Abstract
The CD2-associated protein (CD2AP) rs9349407 polymorphism was first identified to be significantly associated with Alzheimer’s disease (AD) in European ancestry by genome-wide association studies (GWAS). However, the following studies reported no association in Chinese, Japanese, African-American, Canadian, and European populations. We think that these negative results may have been caused by either relatively small sample sizes compared with those used for the previous GWAS in European ancestry or the genetic heterogeneity of the rs9349407 polymorphism in different populations. Here, we reevaluated this association using the relatively large-scale samples from 15 previous studies (N = 54,936; 23,777 cases and 31,159 controls) by searching the PubMed, AlzGene, and Google Scholar databases. Using an additive genetic model, we did not identify a significant heterogeneity among the 15 studies. Using meta-analysis, we observed a significant association between the rs9349407 polymorphism and AD with P = 8.78E-07, odds ratio (OR) = 1.08, 95 % confidence interval (CI) 1.05–1.12. To our knowledge, this is the first meta-analysis to investigate the association between rs9349407 polymorphism and AD in East Asian, American, Canadian, and European populations. Our analysis further supports previous findings that the CD2AP rs9349407 polymorphism contributes to AD susceptibility. We believe that our findings will be very useful for future genetic studies on AD.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Alzheimer’s disease (AD) is a complex and the most common neurodegenerative disease in the elderly [1, 2]. It is estimated that genetic factors cause about 60–80 % of AD risk [3]. However, the specific genes influencing AD are largely unknown. Much effort has been put into identifying the genetic determinants of this disease. Genome-wide association studies (GWAS) have recently provided rapid insights into the genetics of AD and identified some common AD susceptibility variants or genes, which include complement receptor 1 (CR1); bridging integrator 1 (BIN1); clusterin (CLU); phosphatidylinositol binding clathrin assembly protein (PICALM); membrane-spanning 4-domains, subfamily A, member 4 (MS4A4)/membrane-spanning 4-domains, subfamily A, member 6E (MS4A6E); CD2-associated protein (CD2AP); CD33 molecule (CD33); EPH receptor A1 (EPHA1); and ATP-binding cassette transporter A7 (ABCA7) [4].
Two large-scale AD GWAS identified CD2AP rs9349407 polymorphism to be significantly associated with AD susceptibility in European ancestry with P = 8.6E-09, odds ratio (OR) = 1.11, 95 % confidence interval (CI) 1.07–1.15 [5, 6]. These two studies show that the carriers of rs9349407 risk variant have an about 11 % additional increased risk for AD [5, 6].
Considering the different genetic architecture, AD incidence rates, and environmental exposures across different ethnic populations, it is important to investigate whether rs9349407 polymorphism is associated with AD risk in other ethnic populations. The following studies also investigated this finding in Chinese, Japanese, African-American, European-American, European, and Canadian populations (Table 1). However, these studies reported consistent and inconsistent results (Table 1).
Recent studies investigated the mechanisms of rs9349407 in AD pathogenesis and indicated that rs9349407 was associated with neuritic plaque burden [7]. Based on recent findings above, we hypothesized that these negative results may have been caused by either relatively small sample sizes compared with those used for the previous GWAS in European ancestry or the genetic heterogeneity of the rs9349407 polymorphism in different populations. Here, we reevaluated this association using the relatively large-scale samples from 15 previous studies (N = 54,936; 23,777 cases and 31,159 controls) by searching the PubMed, AlzGene, and Google Scholar databases.
Methods and Materials
Literature Search
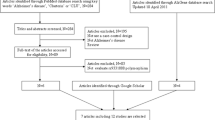
Guiyou Liu searched the PubMed (http://www.ncbi.nlm.nih.gov/pubmed) and AlzGene (http://www.alzgene.org/) databases to select all possible studies with key words including “Alzheimer’s disease” and “CD2AP”. The literature search was updated on June 10, 2014. Meanwhile, we used the Google Scholar (http://scholar.google.com/) to query the articles citing the studies and all references in these studies identified by the PubMed and AlzGene databases. We selected only published articles written in English.
Inclusion Criteria
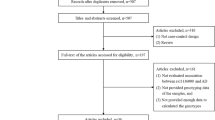
We selected the studies meeting the following criteria: (1) the study was conducted by a case-control design; (2) the study evaluated the association between rs9349407 polymorphism and AD; (3) the study provided the numbers of rs9349407 genotypes or (4) the study must provide sufficient data to calculate the numbers of rs9349407 genotypes or (5) the study provided an OR with 95 % CI as well as the P value or (6) the study must provide sufficient data to calculate the OR and 95 % CI (Fig. 1).
Data Extraction
Guiyou Liu extracted the following information from each study: (1) the name of the first author, (2) the year of publication, (3) the population and ethnicity, (4) the numbers of AD cases and controls, (5) the genotype numbers of rs9349407 polymorphism in cases and controls, (6) the genotyping platform, (7) the OR with 95 % CI or (8) sufficient data to calculate the OR and 95 % CI, and (9) the inclusion criteria for Alzheimer’s disease patients and controls. All relevant calculations were completed using the program R (http://www.r-project.org/).
Quality Evaluation
Here, the criteria proposed by Clark and Baudouin were used to evaluate the quality of selected genetic association studies [8]. This scoring system consists of ten components. A component of the criteria was scored as 1 if present or 0 if absent. A final quality score was obtained by summing each component, resulting in a scoring range of 0–10 for case-control association studies [8]. Selected studies were scored as “good” if the score was greater than or equal to 8, “mediocre” if the score was 5–7, and “poor” if the score was less than 4 [9]. Two authors performed the quality evaluation independently using the criteria proposed by Clark and Baudouin. A third author adjudicated any differences between the two authors.
Genetic Model
Here, we selected the additive genetic model for further meta-analysis. The CD2AP rs9349407 polymorphism has two alleles including C and G. C is the minor allele. We assume that C is the high-risk allele and G is the lower-risk allele. The additive model can be described as C allele versus G allele [10].
Heterogeneity Test
We evaluated the genetic heterogeneity among the studies included using Cochran’s Q test, which approximately follows a χ 2 distribution with k-1 degrees of freedom (k stands for the number of studies for analysis). \( {I}^2=\raisebox{1ex}{$\left( Q-\left( k-1\right)\right)$}\!\left/ \!\raisebox{-1ex}{$ Q$}\right.\times 100\% \), which ranges from 0 to 100 %, was also used [11]. Low, moderate, large, and extreme heterogeneity corresponded to 0–25, 25–50, 50–75, and 75–100 %, respectively [11]. The significance levels for heterogeneity are defined to be with P < 0.01 and I 2 > 50 %.
Meta-Analysis
If there is no significant heterogeneity among the included studies, the pooled OR is calculated by the fixed effect model (Mantel-Haenszel); otherwise, the OR is calculated by random effect model (DerSimonian-Laird). Z test is used to determine the significance of OR. All statistical tests for heterogeneity and meta-analysis were computed using R package for meta-analysis (http://cran.r-project.org/web/packages/meta/index.html).
Sensitivity and Publication Bias Analyses
We evaluated the relative influence of each study by omitting each study at a time. Meanwhile, we used funnel plots to evaluate the potential publication bias [12]. Begg’s and Egger’s tests were used to evaluate the asymmetry of the funnel plot [12]. The significance level for publication bias test is 0.01.
Results
Literature Search
Twenty-five, two, and three articles were identified through PubMed, AlzGene, and Google Scholar search, respectively. Finally, after excluding those studies with overlapping samples, seven articles including 15 independent studies were included for our following analysis. More detailed information about the inclusion or exclusion of selected studies and quality evaluation is provided in the supplementary materials. The main characteristics of the included studies are described in Table 1.
Heterogeneity Test and Meta-analysis
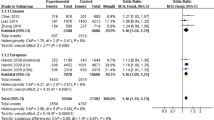
We evaluated the genetic heterogeneity of rs9349407 polymorphism among the selected studies. We did not identify a significant heterogeneity among these studies with P = 0.098 and I 2 = 33.8 %. We further calculated the overall OR by the fixed effect model. Our results showed a significant association between rs9349407 polymorphism and AD with P = 8.78E-07, OR = 1.08, 95 % CI 1.05–1.12. Detailed results are described in Fig. 2.
Sensitivity Analysis and Publication Bias Analysis
By excluding any one study, we identified that the association between rs9349407 polymorphism and AD did not vary substantially (detailed data not shown). Take Naj et al. (Alzheimer’s Disease Genetic Consortium (ADGC)—stage 1) [5], for example, as it includes more sample than other studies. After excluding this study, we did not identify a significant heterogeneity among other studies using the additive model (P = 0.2566 and I 2 = 18 %). Meta-analysis using the fixed effect model showed a significant association between rs9349407 polymorphism and AD (P = 8.60E-03, OR = 1.05, 95 % CI 1.00–1.10). The funnel plots are symmetrical inverted funnels for models (Fig. 3), suggesting no significant publication bias for the additive model (Begg’s test, P = 0.02 and Egger’s test, P = 0.02).
Discussion
Recent GWAS identified rs9349407 polymorphism to be significantly associated with AD in European ancestry [5, 6]. The following studies investigated this association and reported inconsistent results. We think that these inconsistent results may have been caused by either relatively small sample sizes compared with those used for the previous GWAS in European ancestry or the genetic heterogeneity of rs9349407 polymorphism in different populations.
Shulman et al. investigate whether AD susceptibility loci from GWAS affect neuritic plaque pathology [7]. They performed a candidate polymorphism analysis in a joint clinicopathologic cohort, including 725 deceased subjects from the Religious Orders Study and the Rush Memory and Aging Project, followed by targeted validation in an independent neuroimaging cohort, including 114 subjects from multiple clinical and research centers. Their results indicated that rs9349407 was associated with neuritic plaque burden [7]. This finding enhances the understanding of AD risk factors by relating validated susceptibility alleles to increased neuritic plaque pathology [7].
Considering the important role of rs9349407 polymorphism in AD, we reevaluated this association using the relatively large-scale samples from 15 studies. We did not identify a significant heterogeneity of rs9349407 polymorphism pooled populations. We observed a significant association between the rs9349407 polymorphism and AD in pooled populations. The sensitivity analysis and publication bias analysis indicated that our results were robust and no publication bias was observed.
Recently, Carrasquillo et al. performed a meta-analysis to investigate the association between rs9349407 polymorphism and AD. Our study is different from the previous study conducted by Carrasquillo et al. [13]. Carrasquillo et al. used the samples from three articles [5, 6, 13]. Here, we selected seven articles and performed an updated analysis. To our knowledge, this is the first meta-analysis to investigate the association between rs9349407 polymorphism and AD in East Asian, American, Canadian, and European populations. Our analysis further supports previous findings that the rs9349407 polymorphism is associated with AD susceptibility. We believe that our findings will be very useful for future genetic studies on AD.
In addition to the CR1 rs9349407 polymorphism, common variants in another eight AD susceptibility genes were also reported in European population. Interestingly, these variants were successfully replicated by analyzing large-scale dataset, such as PICALM rs3851179, BIN1 rs744373, CLU rs11136000 and rs2279590, ABCA7 rs3764650, and CR1 rs6656401 polymorphisms [4, 11, 14–18]. These results indicate that analyzing relatively large-scale samples is effective to identify the significant association between these common variants and AD.
Despite these interesting results, there is also a limitation in our study. Here, we investigated the association between CD2AP rs9349407 polymorphism and AD using the additive model. It is reported that most meta-analyses used an additive genetic model [19]. In general, this model performs well when the true underlying genetic model is uncertain [19]. It was also important to analyze the association using dominant model and recessive model [10]. However, the dominant and recessive models required exact genotype numbers of all studies used in our analysis. We attempted to obtain these genotype numbers but were not successful. Future studies using genotype data are required to replicate our findings.
References
Liu G, Jiang Y, Wang P, Feng R, Jiang N, Chen X, Song H, Chen Z (2012) Cell adhesion molecules contribute to Alzheimer’s disease: multiple pathway analyses of two genome-wide association studies. J Neurochem 120:190–198
Liu G, Yao L, Liu J, Jiang Y, Ma G, Chen Z, Zhao B, Li K (2014) Cardiovascular disease contributes to Alzheimer’s disease: evidence from large-scale genome-wide association studies. Neurobiol Aging 35:786–792
Lambert JC, Grenier-Boley B, Chouraki V, Heath S, Zelenika D, Fievet N, Hannequin D, Pasquier F, Hanon O, Brice A, Epelbaum J, Berr C, Dartigues JF, Tzourio C, Campion D, Lathrop M, Amouyel P (2010) Implication of the immune system in Alzheimer’s disease: evidence from genome-wide pathway analysis. J Alzheimers Dis 20:1107–1118
Liu G, Wang H, Liu J, Li J, Li H, Ma G, Jiang Y, Chen Z, Zhao B, Li K (2014) The CLU gene rs11136000 variant is significantly associated with Alzheimer’s disease in Caucasian and Asian populations. Neuromol Med 16:52–60
Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD, Jarvik GP, Crane PK, Larson EB, Bird TD, Boeve BF, Graff-Radford NR, De Jager PL, Evans D, Schneider JA, Carrasquillo MM, Ertekin-Taner N, Younkin SG, Cruchaga C, Kauwe JS, Nowotny P, Kramer P, Hardy J, Huentelman MJ, Myers AJ, Barmada MM, Demirci FY, Baldwin CT, Green RC, Rogaeva E, St George-Hyslop P, Arnold SE, Barber R, Beach T, Bigio EH, Bowen JD, Boxer A, Burke JR, Cairns NJ, Carlson CS, Carney RM, Carroll SL, Chui HC, Clark DG, Corneveaux J, Cotman CW, Cummings JL, DeCarli C, DeKosky ST, Diaz-Arrastia R, Dick M, Dickson DW, Ellis WG, Faber KM, Fallon KB, Farlow MR, Ferris S, Frosch MP, Galasko DR, Ganguli M, Gearing M, Geschwind DH, Ghetti B, Gilbert JR, Gilman S, Giordani B, Glass JD, Growdon JH, Hamilton RL, Harrell LE, Head E, Honig LS, Hulette CM, Hyman BT, Jicha GA, Jin LW, Johnson N, Karlawish J, Karydas A, Kaye JA, Kim R, Koo EH, Kowall NW, Lah JJ, Levey AI, Lieberman AP, Lopez OL, Mack WJ, Marson DC, Martiniuk F, Mash DC, Masliah E, McCormick WC, McCurry SM, McDavid AN, McKee AC, Mesulam M, Miller BL, Miller CA, Miller JW, Parisi JE, Perl DP, Peskind E, Petersen RC, Poon WW, Quinn JF, Rajbhandary RA, Raskind M, Reisberg B, Ringman JM, Roberson ED, Rosenberg RN, Sano M, Schneider LS, Seeley W, Shelanski ML, Slifer MA, Smith CD, Sonnen JA, Spina S, Stern RA, Tanzi RE, Trojanowski JQ, Troncoso JC, Van Deerlin VM, Vinters HV, Vonsattel JP, Weintraub S, Welsh-Bohmer KA, Williamson J, Woltjer RL, Cantwell LB, Dombroski BA, Beekly D, Lunetta KL, Martin ER, Kamboh MI, Saykin AJ, Reiman EM, Bennett DA, Morris JC, Montine TJ, Goate AM, Blacker D, Tsuang DW, Hakonarson H, Kukull WA, Foroud TM, Haines JL, Mayeux R, Pericak-Vance MA, Farrer LA, Schellenberg GD (2011) Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet 43:436–441
Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, Abraham R, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Jones N, Stretton A, Thomas C, Richards A, Ivanov D, Widdowson C, Chapman J, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Beaumont H, Warden D, Wilcock G, Love S, Kehoe PG, Hooper NM, Vardy ER, Hardy J, Mead S, Fox NC, Rossor M, Collinge J, Maier W, Jessen F, Ruther E, Schurmann B, Heun R, Kolsch H, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Gallacher J, Hull M, Rujescu D, Giegling I, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Pankratz VS, Sando SB, Aasly JO, Barcikowska M, Wszolek ZK, Dickson DW, Graff-Radford NR, Petersen RC, van Duijn CM, Breteler MM, Ikram MA, DeStefano AL, Fitzpatrick AL, Lopez O, Launer LJ, Seshadri S, Berr C, Campion D, Epelbaum J, Dartigues JF, Tzourio C, Alperovitch A, Lathrop M, Feulner TM, Friedrich P, Riehle C, Krawczak M, Schreiber S, Mayhaus M, Nicolhaus S, Wagenpfeil S, Steinberg S, Stefansson H, Stefansson K, Snaedal J, Bjornsson S, Jonsson PV, Chouraki V, Genier-Boley B, Hiltunen M, Soininen H, Combarros O, Zelenika D, Delepine M, Bullido MJ, Pasquier F, Mateo I, Frank-Garcia A, Porcellini E, Hanon O, Coto E, Alvarez V, Bosco P, Siciliano G, Mancuso M, Panza F, Solfrizzi V, Nacmias B, Sorbi S, Bossu P, Piccardi P, Arosio B, Annoni G, Seripa D, Pilotto A, Scarpini E, Galimberti D, Brice A, Hannequin D, Licastro F, Jones L, Holmans PA, Jonsson T, Riemenschneider M, Morgan K, Younkin SG, Owen MJ, O’Donovan M, Amouyel P, Williams J (2011) Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet 43:429–435
Shulman JM, Chen K, Keenan BT, Chibnik LB, Fleisher A, Thiyyagura P, Roontiva A, McCabe C, Patsopoulos NA, Corneveaux JJ, Yu L, Huentelman MJ, Evans DA, Schneider JA, Reiman EM, De Jager PL, Bennett DA (2013) Genetic susceptibility for Alzheimer disease neuritic plaque pathology. JAMA Neurol 70:1150–1157
Clark MF, Baudouin SV (2006) A systematic review of the quality of genetic association studies in human sepsis. Intensive Care Med 32:1706–1712
Lv H, Jiang Y, Li J, Zhang M, Shang Z, Zheng J, Wu X, Liu P, Zhang R, Yu H (2014) Association between polymorphisms in the promoter region of interleukin-10 and susceptibility to inflammatory bowel disease. Mol Biol Rep 41(3):1299–1310
Lewis CM, Knight J (2012) Introduction to genetic association studies. Cold Spring Harb Protocol 2012:297–306
Liu G, Zhang S, Cai Z, Ma G, Zhang L, Jiang Y, Feng R, Liao M, Chen Z, Zhao B, Li K (2013) PICALM gene rs3851179 polymorphism contributes to Alzheimer’s disease in an Asian population. Neuromol Med 15:384–388
Jiang Y, Zhang R, Zheng J, Liu P, Tang G, Lv H, Zhang L, Shang Z, Zhan Y, Lv W, Shi M (2012) Meta-analysis of 125 rheumatoid arthritis-related single nucleotide polymorphisms studied in the past two decades. PLoS One 7:e51571
Carrasquillo MM, Belbin O, Hunter TA, Ma L, Bisceglio GD, Zou F, Crook JE, Pankratz VS, Sando SB, Aasly JO, Barcikowska M, Wszolek ZK, Dickson DW, Graff-Radford NR, Petersen RC, Passmore P, Morgan K, Younkin SG (2011) Replication of EPHA1 and CD33 associations with late-onset Alzheimer’s disease: a multi-centre case-control study. Mol Neurodegener 6:54
Liu G, Zhang S, Cai Z, Li Y, Cui L, Ma G, Jiang Y, Zhang L, Feng R, Liao M, Chen Z, Zhao B, Li K (2013) BIN1 gene rs744373 polymorphism contributes to Alzheimer’s disease in East Asian population. Neurosci Lett 544:47–51
Liu G, Zhang L, Feng R, Liao M, Jiang Y, Chen Z, Zhao B, Li K (2013) Lack of association between PICALM rs3851179 polymorphism and Alzheimer’s disease in Chinese population and APOEepsilon4-negative subgroup. Neurobiol Aging 34:1310. e1319–1310. e1310
Liu G, Li F, Zhang S, Jiang Y, Ma G, Shang H, Liu J, Feng R, Zhang L, Liao M, Zhao B, Li K (2014) Analyzing large-scale samples confirms the association between the ABCA7 rs3764650 polymorphism and Alzheimer’s disease susceptibility. Mol Neurobiol. doi:10.1007/s12035-014-8670-4
Shen N, Chen B, Jiang Y, Feng R, Liao M, Zhang L, Li F, Ma G, Chen Z, Zhao B, Li K, Liu G (2014) An updated analysis with 85,939 samples confirms the association between CR1 rs6656401 polymorphism and Alzheimer’s disease. Mol Neurobiol. doi:10.1007/s12035-014-8761-2
Zhang S, Zhang D, Jiang Y, Wu L, Shang H, Liu J, Feng R, Liao M, Zhang L, Liu Y, Liu G, Li K (2014) CLU rs2279590 polymorphism contributes to Alzheimer’s disease susceptibility in Caucasian and Asian populations. J Neural Transm. doi:10.1007/s00702-014-1260-9
Gogele M, Minelli C, Thakkinstian A, Yurkiewich A, Pattaro C, Pramstaller PP, Little J, Attia J, Thompson JR (2012) Methods for meta-analyses of genome-wide association studies: critical assessment of empirical evidence. Am J Epidemiol 175:739–749
Tan L, Yu JT, Zhang W, Wu ZC, Zhang Q, Liu QY, Wang W, Wang HF, Ma XY, Cui WZ (2013) Association of GWAS-linked loci with late-onset Alzheimer’s disease in a northern Han Chinese population. Alzheimers Dement 9:546–553
Miyashita A, Koike A, Jun G, Wang LS, Takahashi S, Matsubara E, Kawarabayashi T, Shoji M, Tomita N, Arai H, Asada T, Harigaya Y, Ikeda M, Amari M, Hanyu H, Higuchi S, Ikeuchi T, Nishizawa M, Suga M, Kawase Y, Akatsu H, Kosaka K, Yamamoto T, Imagawa M, Hamaguchi T, Yamada M, Moriaha T, Takeda M, Takao T, Nakata K, Fujisawa Y, Sasaki K, Watanabe K, Nakashima K, Urakami K, Ooya T, Takahashi M, Yuzuriha T, Serikawa K, Yoshimoto S, Nakagawa R, Kim JW, Ki CS, Won HH, Na DL, Seo SW, Mook-Jung I, St George-Hyslop P, Mayeux R, Haines JL, Pericak-Vance MA, Yoshida M, Nishida N, Tokunaga K, Yamamoto K, Tsuji S, Kanazawa I, Ihara Y, Schellenberg GD, Farrer LA, Kuwano R (2013) SORL1 is genetically associated with late-onset Alzheimer’s disease in Japanese, Koreans and Caucasians. PLoS One 8:e58618
Omoumi A, Fok A, Greenwood T, Sadovnick AD, Feldman HH, Hsiung GY (2014) Evaluation of late-onset Alzheimer disease genetic susceptibility risks in a Canadian population. Neurobiol Aging 35:936. e935–936. e912
Logue MW, Schu M, Vardarajan BN, Buros J, Green RC, Go RC, Griffith P, Obisesan TO, Shatz R, Borenstein A, Cupples LA, Lunetta KL, Fallin MD, Baldwin CT, Farrer LA (2011) A comprehensive genetic association study of Alzheimer disease in African Americans. Arch Neurol 68:1569–1579
Acknowledgments
This work was supported by funding from the National Nature Science Foundation of China (grant numbers 81300945, 31301938, 31200934, 31171219, 81271213, 81070878, 81271214, and 81261120404), the Natural Science Foundation of Guangdong Province, China (No. S2012010008222), and the Science and Technology Innovation Fund of Guangdong Medical College (No. STIF 201101).
Conflict of Interest
The authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Hongyuan Chen and Guihua Wu contributed equally to this work.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Supplementary Table 1
(DOC 117 kb)
Rights and permissions
About this article
Cite this article
Chen, H., Wu, G., Jiang, Y. et al. Analyzing 54,936 Samples Supports the Association Between CD2AP rs9349407 Polymorphism and Alzheimer’s Disease Susceptibility. Mol Neurobiol 52, 1–7 (2015). https://doi.org/10.1007/s12035-014-8834-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-014-8834-2