Abstract
Large-scale genome-wide association studies (GWAS) identified three single nucleotide polymorphisms rs11136000, rs2279590, and rs9331888 in CLU gene to be significantly associated with Alzheimer’s disease (AD) in Caucasian ancestry. Both rs11136000 and rs2279590 variants were successfully replicated in Asian population. However, previous studies reported either a weak association or no association between rs9331888 polymorphism and AD in Asian population. Here, we searched the PubMed, AlzGene, and Google Scholar databases. We selected 12 independent studies that evaluated the association between the rs9331888 polymorphism and AD using a case-control design. Using an additive model, we did not identify significant heterogeneity among these 12 studies. We observed significant association between rs9331888 polymorphism and AD in pooled populations (P = 2.26E − 07, odds ratio (OR) = 1.10, 95 % confidence interval (CI) 1.06–1.14). In subgroup analysis, we did not identify significant heterogeneity in both Asian and Caucasian populations. We identified significant association in Caucasian population (P = 1.67E − 08, OR = 1.13, 95 % CI 1.08–1.18) but not in East Asian population (P = 0.49, OR = 1.02, 95 % CI 0.96–1.10).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Alzheimer’s disease (AD) is a complex and most common neurodegenerative disease in the elderly [1]. Much effort has been put into the identification of common AD variants. Since 2009, large-scale genome-wide association studies (GWAS) have been conducted, and reported several AD susceptibility genes including CR1, BIN1, CLU, PICALM, MS4A4/MS4A6E, CD2AP, CD33, EPHA1 and ABCA7 [2–4], SORL1 [5], and TREM2 [6]. A recent large meta-analysis of AD GWAS in individuals of European ancestry identified 11 new AD genetic risk factors, which include HLA-DRB5/DRB1, PTK2B, SLC24A4-0RING3, DSG2, INPP5D, MEF2C, NME8, ZCWPW1, CELF1, FERMT2, and CASS4 [4].
Lambert et al. in 2009 identified three single nucleotide polymorphisms (SNPs), including rs11136000, rs2279590, and rs9331888 in CLU gene, to be significantly associated with AD by large-scale GWAS in Caucasian ancestry [7]. They found that all these three CLU markers are within a linkage disequilibrium (LD) block [7]. They reported three common haplotypes including TTC, CCC, and CCG, among which TTC is the most frequent haplotype [7]. The SNPs as ordered from left to right (5′ to 3′) are rs2279590, rs11136000, and rs9331888. Compared to TTC, both CCC and CCG haplotypes were significantly associated with an increased AD risk [7].
Based on this finding, the rs9331888 polymorphism may be a genetic risk factor for AD. However, inconsistent results regarding this variant have been reported in Asian populations. These studies reported weak associations or no association between the rs9331888 polymorphism and AD in Chinese, Japanese, and Korean populations [5, 8–11]. Here, we searched the PubMed, AlzGene, and Google Scholar databases. We selected 12 independent studies (six studies in Asian population and six studies in Caucasian population) that evaluated the association between the rs9331888 polymorphism and AD using a case-control design. We first evaluated the genetic heterogeneity of rs9331888 polymorphism in Caucasian and Asian populations, and then performed a meta-analysis using an additive genetic model.
Methods and Materials
Literature Search
We selected PubMed, AlzGene, and Google Scholar databases for literature search. In stage 1, we used the “Alzheimer’s disease,” “Clusterin,” or ”CLU” as the keywords to select all possible studies in PubMed and AlzGene databases. In stage 2, we query the articles citing the studies and all references in stage 1 in the Google Scholar database. The literature search was updated on November 25, 2014.
Inclusion Criteria
All the selected studies meet the following criteria: (1) evaluated the association between rs9331888 polymorphism and AD using a case-control design, (2) provided the numbers of rs9331888 genotypes or allele, (3) provided sufficient data to calculate the numbers of rs9331888 genotypes or allele, (4) provided an odds ratio (OR) with 95 % confidence interval (CI) in allele genetic model, or (5) provided sufficient data to calculate the OR and 95 % CI.
Data Extraction
The following information is extracted from all the selected studies: (1) first author name, (2) publication year, (3) population and ethnicity, (4) numbers of AD cases and controls, (5) genotyping platform, (6) numbers of rs9331888genotypes or (7) to calculate the numbers of rs9331888 genotypes, (8) OR with 95 % CI or (9) to calculate the OR and 95 % CI, and (10) minor allele frequency.
Genetic Model
The CLU rs9331888 polymorphism includes C and G alleles. C is the minor allele. C is taken as the high-risk allele, and G is the lower-risk allele. Following analyses are based on the additive genetic model (allele model), which can be described as C allele versus G allele [12, 13].
Heterogeneity Test
Genetic heterogeneity among all the selected studies is evaluated by Cochran’s Q test, approximately following a χ 2 distribution with k − 1 degrees of freedom (k stands for the number of studies for analysis). Another measure \( {I}^2=\raisebox{1ex}{$\left(Q-\left(k-1\right)\right)$}\!\left/ \!\raisebox{-1ex}{$Q$}\right.\times 100\% \) ranging from 0 to 100 % was also used [14]. I 2 is a measure of heterogeneity and a statistic that indicates the percentage of variance in a meta-analysis that is attributable to study heterogeneity [15]. Zero to 25, 25–50, 50–75, and 75–100 % corresponded to low, moderate, large, and extreme heterogeneity [14]. The tests with P < 0.01 and I 2 > 50 % indicate significant heterogeneity.
Meta-Analysis
The pooled OR is calculated by the fixed effect model (Mantel-Haenszel), if there is no significant heterogeneity among the included studies. Otherwise, the OR is calculated by random effect model (DerSimonian-Laird). Z test is used to determine the significance of OR. All statistical tests for heterogeneity and meta-analysis were computed using R package.
Sensitivity Analysis
We omit each study at a time to assess the influence of each individual study on the pooled OR.
Publication Bias Analysis
A funnel plot introduced by Light and Pillemer in 1984 and discussed in detail by Egger and colleagues [16, 17] is commonly used to check for the existence of publication bias. In the absence of publication bias, it assumes that the largest studies will be plotted near the average, and smaller studies will be spread evenly on both sides of the average, creating a roughly funnel-shaped distribution. Deviation from this shape can indicate publication bias. Meanwhile, the regression-based approach proposed by Egger and colleagues and the rank correlation test provided by Begg and Mazumdar are widely used to test for publication bias, which evaluate the asymmetry of the funnel plot to provide statistical evidence, with a P < 0.01 indicating that there was a significant publication bias [18].
Results
Literature Search
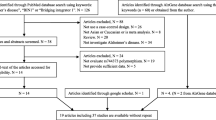
We selected a total of 288 articles from the PubMed (n = 284) and AlzGene (n = 4) databases. Finally, seven articles including 12 independent studies (six studies in Asian population and six studies in Caucasian population) were selected for following analysis. We provided the detailed information in Fig. 1. The main characteristics of the included studies are described in Table 1.
Heterogeneity Test and Meta-Analysis
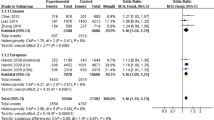
We identified no significant heterogeneity among the selected studies (P = 0.032 and I 2 = 48 %). We calculated the overall OR by the fixed effect model. We reported significant association between rs9331888 and AD with P = 2.26E − 07, OR = 1.10, 95 % CI 1.06–1.14. Detailed results are described in Fig. 2.
Heterogeneity Test and Subgroup Analysis
We performed a subgroup analysis in East Asian and Caucasian populations, respectively. We did not identify significant heterogeneity East Asian (P = 0.16 and I 2 = 37.7 %) and Caucasian populations (P = 0.18 and I 2 = 34.4 %). The meta-analysis further showed significant association between rs9331888 and AD Caucasian population (P = 1.67E − 08, OR = 1.13, 95 % CI 1.08–1.18) but not in East Asian population (P = 0.49, OR = 1.02, 95 % CI 0.96–1.10).
Sensitivity Analysis and Publication Bias Analysis
The sensitivity analysis showed that the association between rs9331888 and AD did not vary substantially. The funnel plot is symmetrically inverted funnel (Fig. 3). Begg and Egger’s tests provide statistical evidence of funnel plot symmetry (Begg’s test, P = 0.23, and Egger’s test, P = 0.23), which suggest no significant publication bias.
Discussion
Large-scale GWAS identified significant association between rs9331888 and AD [7]. Here, based on the inconsistent results reported in Asian and Caucasian populations, we first evaluated the genetic heterogeneity of the rs9331888 in Asian and Caucasian populations. We then performed a meta-analysis. We did not identify significant heterogeneity among all the selected studies. Our results showed significant association between rs9331888 and AD with P = 2.26E − 07. We further evaluated the effect of the rs9331888 polymorphism on AD risk in different ethnic backgrounds. The results showed that rs9331888 contributed to AD in Caucasian but not Asian populations.
The influence of CLU on clinical and pathological measures of AD cases was investigated. Evidence indicates that CLU significantly reduces CSF Aβ1-42 in AD [19] and affected white matter microstructure in young adults [20]. Evidence also shows that CLU influenced longitudinal changes in brain function and faster cognitive decline [21, 22]. Emerging evidence suggests that CLU contributes to AD through lipid metabolism [23]. Xing et al. tested the association of the rs9331888 polymorphism with blood clusterin levels using 104 AD subjects and 104 healthy controls [24]. They identified that blood clusterin levels were significantly elevated in AD patients compared with controls [24]. The rs9331888 variant was associated with low clusterin mRNA and protein levels in both groups [24]. Szymanski et al. tested the quantitative effects of CLU SNPs on individual alternative transcripts [25]. In 190 temporal lobe samples without pathology, they found that the risk allele of the rs9331888 polymorphism increased the relative abundance of transcript NM_203339 [25]. Using an independent set of 115 AD and control samples, they replicated this result and further observed that multiple CLU transcripts are at higher levels in AD compared with controls [25]. The rs9331888 polymorphism is located in the first exon of transcript NM_203339 and is one of the functional DNA variants underlying this association between CLU and AD [25].
In addition to the rs9331888 polymorphism, another two CLU variants rs11136000 and rs2279590 were identified by large-scale GWAS in Caucasian population [7]. Evidence showed that CLU rs11136000 polymorphism affected the microstructure of white matter in young adults [20]. The healthy carriers of the variant exhibited altered coupling between the hippocampus and prefrontal cortex during memory processing [26]. The rs11136000 genotype of an individual affected longitudinal changes in brain function in asymptomatic individuals and was associated with a faster cognitive decline in the presymptomatic stages of disease progression [27]. The rs11136000 polymorphism significantly influenced the expression levels of CLU in the temporal cortex [28]. CLU rs2279590 polymorphism has been reported to be significantly associated with type 2 diabetes [29].
CLU rs11136000 and rs2279590 variants were reported in Caucasian population. Interestingly, both variants were successfully replicated in Asian population by analyzing large-scale dataset [2, 30]. Here, the CLU rs9331888 polymorphism is not associated with AD in East Asian population. However, Yu et al. identified three haplotypes TTC, CCC, and CCG composed of rs2279590, rs11136000, and rs9331888, which are associated with AD susceptibility in Chinese population [9].
Here, we investigated the association between rs9331888 and AD using an additive model. However, the exact genotype numbers of rs9331888 in most studies are not available. Thus, we did not use the dominant and recessive models. Future studies with genotype data are required to verify our findings. To our knowledge, this is the first study to investigate rs9331888 polymorphism in Asian population by a meta-analysis. Our analysis supports previous findings that CLU rs9331888 polymorphism contributes to AD susceptibility.
References
Liu G, Yao L, Liu J, Jiang Y, Ma G, Chen Z, Zhao B, Li K (2014) Cardiovascular disease contributes to Alzheimer’s disease: evidence from large-scale genome-wide association studies. Neurobiol Aging 35:786–792
Liu G, Wang H, Liu J, Li J, Li H, Ma G, Jiang Y, Chen Z et al (2014) The CLU gene rs11136000 variant is significantly associated with Alzheimer’s disease in caucasian and asian populations. Neuromolecular Med 16:52–60
Reitz C, Jun G, Naj A, Rajbhandary R, Vardarajan BN, Wang LS, Valladares O, Lin CF et al (2013) Variants in the ATP-binding cassette transporter (ABCA7), apolipoprotein E 4, and the risk of late-onset Alzheimer disease in African Americans. JAMA 309:1483–1492
Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, DeStafano AL, Bis JC et al (2013) Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet 45:1452–1458
Miyashita A, Koike A, Jun G, Wang LS, Takahashi S, Matsubara E, Kawarabayashi T, Shoji M et al (2013) SORL1 is genetically associated with late-onset Alzheimer’s disease in Japanese, Koreans and Caucasians. PLoS One 8:e58618
Guerreiro R, Wojtas A, Bras J, Carrasquillo M, Rogaeva E, Majounie E, Cruchaga C, Sassi C et al (2013) TREM2 variants in Alzheimer’s disease. N Engl J Med 368:117–127
Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D et al (2009) Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet 41:1094–1099
Chen LH, Kao PY, Fan YH, Ho DT, Chan CS, Yik PY, Ha JC, Chu LW et al (2012) Polymorphisms of CR1, CLU and PICALM confer susceptibility of Alzheimer’s disease in a southern Chinese population. Neurobiol Aging 33(210):e211–217
Yu JT, Li L, Zhu QX, Zhang Q, Zhang W, Wu ZC, Guan J, Tan L (2010) Implication of CLU gene polymorphisms in Chinese patients with Alzheimer’s disease. Clin Chim Acta 411:1516–1519
Lu SJ, Li HL, Sun YM, Liu ZJ, Yang P, Wu ZY (2014) Clusterin variants are not associated with southern Chinese patients with Alzheimer’s disease. Neurobiol Aging 35(2656):e2659–2611
Komatsu M, Shibata N, Kuerban B, Ohnuma T, Baba H, Arai H (2011) Genetic association between clusterin polymorphisms and Alzheimer’s disease in a Japanese population. Psychogeriatrics 11:14–18
Liu G, Li F, Zhang S, Jiang Y, Ma G, Shang H, Liu J, Feng R et al (2014) Analyzing large-scale samples confirms the association between the ABCA7 rs3764650 polymorphism and Alzheimer’s disease susceptibility. Mol Neurobiol 50(3):757–64. doi:10.1007/s12035-014-8670-4
Chen H, Wu G, Jiang Y, Feng R, Liao M, Zhang L, Ma G, Chen Z et al (2014) Analyzing 54,936 samples supports the association between CD2AP rs9349407 polymorphism and Alzheimer’s disease susceptibility. Neurobiol, Mol. doi:10.1007/s12035-014-8834-2
Liu G, Zhang S, Cai Z, Ma G, Zhang L, Jiang Y, Feng R, Liao M et al (2013) PICALM gene rs3851179 polymorphism contributes to Alzheimer’s disease in an Asian population. Neuromolecular Med 15:384–388
Higgins JP, Thompson SG, Deeks JJ, Altman DG (2003) Measuring inconsistency in meta-analyses. BMJ 327:557–560
Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315:629–634
Sterne JA, Egger M (2001) Funnel plots for detecting bias in meta-analysis: guidelines on choice of axis. J Clin Epidemiol 54:1046–1055
Song F, Khan KS, Dinnes J, Sutton AJ (2002) Asymmetric funnel plots and publication bias in meta-analyses of diagnostic accuracy. Int J Epidemiol 31:88–95
Elias-Sonnenschein LS, Helisalmi S, Natunen T, Hall A, Paajanen T, Herukka SK, Laitinen M, Remes AM et al (2013) Genetic loci associated with Alzheimer’s disease and cerebrospinal fluid biomarkers in a Finnish case-control cohort. PLoS One 8:e59676
Braskie MN, Jahanshad N, Stein JL, Barysheva M, McMahon KL, de Zubicaray GI, Martin NG, Wright MJ et al (2011) Common Alzheimer’s disease risk variant within the CLU gene affects white matter microstructure in young adults. J Neurosci 31:6764–6770
Thambisetty M, Beason-Held LL, An Y, Kraut M, Nalls M, Hernandez DG, Singleton AB, Zonderman AB et al (2013) Alzheimer risk variant CLU and brain function during aging. Biol Psychiatry 73:399–405
Sweet RA, Seltman H, Emanuel JE, Lopez OL, Becker JT, Bis JC, Weamer EA, DeMichele-Sweet MA et al (2012) Effect of Alzheimer’s disease risk genes on trajectories of cognitive function in the cardiovascular health study. A J Psychiatry 169:954–962
Yu JT, Tan L (2012) The role of clusterin in Alzheimer’s disease: pathways, pathogenesis, and therapy. Mol Neurobiol 45:314–326
Xing YY, Yu JT, Cui WZ, Zhong XL, Wu ZC, Zhang Q, Tan L (2012) Blood clusterin levels, rs9331888 polymorphism, and the risk of Alzheimer’s disease. J Alzheimers Dis 29:515–519
Szymanski M, Wang R, Bassett SS, Avramopoulos D (2011) Alzheimer’s risk variants in the clusterin gene are associated with alternative splicing. Transl Psychiatry 1:e18
Erk S, Meyer-Lindenberg A, Opitz von Boberfeld C, Esslinger C, Schnell K, Kirsch P, Mattheisen M, Muhleisen TW et al (2011) Hippocampal function in healthy carriers of the CLU Alzheimer’s disease risk variant. J Neurosci 31:18180–18184
Thambisetty M, Beason-Held LL, An Y, Kraut M, Nalls M, Hernandez DG, Singleton AB, Zonderman AB et al (2013) Alzheimer risk variant CLU and brain function during aging. Biol Psychiatry 73:399–405
Allen M, Zou F, Chai HS, Younkin CS, Crook J, Pankratz VS, Carrasquillo MM, Rowley CN et al (2012) Novel late-onset Alzheimer disease loci variants associate with brain gene expression. Neurology 79:221–228
Daimon M, Oizumi T, Karasawa S, Kaino W, Takase K, Tada K, Jimbu Y, Wada K et al (2011) Association of the clusterin gene polymorphisms with type 2 diabetes mellitus. Metabolism 60:815–822
Zhang S, Zhang D, Jiang Y, Wu L, Shang H, Liu J, Feng R, Liao M et al (2014) CLU rs2279590 polymorphism contributes to Alzheimer’s disease susceptibility in Caucasian and Asian populations. J Neural Transm. doi:10.1007/s00702-014-1260-9
Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD et al ((2011) Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet 43:436
Acknowledgments
This work was supported by funding from the National Nature Science Foundation of China (Grant Nos. 81300945, 31200934, 31301938, 81471294, 31171219, 81271213, and 81271214) and Harbin Science and Technology Bureau (2014RFXGJ042).
Conflict of Interest
The authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Shuyan Zhang, Xuling Li and Guoda Ma contributed equally to this work.
Rights and permissions
About this article
Cite this article
Zhang, S., Li, X., Ma, G. et al. CLU rs9331888 Polymorphism Contributes to Alzheimer’s Disease Susceptibility in Caucasian But Not East Asian Populations. Mol Neurobiol 53, 1446–1451 (2016). https://doi.org/10.1007/s12035-015-9098-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-015-9098-1