabilities [30, 31]. The mechanisms contributing to sex differences in arc promotor methylation and gene expression remain unclear. If altered promoter methylation of arc is responsible for the observed increase in Arc mRNA expression, it would suggest that there is more significance placed on the 5-upstream region of arc promoter over 5-UTR and exon 1. Hypomethylation of the 5-upstream promotor region does not appear to be transmitted to offspring which could, at least in part, be explained by nonheritability of aberrant somatic cell methylation. Lack of transgenerational arc promoter hypomethylation and observed transgenerational upregulation of Arc mRNA in Gen1 males indicates other potential mechanisms at play.
For instance, recent findings suggest that promoter DNA methylation associated lack of gene activity could be mediated through attraction or repulsion of regulatory transcription factors. DNA methylation-mediated regulation factors are dependent upon genomic context, such as CpG variations, TF sites, and transcriptional regulators [38]. Additionally, histone modification can act as epigenetic regulators. However, neuronal activity controls arc transcription, Arc mRNA trafficking and accumulation, and Arc protein translation, localization, and stability [39]. Proper hippocampus function relies upon neuronal activity–induced transcription factors to make discrete changes in gene expression [40]. The increase in Arc mRNA expression could be the result of increased subicular neuronal activity by synaptic reorganization or synaptic disruption in the hippocampus.
The development of neural networks requires appropriate response to stimuli and is essential to the subicular processing and integration of information that is relayed to other cortical areas. IEGs are thought to function as experience-dependent modifiers of the underlying synaptic circuitry [41]. Arc, specifically, is a member of IEGs that responds to experience-driven stimuli via rapid and selective upregulation in specific neural ensembles [34]. We report an upregulation of Arc mRNA in sevoflurane-exposed Gen0 of both sexes and male offspring of Gen0 females. Moreover, Arc mRNA plays a crucial role in maintenance phase of LTP, consolidation of memory, and enduring synaptic plasticity as well as in synaptic plasticity of activity-induced synaptic weakening [30, 31]. Synaptic weakening encompasses the homeostatic downscaling of synapses, metabotropic glutamate receptor–induced long-term synaptic depression (mGluR-LTD), and synapse elimination [34]. Arc’s role as an activity-dependent synapse eliminator allows for refinement of circuit connections via the removal of redundant or inappropriate connections. Sevoflurane-induced transgenerational upregulation of Arc mRNA could potentially be the result of increase neuronal activity within the subiculum signaled from the hippocampus due to synaptic disruption. GluA1 C-terminal palmitoylation-deficient (GluA1C811S) mice revealed increased Arc expression as the potential cause for unstable neural circuits and excess excitation [42]. It is thought that abnormal consolidation of Arc positive neural ensembles during learning may lead to cognitive deficits such as those seen in autism and intellectual disability [34]. It remains to be examined whether similar correlation could explain anesthesia-induced cognitive impairments.
There are two different types of IEGs. Type 1, like arc, encodes for proteins with direct implications in cell structure and signal transduction. Type 2, like junB, regulates the expression of downstream late-response genes involved in neuronal physiology [27]. Both types of IEGs respond to neuronal stimulation and are essential to regulation of the cells response to received stimuli, playing a central role in controlling mechanisms of plasticity at the neuronal, behavioral, and perceptual level [27]. In our study, we observed both types of IEGs being transgenerationally upregulated, which is possible due to sevoflurane-induced modification of neuronal response to stimulation.
Sevoflurane induces a sexually dimorphic transgenerational upregulation of JunB mRNA, expressed exclusively in male offspring (Gen1 males). Normal expression of JunB has been shown to be essential to genetic stability and is implicated in cell survival, cell proliferation, and programmed cell death and senescence [33]. Research has shown that exposure to sevoflurane results in decreased synaptic density, filopodia length, neuronal spines, and spine density of apical dendrites [11, 43, 44]. Inappropriate transcription of junB and arc may underlie dysregulated synaptic growth. Future studies to investigate the methylation status of the junB gene and the impact of increased IEG expression following sevoflurane exposure are warranted by our findings.
In conclusion, we report that postnatal exposure to sevoflurane causes transgenerational epigenetic changes that are expressed as global subicular-neuronal hypomethylation in Gen0 offspring. This hypomethylation has the potential to directly and indirectly influence, via regulation of targets transcription, the upregulation of Arc and JunB mRNA. These transgenerational modifications to components crucial to synaptic plasticity could partially contribute to morphological and cognitive deficits known to occur with neonatal sevoflurane exposure. However, further studies are required to directly link observed epigenetic modifications and dysregulation of IEGs to anesthesia-induced neurotoxicity and synaptic morphological and myriad behavioral deficits [8, 10, 11, 21, 45,46,47].
Abbreviations
- Gen0:
-
Generation 0
- Gen1:
-
Generation 1
- GA:
-
General anesthesia
- 5-mC:
-
5-Methylcytosine
- 5-hmC:
-
5-Hydroxymethylcytosine
- CpG:
-
Cytosine-phosphate-guanine
- IEGs:
-
Immediate early genes
References
U.S. Food & Drug Administration. (2016) FDA Drug Safety Communication: FDA review results in new warnings about using general anesthetics and sedation drugs in young children and pregnant women. https://www.fda.gov/drugs/drug-safety-and-availability/fda-drug-safety-communication-fdareview-results-new-warnings-about-using-general-anesthetics-and. Accessed 01 Dec 2018
Fan CH, Peng B, Zhang FC (2018) The postoperative effect of sevoflurane inhalational anesthesia on cognitive function and inflammatory response of pediatric patients. Eur Rev Med Pharmacol Sci 22:3971–3975
Wilder RT, Flick RP, Sprung J, Katusic SK, Barbaresi WJ, Mickelson C, Gleich SJ, Schroeder DR et al (2009) Early exposure to anesthesia and learning disabilities in a population-based birth cohort. Anesthesiology 110:796–804. https://doi.org/10.1097/01.anes.0000344728.34332.5d
Flick RP, Katusic SK, Colligan RC, Wilder RT, Voigt RG, Olson MD, Sprung J, Weaver AL et al (2011) Cognitive and behavioral outcomes after early exposure to anesthesia and surgery. Pediatrics 128:273–292. https://doi.org/10.1542/peds.2011-0351
Shen X, Xiao Y, Li W, Chen K, Yu H (2018) Sevoflurane anesthesia during pregnancy in mice induces hearing impairment in the offspring. Drug Des Devel Ther 12:1827–1836
Ozer A, Ceribasi S, Ceribasi A et al (2017) Effects of sevoflurane on apoptosis, BDNF and cognitive functions in neonatal rats. Bratisl Med J 118:80–84. https://doi.org/10.4149/BLL
Shih J, May L d V, Gonzalez HE et al (2012) Delayed environmental enrichment reverses sevoflurane-induced memory impairment in rats. Anesthesiology 116:586–602. https://doi.org/10.1038/jid.2014.371
Satomoto M, Satoh Y, Terui K, Miyao H, Takishima K, Ito M, Imaki J (2009) Neonatal exposure to sevoflurane induces abnormal social behaviors and deficits in fear conditioning in mice. Anesthesiology 110:628–637. https://doi.org/10.1097/ALN.0b013e3181974fa2
Zhang DX, Jiang S, Yu LN et al (2015) The effect of sevoflurane on the cognitive function of rats and its association with the inhibition of synaptic transmission. Int J Clin Exp Med 8:20853–20860 https://doi.org/10.1016/j.anai.2009.10.002
Shen X, Dong Y, Xu Z, Wang H, Miao C, Soriano SG, Sun D, Baxter MG et al (2013) Selective anesthesia-induced neuroinflammation in developing mouse brain and cognitive impairment. Anesthesiology 118:502–515. https://doi.org/10.1097/ALN.0b013e3182834d77
Tao G, Luo Y, Xue Q, Li G, Tan Y, Xiao J, Yu B (2016) Docosahexaenoic acid rescues synaptogenesis impairment and long-term memory deficits caused by postnatal multiple sevoflurane exposures. Biomed Res Int 2016:1–7. https://doi.org/10.1155/2016/4062579
Chalon J, Tang CK, Ramanathan S et al (1981) Exposure to halothane and enflurane affects learning function of murine progeny. Anesth Analg 60:794–797. https://doi.org/10.1097/IAE.0b013e3181dde5f5
Massara LD, Osuru HP, Ph D et al (2016) General anesthesia causes epigenetic histone modulation of c-Fos and brain-derived neurotrophic factor, target genes important for neuronal development in the immature rat hippocampus. Am Sciety Anesthesiol 124:1311–1327
Jia M, Ji M, Yang J (2017) Epigenetic regulation of general anesthesia-induced neonatal neurodegeneration. Oncotarget 8:5652–5653
L sha J, Jia M, Sun J et al (2016) Hypermethylation of hippocampal synaptic plasticity-related genes is involved in neonatal sevoflurane exposure-induced cognitive impairments in rats. Neurotox Res 29:243–255. https://doi.org/10.1007/s12640-015-9585-1
Joksimovic SM, Osuru HP, Oklopcic A, Beenhakker MP, Jevtovic-Todorovic V, Todorovic SM (2018) Histone deacetylase inhibitor entinostat (MS-275) restores anesthesia-induced alteration of inhibitory synaptic transmission in the developing rat hippocampus. Mol Neurobiol 55:222–228. https://doi.org/10.1007/s12035-017-0735-8
Oliveira AMM (2016) DNA methylation: a permissive mark in memory formation and maintenance. Learn Mem 23:587–593. https://doi.org/10.1101/lm.042739.116
Liu L, van Groen T, Kadish I, Tollefsbol TO (2009) DNA methylation impacts on learning and memory in aging. Neurobiol Aging 30:549–560. https://doi.org/10.1016/j.neurobiolaging.2007.07.020
Smeester L, Rager JE, Bailey KA et al (2017) An overview of epigenetic assays. J Neurosci 8:e0163690. https://doi.org/10.1371/journal.pone.0163690
Ju LS, Yang JJ, Morey TE, Gravenstein N, Seubert CN, Resnick JL, Zhang J (2018) Role of epigenetic mechanisms in transmitting the effects of neonatal sevoflurane to the next generation of male, but not female, rats. Br J Anaesth 121:406–416
Sanchez V, Feinstein SD, Lunardi N, Joksovic PM, Boscolo A, Todorovic SM, Jevtovic-Todorovic V (2011) General anesthesia causes long-term impairment of mitochondrial morphogenesis and synaptic transmission in developing rat brain. Anesthesiology 115:992–1002. https://doi.org/10.1097/OPX.0b013e3182540562.The
O’Mara S (2005) The subiculum: what it does, what it might do, and what neuroanatomy has yet to tell us. J Anat 207:271–282. https://doi.org/10.1111/j.1469-7580.2005.00446.x
Behr J, Wozny C, Fidzinski P, Schmitz D (2009) Synaptic plasticity in the subiculum. Prog Neurobiol 89:334–342. https://doi.org/10.1016/J.PNEUROBIO.2009.09.002
McNaughton N (2006) The role of the subiculum within the behavioural inhibition system. Behav Brain Res 174:232–250. https://doi.org/10.1016/j.bbr.2006.05.037
Lynch MA (2004) Long-term potentiation and memory. Physiol Rev 84:87–136. https://doi.org/10.1098/rstb.2002.1230
Tischmeyer W, Grimm R (1999) Activation of immediate early genes and memory formation. Cell Mol Life Sci 55:564–574
Sommerlandt FMJ, Brockmann A, Rössler W, Spaethe J (2018) Immediate early genes in social insects: a tool to identify brain regions involved in complex behaviors and molecular processes underlying neuroplasticity. Cell Mol Life Sci 76:1–15. https://doi.org/10.1007/s00018-018-2948-z
Srivas S, Thakur MK (2017) Epigenetic regulation of neuronal immediate early genes is associated with decline in their expression and memory consolidation in scopolamine-induced amnesic mice. 5107–5119. https://doi.org/10.1007/s12035-016-0047-4
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3:1101–1108. https://doi.org/10.1038/nprot.2008.73
Penner MR, Roth TL, Chawla MK et al (2011) Age-related changes in Arc transcription and DNA methylation within the hippocampus. Neurobiol Aging 32:2198–2210. https://doi.org/10.1038/jid.2014.371
Dyrvig M, Hansen HH, Christiansen SH, Woldbye DPD, Mikkelsen JD, Lichota J (2012) Epigenetic regulation of Arc and c-Fos in the hippocampus after acute electroconvulsive stimulation in the rat. Brain Res Bull 88:507–513. https://doi.org/10.1016/j.brainresbull.2012.05.004
Zhang L y, Li P l, Wang T z, Zhang X c (2015) Prognostic values of 5-hmC, 5-mC and TET2 in epithelial ovarian cancer. Arch Gynecol Obstet 292:891–897. https://doi.org/10.1007/s00404-015-3704-3
Hicks MJ, Hu QP, Macrae E, DeWille J (2015) Mitogen-activated protein kinase signaling controls basal and oncostatin M-mediated JUNB gene expression. Mol Cell Biochem 403:115–124. https://doi.org/10.1007/s11010-015-2342-1
Wilkerson JR, Albanesi JP, Huber KM (2018) Roles for Arc in metabotropic glutamate receptor-dependent LTD and synapse elimination: Implications in health and disease. Semin Cell Dev Biol 77:51–62. https://doi.org/10.1016/j.semcdb.2017.09.035
Alberini CM (2014) Transcription factors in long-term memory and synaptic plasticity. Physiol Rev 89:1–46. https://doi.org/10.1152/physrev.00017.2008.Transcription
Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y et al (2013) Global epigenomic reconfiguration during mammalian brain development. Science (80- ) 341:1237905. https://doi.org/10.1126/science.1237905
Bogdanović O, Veenstra GJC (2009) DNA methylation and methyl-CpG binding proteins: developmental requirements and function. Chromosoma 118:549–565. https://doi.org/10.1007/s00412-009-0221-9
Ambrosi C, Manzo M, Baubec T (2017) Dynamics and context-dependent roles of DNA methylation. J Mol Biol 429:1459–1475. https://doi.org/10.1016/j.jmb.2017.02.008
Korb E, Finkbeiner S (2011) Arc in synaptic plasticity: from gene to behavior. Trends Neurosci 34:591–598. https://doi.org/10.1016/j.tins.2011.08.007
Eagle AL, Gajewski PA, Robison AJ (2016) Role of hippocampal activity-induced transcription in memory consolidation. Rev Neurosci 27:559–573. https://doi.org/10.1515/revneuro-2016-0010.Role
Clayton DF (2000) The genomic action potential. Neurobiol Learn Mem 74:185–216. https://doi.org/10.1006/nlme.2000.3967
Itoh, Masayuki & Okuno, Hiroyuki & Yamada, Daisuke & Yamashita, Mariko & Abe, Manabu & Natsume, Rie & Kaizuka, Toshie & Sakimura, Kenji & Hoshino, Mikio & Mishina, Masayoshi & Wada, Keiji & Sekiguchi, Masayuki & Hayashi, Takashi. (2018). Perturbed expression pattern of the immediate early gene Arc in the dentate gyrus of GluA1 C‐terminal palmitoylation‐deficient mice. Neuropsychopharmacology Reports. 39.https://doi.org/10.1002/npr2.12044
Xiao H, Liu B, Chen Y, Zhang J (2016) Learning, memory and synaptic plasticity in hippocampus in rats exposed to sevoflurane. Int J Dev Neurosci 48:38–49. https://doi.org/10.1016/j.ijdevneu.2015.11.001
Zimering JH, Dong Y, Fang F, Huang L, Zhang Y, Xie Z (2016) Anesthetic sevoflurane causes rho-dependent filopodial shortening in mouse neurons. PLoS One 11:1–15. https://doi.org/10.1371/journal.pone.0159637
Jevtovic-Todorovic V, Hartman RE, Izumi Y, Benshoff ND, Dikranian K, Zorumski CF, Olney JW, Wozniak DF (2003) Early exposure to common anesthetic agents causes widespread neurodegeneration in the developing rat brain and persistent learning deficits. J Neurosci 23:876–882. https://doi.org/10.1523/JNEUROSCI.23-03-00876.2003
Loepke AW, Istaphanous GK, McAuliffe JJ et al (2009) The effects of neonatal isoflurane exposure in mice on brain cell viability, adult behavior, learning, and memory. Anesth Analg 108:90–104. https://doi.org/10.1017/S0031182016001955
Paule MG, Li M, Allen RR, Liu F, Zou X, Hotchkiss C, Hanig JP, Patterson TA et al (2011) Ketamine anesthesia during the first week of life can cause long-lasting cognitive deficits in rhesus monkeys. Neurotoxicol Teratol 33:220–230. https://doi.org/10.1016/j.ntt.2011.01.001
Funding
Supported in part by funds from the Department of Anesthesiology at the University of Colorado Anschutz Medical campus and R0144517, R0144517-S, R01 GM118197, R01 GM118197, and R21 HD080281, March of Dimes National Award, CU Medicine Endowment.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
These experiments were approved by the Animal Use and Care Committee of the University of Colorado Anschutz Medical Campus, the Office of Laboratory Animal Resources (OLAR), Aurora, CO. All experiments were conducted in accordance with Public Health Service’s Policy on Humane Care and Use of Laboratory Animals.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Chastain-Potts, S.E., Tesic, V., Tat, Q.L. et al. Sevoflurane Exposure Results in Sex-Specific Transgenerational Upregulation of Target IEGs in the Subiculum. Mol Neurobiol 57, 11–22 (2020). https://doi.org/10.1007/s12035-019-01752-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-019-01752-0