Abstract
A number of single-nucleotide polymorphisms within the 3′-UTR of genes have been shown to relate to the occurrence of cancers. In this study, by using polymerase chain reaction–restriction fragment length analysis method, we determined an SNP rs1599795 in the 3′-UTR of CD80 gene in 183 gastric cancer patients and 348 healthy controls. Statistical analysis results showed that SNP rs1599795 genotypes were significantly correlated with the risk of gastric cancer. Compared with the AA homozygotes, the TA heterozygotes were significantly more prevalent in the patients (OR 1.44, 95 % CI 0.98–2.11) with a larger tumor size (P = 0.001), deeper infiltration (P = 1.5 × 10−5), higher possibility of lymph node metastasis (P = 0.003), and more in the late stage (TNM stage III and IV; P = 0.003); the TT homozygotes had larger tumor size (P = 0.001) and lower degree of differentiation (P = 2.2 × 10−4). Dual-luciferase reporter assays showed that miR-132-3p, miR-212-3p, and miR-361-5p inhibited the expression of CD80 through binding with the CD80 3′-UTR, and this inhibitory role of miR-132-3p, miR-212-3p, and miR-361-5p was impacted by rs1599795. Our findings have shown that the SNP rs1599795 in CD80 3′-UTR, through disrupting the regulatory role of miR-132-3p, miR-212-3p, and miR-361-5p in CD80 expression, contributed to the occurrence of gastric cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Although with the continuous improvement of diagnostic techniques and surgical treatment, the survival rate of patients with gastric cancer has been improved [1]; gastric cancer is still the second cause of deaths from cancers worldwide [2]. It is estimated that 40 % of gastric cancer occurred in China [2]. The occurrence and development of gastric cancer is a multiple-factor and multiple-stage process involving genetic and epigenetic alterations of protein-coding proto-oncogenes and tumor suppressor genes [3].
CD80, an important co-stimulatory ligand, binding to its corresponding receptor CD28 or CTLA-4 [4], is essential for the activation and regulation of T-cell immunity [5]. CD80 is expressed on a variety of normal epithelial tissue, such as skin, respiratory tract, gastrointestinal tract, and genitourinary tract [6]. However, low expression of CD80 has been found in the majority of tumors, which cannot induce effective anti-tumor immunity response, resulting in tumor immune escape [7]. Almost all patients with benign gastric mucosa and primary gastric carcinoma showed high levels of expression of the CD80; however, the percentage of CD80-positive cells of well-differentiated carcinoma and normal mucosa was significantly higher than that of poorly differentiated primary carcinomas. Furthermore, almost all of the metastatic carcinoma cells revealed consistently very low or undetectable levels of expression of the CD80 molecule [8]. The expression of the CD80 mRNA was observed in the process of the development of cancer, but the protein level of CD80 decreased along with the development of cancer [9], indicating that there are some posttranscriptional regulation mechanism mediated in the expression of CD80 gene.
MicroRNA (miRNAs), a class of small noncoding RNAs of about 22 nucleotides, play a posttranscriptional regulation role in gene expression by binding to the 3′-untranslated regions (UTRs) of target messenger RNAs (mRNAs), leading to translational repression or gene silencing [9, 10]. miRNA genes represent only a small part of human genome, but they can regulate almost one-third of all the human genes [11]. Recent studies have shown that miRNAs were associated with a variety of physiological and pathological processes, including cell differentiation, value-added, apoptosis, insulin secretion, cholesterol synthesis, and tumorigenesis [12]. miRNA can inhibit the translation of a target gene by binding to the target mRNA with 6–8 complementary nucleotides, which is usually restricted to the 2–8 nucleotides of the 5′-end of the miRNA, and this sequence is referred to as “seed region” [13]. If variation occurs in this region, it will seriously affect the regulatory role of miRNA. Single-nucleotide polymorphisms (SNPs), the most common human genetic variations, have been proved to be significantly related to the occurrence of diseases including gastric cancer. More and more studies have provided evidences that SNPs located in the miRNA (miRSNPs) binding sites through affecting the binding of miRNAs with the target genes resulted in reduction or increase in the target mRNA translation, and thus being associated with the susceptibility to gastric cancers [10]. Wu et al. [14] pointed out that the rs2910164 polymorphism in the sequence of miR-146a precursor may influence the susceptibility to gastric cancer in the Chinese population. We have also found that SNPs rs4143815 and rs4819388 in the 3′-UTRs of B7-H1 and B7-H2 genes were significantly related to the occurrence of gastric cancer [15, 16]. In this study, we used the case–control method to investigate the relationship between miRSNPs in the 3′-UTR of CD80 gene and the risk of gastric cancer. In addition, we also assessed the effect of the miRSNPs on the regulatory role of miRNAs in the expression of the CD80 protein.
Materials and methods
Study subjects
A total of 531 genetically unrelated subjects including 348 normal controls and a total of 183 gastric cancer patients participated in this study after giving written informed consent. They were recruited from the First Affiliated Hospital of Soochow University between March 2007 and May 2009. All subjects were Han Chinese and were raised in Suzhou. They were histologically confirmed by two pathologists. Individuals those were previously cancerous or had metastasizing cancer from other origins were excluded. For gastric cancer patients, the clinicopathological variables, including tumor markers, tumor location, histological type, tumor size, tumor area, differentiation grade, depth of tumor infiltration, lymph node metastasis, distant metastasis, and TNM stage, were obtained from the medical records. None of the patients had undergone radiotherapy or chemotherapy before the surgery. The variables of depth in tumor infiltration, lymph node metastasis, distant metastasis, and TNM stage were examined and staged according to the American Joint Commission for Cancer Staging in 2002. The controls have no gastrointestinal disorders and personal or familial history of cancers, which was traced back ≥3 generations and laterally to second- and third-degree relatives. They were obtained from the hospital of patients’ routine health examinations. The research protocol was approved by the institutional review board of Soochow University.
In the patients with gastric cancer, 92 and 91 cancers were occurred at the antrum and others, respectively; 56 and 104 cases were moderately and poorly differentiated, respectively; 11, 19, 145, and 8 tumors were at T1, T2, T3, and T4 stages, respectively; 140 and 32 patients were with lymph node metastasis and distant metastasis; 19, 24, 100, and 40 patients were at TNM I, II, III, and IV stages, respectively (Table 1).
Genomic DNA samples
The whole blood samples from patients and controls were collected and stored in Vacutainer® tubes (BD Franklin Lakes, NJ) containing anticoagulant of EDTA. Total genomic DNA was extracted from the whole blood according to the phenol/chloroform method. The purity and concentration of the extracted DNA were determined by UV–Vis spectrophotometer. The extracted DNA was stored at 4 °C in TE buffer (10 mmol/l Tris–HCl, 1 mmol/l EDTA, pH 8.0).
Selection of miRSNPs
First, the SNPs in the 3′-UTR of the CD80 gene were obtained from the published databases of NCBI dbSNP BUILED 129 (http://www.ncbi.nlm.nih.gov/SNP) and ENSEMBL v58 (http://www.ensembl.org/). From the HapMap, the reported SNP allele frequency selected a minor allele frequency (MAF) of not less than 10 % of the SNPs. Then, the online softwares of Diana-Micro (http://diana.cslab.ece.ntua.gr/microT/), MicroInspector (http://miRNA.imbb.forth.gr/microinspector/), miR-anda (http://www.microrna.org/), miRNAMap (http://miRNAmap.mbc.nctu.edu.tw/), RNAhybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/), and Target Scan v5.1 (http://targetscan.org/) were used to predict the possible miRNA binding sites in CD80 3′-UTR. The SNPs located in putative miRNA binding sites were regarded as candidate miRSNPs.
SNP genotyping
The genotypes of the SNP rs1599795 (A>T) were determined by using polymerase chain reaction–restriction fragment length polymorphism (PCR–RFLP) analysis, which were validated by Sanger’s DNA sequencing method (Gene Script, Nanjing, China). Specific measurement procedure is as follows:
The PCR was performed in 25 μl containing 10 mM Tris–HCl (pH 8.0), 50 mM KCl, 1.5 mM MgCl2, 0.2 mM of each dNTP, 0.4 μM of forward primer (5′-GGTGTTTACCCAGTATTC C-3′) and reverse primer (5′-CAAATGAGGTCAAACTAG C-3′), 1 μl of genomic DNA (~100 ng), and 0.5 U Taq DNA Polymerase (MBI). The PCR cycling was performed in a PTC-200 DNA Tetrad thermal cycler (Bio-Rad) as follows: 94 °C for 5 min, followed by 30 cycles of 94 °C for 20 s, 56 °C for 30 s, and 72 °C for 30 s. Final extension was completed at 72 °C for 7 min. Then, 10 μl of the PCR products was digested with 10 U of restriction enzyme TaqI for 8 h at 37 °C in 20 μl of mixture. After heat inactivation for 15 min at 70 °C, 10 μl of the digested DNA fragments was analyzed at room temperature by electrophoresis on a 2 % agarose (LP0028A, Oxoid, Hampshire, England) gel containing 0.5 mg/ml ethidium bromide at 100 V for 20 min in 1× TAE buffer. Images of the gels were taken using GeneGenius BioImaging Systems (SynGene, Cambridge, England). The SNP genotypes of TT, TA, and AA were determined according to the size of the electrophoretic band patterns of 196/43, 239/196/43, and 239 bp, respectively.
Cell culture
The CHO cells were purchased from American Type Culture Collection (Manassas, VA). They were cultured in RPMI 1640 medium (Invitrogen) supplemented with 10 % fetal bovine serum (GBICO BRL, Rockville, MD, USA) at 37 °C in a humidified incubator with 5 % CO2. Cells in the logarithmic growth phase were used for experiments.
Luciferase reporter assays
To generate CD80/3′-UTR/pGL3 luciferase reporter constructs, the CD80 3′-UTR was amplified with the primers of 5′-GGCTAGTCTAGATAGCTCTGGTGACCTTGATC-3′ (forward) and 5′-GGCTAGTCTAGACTTCCCTTAGTATTGCTGAC-3′ (reverse). They were then cloned in the downstream of the firefly luciferase gene in pGL3-Control vector (Promega) according to the manufacturer’s protocol. Positive clones were selected by restriction digestion using XbaI (MBI) and confirmed by DNA sequencing method (GenScript, Nanjing, China).
To generate luciferase reporter constructs containing the potential binding site for miR-132-3p, miR-212-3p, and miR-361-5p, two pairs of nucleotide oligos of 5′-CATATTGGACTGATAATCTCTTTAAAT G-3′ (forward; the allele is underlined) and 5′-CATTTAAAGAGATTATCAGTCCAATAT G-3′ (reverse), and 5′-CATATTGGACTGTTAATCTCTTTAAAT G-3′ (forward; the allele is underlined) and 5′-pCATTTAAAGAGATTAACAGTCCAATAT G-3′ (reverse) were annealed and cloned downstream of the firefly luciferase gene in pGL3-Control vector according to the manufacturer’s protocol, respectively. Positive clones were confirmed by DNA sequencing method.
For luciferase reporter assays, the CHO cells were plated at 5 × 104 cells per well in 24-well plate and were transfected with lipofectamine 2000 (Invitrogen) according to the manufacturer’s recommendations after 24 h. Each co-transfection reaction contained 200 ng of pGL3 construct, 200 ng of pRL-TK plasmid (Promega) that served as a normalizing control, and 50 μM of miR-132-3p, miR-212-3p, or miR-361-5p mimic (GenePharma Inc., Shanghai, China). The negative controls were performed with an unrelated miRNA sequence (5′-UUCUCCGAACGUGUCACGUTT-3′). The transfected CHO cells were maintained in RPMI 1640 medium with 10 % FBS. Each transfection was carried out in triplicate. After 24 h of incubation, cells were collected and analyzed for luciferase activity with the dual-luciferase reporter assay system (Promega).
Statistical analysis
Chi-square test was used to evaluate the differences in frequency distributions of each allele and genotype of the miRSNPs between the cases and controls. Chi-square test was also used to assess the difference between the miRSNPs and the clinicopathological features. The crude and adjusted odds ratios (ORs) and 95 % confidence intervals (CIs) were obtained to assess the association of the miRSNPs with gastric cancer risk using unconditional univariate and multivariate logistic regression models, respectively. Hardy–Weinberg equilibrium of the genotype distribution among control groups was tested by a goodness-of-fit chi-square test. The genotypes were further stratified by subgroups of the clinicopathological variables. All of the statistical analyses were carried out by using STATA software (version 10.0; StataCorp LP, TX, USA) and were done by two persons independently in a blind fashion. For all tests, P < 0.05 was considered statistically significant.
Results
The miRSNPs in the CD80 gene 3′-UTR
First, we found 21 SNPs from the SNP databases NCBI db SNP BUILED 129 and ENSEMBL v58 in the 3′-UTR of CD80 gene, as shown in Table 2. The allele frequencies in the Chinese population (http://www.hapmap.org/index.html.en) are shown in Table 2. Among them, the minor allele frequencies (MAF) of the SNPs rs1599795, rs1599796, rs17281703, rs57271503, and rs7628626 are greater than 10 %.
Then, we used bioinformatic softwares Diana-Micro, MicroInspector, miR-anda, miRNAMap, RNAhybrid, and Target Scan to predict miRNAs that can bind to the CD80 3′-UTR. In the SNPs with MAF of more than 10 %, we found that the SNPs rs1599795 and rs1599796 were located in the miRNA binding sites. Based on the genotype distribution of these two SNPs in 45 healthy Chinese published by HapMap (http://www.hapmap.com), statistical analysis results showed approximately full linkage disequilibrium between the two SNPs. Furthermore, we found that the SNP rs1599795 was located in the binding sites of miR-132-3p, miR-212-3p, and miR-361-5p (Fig. 1). Thus, we investigated the association of this SNP with the risk of gastric cancer.
To study the relationship between the SNP and gastric cancer, we used polymerase chain reaction–restriction fragment length polymorphism analysis method for the determination of the SNP genotype distribution in 183 gastric cancer patients and 348 controls. The genotyping results of 5 samples and typical DNA sequencing results are shown in Fig. 2a, b.
Correlation of rs1599795 with gastric cancer
The SNP rs1599795 genotype frequencies and its correlation with gastric cancer are shown in Table 3. Chi-square (χ2) statistical analysis results showed that the genotypes of rs1599795 were in Hardy–Weinberg equilibrium distribution pattern in the healthy control group (P = 0.48). Further, logistic regression analysis results revealed that a significant increased risk of gastric cancer was associated with the T-allele carriers (OR 1.48, 95 % CI 1.03–2.12; P = 0.036) as compared with the AA genotype.
Then, we did stratified analysis of the association of the rs1599795 genotypes with the clinicopathological parameters of gastric cancer (Table 4). We found a significant association of the rs1599795 genotypes with the tumor size, tumor differentiation, tumor location, depth of invasion, lymph node metastasis, distant metastasis, and TNM staging. Compared with the AA homozygote, the carriers of TA genotype were more likely to have the gastric cancer in the antrum outside parts (P = 0.019), with larger tumor sizes (P = 0.001), more in the late TNM stages (stages III and IV; P = 0.003), deeper infiltration (P = 1.5 × 10−5), and higher possibility of lymph node metastasis (P = 0.003). Furthermore, the TT homozygous patients had larger tumor sizes (P = 0.001), lower differentiation degree of tumor cells (P = 2.2 × 10−4), and less possibility of distant metastases (P = 0.006) than the AA homozygous patients.
In addition, statistical analysis results showed that the A-allele frequency was significantly higher in the patients than that in the control group (35 vs. 28.6 %; P = 0.036), indicating that the A allele is associated with the incidence of gastric cancer.
The effect of rs1599795 on the regulatory role of miR-132-3p, miR-212-3p, and miR-361-5p in CD80 expression
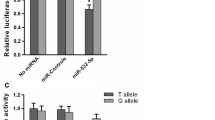
Since the SNP rs1599795 was predicted to locate in the binding site of miR-132-3p, miR-212-3p, and miR-361-5p, we hypothesized that the expression of CD80 is regulated by these microRNAs, which can be impacted by rs1599795. To test whether or not the inhibitory role of miR-132-3p, miR-212-3p, and miR-361-5p is impacted by rs1599795, we constructed pGL3 vectors including A-allele- and T-allele-specific binding sequences, and then co-transfected it with miR-132-3p, miR-212-3p, or miR-361-5p in CHO cells. We found that the expression of T-allele-specific pGL3 construct was significantly suppressed by miR-132-3p (P = 0.003; Fig. 3b) and miR-212-3p (P = 0.018; Fig. 3b). However, the expression of A-allele-specific pGL3 construct was not inhibited by miR-132-3p (P = 0.458) and miR-212-3p (P = 0.659; Fig. 3a). In addition, the miR-132-3p significantly suppressed the expression of A-allele-specific pGL3 construct (P = 0.006; Fig. 3a), instead of T-allele-specific pGL3 construct (P = 0.092; Fig. 3a). These findings suggested that the inhibitory roles of miR-132-3p, miR-212-3p, and miR-361-5p in the expression of CD80 were affected by rs1599795.
Effect of rs1599795 on the inhibitory role of miR-132-3p, miR-212-3p, and miR-361-5p in CD80 expression. a The expression of A-allele-specific pGL3 construct was obviously inhibited by miR-361-5p (P = 0.006). b The expression of T-allele-specific CD80/3′-UTR/pGL3 luciferase was significantly suppressed by miR-132-3p (P = 0.003) and miR-212-3p (P = 0.018), but not by miR-361-5p (P = 0.092). The negative controls (NC) were performed with an unrelated miRNA sequence (5′-UUCUCCGAACGUGUCACGUTT-3′), respectively
Discussion
In the present study, we investigated the relationship between miRSNPs within the 3′-UTR of CD80 gene and the risk of gastric cancer. We found that an SNP rs1599795, locating in the binding sites of miR-132-3p, miR-212-3p, and miR-361-5p, through disrupting the inhibitory role of miR-132-3p, miR-212-3p, and miR-361-5p on CD80 expression, played an important role in the development of gastric cancer. Compared with the AA genotype, both the TA genotype and T-allele carriers had a substantially higher risk of gastric cancer. Furthermore, the TA heterozygotes were significantly more prevalent in the patients (OR 1.44, 95 % CI 0.98–2.11) with a larger tumor size (P = 0.001), deeper infiltration (P = 1.5 × 10−5), higher possibility of lymph node metastasis (P = 0.003), and more in the late stage (TNM stages III and IV; P = 0.003); the TT homozygotes had larger tumor size (P = 0.001) and lower degree of differentiation (P = 2.2 × 10−4). These findings indicated that the T allele might be a risk factor for the occurrence and development of gastric cancer.
CD80, as an important co-stimulatory ligand, by binding with its receptor CD28, plays an important role in the cell-mediated immune responses [17]. Studies have demonstrated that the transfection of CD80 into tumor cells resulted in remarkable reduction of tumor metastasis [18]. However, most tumors including gastric cancer had no or low expression of CD80, leading to unresponsive T cells for the specific antigens and ineffective anti-tumor immune response [19, 20].
Numerous microRNAs have been reported to be deregulated in cancers; however, the consequence of miRNA deregulation in cancer is still unknown for many miRNAs [21]. As for the miRNAs of interest in this study, miR-132 expression was found to be increased in lung cancer [22–24], endocrine pancreatic tumors [22], and colorectal carcinoma [25]. Increased miR-212 expression was reported in colorectal carcinoma [25] and gastric cancer [26], but it was also shown to be down-regulated in gastric cancers [27, 28]. miR-361 expression is increased in bladder cancer [29]. In this study, we also found T allele of SNP rs1599795 that lead to CD80 lost one miRNA of miR-361-5p inhibition, but intensify inhibition by two miRNAs miR-132-3p and miR-212-3p, thus lead to low expression of CD80, which may be involved in cancer immune evasion. That may be an important part in number of mechanisms regulating expression in CD80.
In summary, we reported the first evidence that the SNP rs1599795 in CD80 3′-UTR was involved in the occurrence of gastric cancer. This SNP was also found to be related to the clinicopathological features of gastric cancer, suggesting it may have important roles in tumor development. Furthermore, we provided a novel insight into the regulatory mechanism of CD80 expression mediated by miR-132-3p, miR-212-3p, and miR-361-5p in gastric cancer.
References
Sunami T, Yashiro M, Chung KH. ICAM-1 (intercellular adhesion molecule-1) gene transfection inhibits lymph node metastasis by human gastric cancer cells. Jpn J Cancer Res. 2000;91:925–33.
Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108.
Wu WK, Lee CW, Cho CH, Fan D, Wu K, Yu J, et al. MicroRNA dysregulation in gastric cancer: a new player enters the game. Oncogene. 2010;29:5761–71.
Bretscher P, Cohn M. A theory of self-nonself discrimination. Science. 1970;169:1042–9.
Bhatia S, Edidin M, Almo SC, Nathenson SG. B7-1 and B7-2: similar costimulatory ligands with different biochemical, oligomeric and signaling properties. Immunol Lett. 2006;104:70–5.
Si L, Si H, Chen Y, Sun Y, Wing Y. B7-1 antigen expression in tumor cells from cancerous human tissues. Anal Quant Cytol Histol. 1999;21:521–6.
Wu TC, Huang AY, Jaffee EM, Levitsky HI, Pardoll DM. A reassessment of the role of B7-1 expression in tumor rejection. J Exp Med. 1995;182:1415–21.
Koyama S, Maruyama T, Adachi S, Nozue M. Expression of costimulatory molecules, B7-1 and B7-2 on human gastric carcinoma. J Cancer Res Clin Oncol. 1998;124:383–8.
Scarpa M, Bortolami M, Cecchetto A, Faggian D, Kotsafti A, Ruffolo C, et al. Mucosal immune environment in colonic carcinogenesis: CD80 up-regulation in colonic dysplasia in ulcerative colitis. Eur J Cancer. 2011;47:611–9.
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97.
Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33.
Carthew RW. Gene regulation by microRNAs. Curr Opin Genet Dev. 2006;16:203–8.
Cai Y, Yu X, Hu S, Yu J. A brief review on the mechanisms of miRNA regulation. Genomics Proteomics Bioinform. 2009;7:147–54.
Wu D, Wang F, Dai WQ, He L, Lu J, Xu L, et al. The miR-146a rs2910164 G > C polymorphism and susceptibility to digestive cancer in Chinese. Asian Pac J Cancer Prev. 2013;14:399–403.
Wang WP, Li F, Mao Y, Zhou H, Sun J, Li R, et al. A miR-570 binding-site polymorphism in the B7-H1 gene is associated with the risk of gastric adenocarcinoma. Hum Genet. 2013;132:641–8.
Yang P, Tang R, Zhu J, Zou L, Wu R, Zhou H, et al. A functional variant at miR-24 binding site in B7-H2 alters susceptibility to gastric cancer in a Chinese Han population. Mol Immunol. 2013;56:98–103.
Linsley PS, Brady W, Grosmaire L, Aruffo A, Damle NK, Ledbetter JA. Binding of the B cell activation antigen B7 to CD28 costimulates T cell proliferation and interleukin 2 mRNA accumulation. J Exp Med. 1991;173:721–30.
Fujii H, Inobe M, Kimura F, Murata J, Murakami M, Onishi Y, et al. Vaccination of tumor cells transfected with the B7-1 (CD80) gene induces the anti-metastatic effect and tumor immunity in mice. Int J Cancer. 1996;66:24–219.
Schwartz RH. A cell culture model for T lymphocyte clonal anergy. Science. 1990;248:1349–56.
Chen XL, Cao XD, Kang AJ, Wang KM, Su BS, Wang YL. In situ expression and significance of B7 costimulatory molecules within tissues of human gastric carcinoma. World J Gastroenterol. 2003;9:1370–3.
Mishra PJ, Banerjee D, Bertino JR. miRSNPs or miR-polymorphisms, new players in microRNA mediated regulation of the cell: introducing microRNA pharmacogenomics. Cell Cycle. 2008;7:853–8.
Park J-K, Henry JC, Jiang J, Esau C, Gusev Y, Lerner MR, et al. miR-132 and miR-212 are increased in pancreatic cancer and target the retinoblastoma tumor suppressor. Biochem Biophys Res Commun. 2011;406:518–23.
Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–61.
Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–98.
Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–36.
Wu WY, Xue XY, Chen ZJ, Han SL, Huang YP, Zhang LF, et al. Potentially predictive microRNAs of gastric cancer with metastasis to lymph node. World J Gastroenterol. 2011;17:3645–51.
Xu L, Wang F, Xu XF, Mo WH, Xia YJ, Wan R, et al. Down-regulation of miR-212 expression by DNA hypermethylation in human gastric cancer cells. Med Oncol. 2011;1:189–96.
Wada R, Akiyama Y, Hashimoto Y, Fukamachi H, Yuasa Y. MiR-212 is downregulated and suppresses methyl-CpG-binding protein MeCP2 in human gastric cancer. Int J Cancer. 2010;127:1106–14.
Song Tao, Xia Wei, Ningsheng Shao X, Zhang CW, Yiguang W, et al. Differential miRNA expression profiles in bladder urothelial carcinomas. Asian Pacific J Cancer Prev. 2010;11:905–11.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 81270031), the Natural Science Foundation of the Higher Education Institutions of Jiangsu Province (No. 12KJB320012), and a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Conflict of interest
The authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Ruirong Wu and Fuchao Li contributed equally to this work.
Rights and permissions
About this article
Cite this article
Wu, R., Li, F., Zhu, J. et al. A functional variant at miR-132-3p, miR-212-3p, and miR-361-5p binding site in CD80 gene alters susceptibility to gastric cancer in a Chinese Han population. Med Oncol 31, 60 (2014). https://doi.org/10.1007/s12032-014-0060-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-014-0060-2