Abstract
There has been few report discussing the expression and function of miR-212 in gastric cancer (GC). The aim of this pilot study was to investigate the expression of miR-212 in both gastric cancer tissues and gastric cancer cells and further explores the possible reasons for this change and the impact on the development of gastric cancer. qRT–PCR was used to detect the expression of miR-212 in primary GC tissues, adjacent normal tissues, gastric cancer cell lines BGC-823, SGC-7901, MKN-45, and normal gastric mucosa cell line GES. The expression of miR-212 was evaluated before and after treatment with methylation inhibitor-5-Aza-2′-deoxycitidine (5-Aza-dC), finally anti-miRNA and dual luciferase reporter assay were used to prove that MYC is a target gene of miR-212. The results showed that a significant reduction of miR-212 expression in GC tissues was observed compared to that in normal tissues (P = 0.002). At the same time, miR-212 expression level in normal gastric mucosa cell line GES was higher than that of in gastric cancer cell lines BGC-823, SGC-7901, and MKN-45 (P = 0.015, 0.008, 0.044, respectively). Computer sequence analysis showed the hypermethylation of CpG islands(CPI) in the promoter regions of miR-212 led to the lower expression of miR-212 in gastric cell strains (BGC-823 and SGC-7901). MiR-212 expression was significantly recovered after treatment with methylation inhibitor 5-Aza-dC (P = 0.016, 0.000, 0.015, respectively). Then, the results of AMOs transfection and dual luciferase reporter assay showed that Myc is a target of miR-212, which will be helpful to verify the function of miR-212 in carcinogenesis. The conclusion could be deduced from the study that decreased expression of miR-212 may be due to hypermethylation of CPI in gastric cancer cells, and miR-212 might act on the progression of gastric cancer through the potential target gene Myc.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
MicroRNAs (miRNAs) are small, noncoding RNA controlling the activity of protein-coding genes by combining 3′-UTR of target mRNA and inhibiting its translation [1]. With the in-depth study of miRNAs, it has been reported that miRNAs had different degrees of change in a variety of tumor tissues, indicating the existence of certain relevance between its expression and tumor occurrence, development, and prognosis [2–5].
Gastric cancer is one of the most common malignant tumors. High rates apply to China, Japan, Vietnam, and other Southeast Asian countries due to traditional eating habits, climate, and geography. In China, 0.3 million deaths and 0.4 million new cases from gastric cancer (GC) per year would rank the third most common cancer [6]. Recently, a large number of studies have shown that microRNAs (miRNAs) may play an important role in GC [1].
Previous studies focused on looking for differentially expressed miRNAs in human GCs compared to noncancerous tissues, they found that miR-96, 136, 212, 218, and 339 were down-regulated in GCs [2, 3]. However, the functions of such down-regulated miRNAs are unclear. In this study, we detected the expression of miR-212 and hoped to ascertain whether or not miRNA silencing is due to DNA methylation. First, we detected the expression of miR-212 in gastric cancer cell lines and GC tissues, and then for its CPI of putative promoter to do MSP, to determine the methylation status of different cells. Then, we identified possible target genes by transfecting the specific antisense oligodeoxyribonucleotides for miR-212 and to determine the relevance of the development of gastric cancer.
Materials and methods
Tissue samples
Gastric cancerous tissues and adjacent normal stomach tissues from 10 GC patients in the tenth hospital Tongji University were harvested from surgical biopsies and kept at −80°C for miRNA extraction. Cancerous tissues and adjacent normal tissues were ascertained by the pathological analysis. It had obtained the patients’ consent and approval of the Ethics Committee of Tongji University.
Cell culture
Human gastric cancer cell lines BGC-823, SGC-7901, MKN-45, and normal gastric mucosa cell line GES were obtained from Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China). Cells were cultured in 25-cm2 cell culture flask at 37°C in a humidified atmosphere of 5% CO2 with RPMI-1640 Medium (Life Technologies, Grand Island, NY, USA) containing 10% fetal calf serum with 50 U/ml penicillin and 50 μg/ml streptomycin. Exponentially growing cells were used for experiments. 5-Aza-CdR was added to the cell culture medias in the 1st and 3nd days at the concentration of 1 μmol/l, and then the cells were harvested for following experiments.
Real-time-polymerase chain reaction (Real-time-PCR)
Total RNAs were extracted by using Trizol in accordance with the manufacturer’s instructions. Reverse transcriptase reactions contained RNA samples, 50 nM stem-loop RT primer (Table 1), 1 × RT buffer (Biosystems), 0.25 mM each of dNTPs, 3.33 U/ml MultiScribe reverse transcriptase (Biosystems), and 0.25 U/ml RNase inhibitor (P/N: N8080119; Applied Biosystems). The 7.5 μl reactions were incubated in an PTC-200 Pletier Thermal Cycler in a 30- or 48-well plate for 30 min at 16°C, 30 min at 42°C, 5 min at 85°C, and then were held at 4°C [7]. Both the concentration of RNAs and cDNA were quantified by ultraviolet spectrophotometer (GeneQuant pro, Amersham).
In each sample, we calculated a ∆Ct (target- reference), which is equal to the difference between threshold cycles for miR-212 (target) and those for U6 RNA (reference), target gene–MYC (target) (sense: 5′- TTCTGTGGAAAAGAGGCAGG-3′, antisense: 5′- TGCGTAGTTGTG CTGATGTG-3′) and GAPDH (reference) (sense: 5′- ACATCGCTCAGACACCATG-3′, antisense: 5′- TGTAGTTGAGGTCAATGAAGGG-3′). The fold-change between gastric cancer cell lines or GC tissues and normal gastric mucosa cell line or noncancerous tissues for miR-212 was calculated with the 2−∆∆Ct method, in which ∆∆Ct = ∆Ct (target- reference) (in tumor cell lines or GC tissues)−∆Ct (target-reference) (in normal gastric mucosa cell line or noncancerous tissues) [8]. The 2−∆∆Ct values were calculated for each sample relative to the normal control for the expression of miR-212. The same method was also used to determine the relative expression levels of miR-212 and MYC between GES transfected with AMO-212, Mutant AMO-212 and NC. Each sample was measured three times.
Sodium bisulfite modification, methylation-specific PCR (MSP)
Fully methylated DNA was prepared by methylating genomic DNA with SssI methylase (Genetimes Technology, China). Bisulfite modification was performed using 2 μg of genomic DNA as previously described [9], and the modified DNA was suspended in 40 μl of Tris–EDTA buffer. An aliquot of 1 μl was used for methylation-specific PCR (MSP) unmethylated (U) sequences (Table 1).
For MSP, the fully methylated DNA was used to determine an annealing temperature that specifically amplifies only methylated DNA. A minimum number of PCR cycles to obtain visible bands were determined using the fully methylated DNA, and four cycles were added for the analysis of gastric cancer cell lines. The primers were designed just upstream of reported transcription start sites within the CpG island (CGI) (Fig. 2).
Targets scan
In order to find the functional explanation of miR-212, we explored the most potential targets of miR-212. Information was collected from three most known target predication databases, TargetScan, PicTar, and mirBase. These approaches allowed us to select the common targets found in three different algorithms.
Transfection of anti-miR-212 oligodeoxyribonucleotides
Anti-miRNA antisense oligodeoxyribonucleotides (AMOs) of miR-212 (5′-GGCCGTGACTGGAGACTGTTA-3′) were chemically synthesized and transfected into normal gastric mucosa cell line GES, missense oligonucleotide named for Mutant AMO-212 as randomized controls and cells nontransfected AMO-212 as blank control group. Using 50 μl serum-free RPMI-1640 medium was diluted 30 pmol miR-212 AMO, and then take 2 μl of the liposome Lipofectamine ™2000 diluted to 50 μl RPMI-1640 culture based in mixing gently incubated at room temperature after 5 min, then diluted Lipofectamine ™2000 separately with AMO-212 and Mutant AMO-212 diluted mixed gently incubated at room temperature after mixing 20 min. Each well by adding complex, gently shake the culture plates for mixing. Placed in 37°C, 5% CO2 incubator overnight incubation, the replacement of the medium containing 10% fetal calf serum continued to train 48 h for follow-up testing.
Western blot
To study the changes of miR-212 targets in protein levels, Western blot analysis was performed to detect MYC and MECP2 proteins in cells transfected with miR-212 inhibitor and NC cell line GES. During the experiment, mouse anti-human monoclonal antibodies (Santa Cruz Biotechnology, at 1:500 dilutions for MYC, 1:300 dilutions for MECP2) were used as the primary antibody, and anti-mouse-HRP conjugate antibody (Santa Cruz Biotechnology, at 1:1500 dilutions) was used as the secondary antibody. The immunocomplexes were detected using the ECL system (Beyotime Biotechnology, China). β-actin was used as the positive control.
Dual luciferase reporter assay
The 47-bp MYC 3-UTR containing the putative target site for miR-212 was synthesized. Luciferase constructs were made by ligating the sequence into the pGL3 (luc2/SV40) firefly luciferase reporter vector (Promega, Madison, WI). BGC-823 cells were co-transfected using HiPerFect (Qiagen) with 4 ng of pGL3 vector containing a 3-UTR sequence or mutant, 4 ng pGL3 (hRluc/TK) renilla luciferase control vector and 10 nM miR-212 mimics molecule. Luciferase activity was measured 24 h after transfection using a Dual-Luciferase Reporter Assay System (Promega). Relative luciferase activity was calculated by normalizing the firefly luminescence as to the renilla luminescence.
Statistical analysis
Data were expressed as mean ± SD. With SPSS 11.0 software, the statistical analysis was performed using one-way ANOVA and student t-test. P < 0.05 was considered statistically significant.
Results
Down-regulation of miR-212 expression in human malignant GC tissues and gastric cancer cell lines
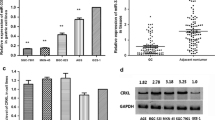
We investigated the expression levels of miR-212 in malignant GC tissues and adjacent normal tissues, gastric cancer cell lines, and normal gastric mucosa cell line using real-time RT–PCR. The results of paired group t-test showed that there was no statistical difference of miR-212 expression in GC tissues compared to normal tissues (data not shown), but after the 10 patients were divided into two groups according to the pathological data, group T1 included the GC patients without metastasis, whereas group T2 were the GC patients that have gastric micrometastasis. Then, the miR-212 mRNA expressions in group T1, T2 were compared to that in the normal group using t-test. The result showed that the expression level of miR-212 in group T1 decreased significantly, whereas that in group T2 increased about twofold (Fig. 1a–b, T1 vs. N, P = 0.002; T2 vs. N, P = 0.003). It also showed that miR-212 expression level in gastric cancer cell lines BGC-823, SGC-7901, and MKN-45 were significantly lower than that in normal gastric mucosa cell line GES (Fig. 1c–d, SGC-7901 vs. GES, P = 0.015; BGC-823 vs. GES, P = 0.008; MKN-45 vs. GES, P = 0.044).
The expression of miR-212 in human malignant GC tissues and gastric cancer cell lines. a 10 GC tissues were collected in the surgical biopsies, miR-212 were detected using real-time RT–PCR. The results were quantified by determining the intensities of the bands compared to that of ß-actin. Every experiments were repeated three times; the data are presented as mean ± SD. b 10 patients were divided into two groups according to the pathological data, that is, group T1 included S1, S2, S4, S5, S6, S7, S9, S10, which were GC without metastasis; group T2 included S3, S8, which were micrometastasis. The expression level of miR-212 in group T1 and T2 were compared to that in normal tissues using t-test, P < 0.05 (*), P < 0.01 (**) were considered a statistically significant difference. c–d The expression of miR-212 in human gastric cancer cell lines. BGC-823, SGC-7901, MKN45 are three kinds of GC cell strains, whereas GES is normal gastric mucosa cell line. All of the cells were cultured, total RNAs were extracted and analyzed by qRT–PCR. The results were quantified by determining the intensities of the bands compared to that of U6 RNA, which is the reference gene. c A representative RT–PCR result from one experiment of three, U6 was used as the control; d The expression of miR-212 was significantly lower in GC cells than that in normal gastric cells. The data are presented as mean ± SD. Significances were calculated using t-test. P < 0.05 (*), P < 0.01 (**) were considered a statistically significant difference
The presence of miR-212 hypermethylation in gastric cancer cell lines
Our analysis showed that miR-212 gene locates in CpG islands (CGI), while 5′-UTR of the gene is also existence of CGI (Fig. 2). So it is tempting to speculate that hypermethylation of CGI leads to silencing of miR-212 gene expression. To confirm this hypothesis, methylation-specific PCR (MSP) was used to detect the CGI of miR-212, and the result showed that hypermethylation of CGI was found in higher level in at least two kinds of gastric cancer cell lines (SGC-7901 and BGC- 823) compared to normal gastric mucosa cell line (Fig. 3c, P = 0.005, P = 0.002, respectively).
The expression of methylation (M) and demethylation (U) of CGI in human gastric cancer cell lines before and after 5-Aza-CdR treatment. BGC-823, SGC-7901, MKN-45 are three kinds of GC cell strains, whereas GES is normal gastric mucosa cell line. SssI methylase was the methylation-positive control. a–b The expression of CGI hypermethylation (M) and demethylation (U) in human gastric cancer cell lines before and after 5-Aza-CdR treatment, respectively. Representive RT–PCR result was shown here; c the relative expression level of M/U. (−) is the results before 5-Aza-CdR treatment, whereas (+) is the results after 5-Aza-CdR treatment. d The relative expression level of miRNA-212 after the 5-Aza-CdR treatment. Every experiment was repeated three times. The data are presented as mean ± SD. The relative expression level of M and U in GC cells were compared to those GES, Significances were calculated using t-test. P < 0.05 (*), P < 0.01 (**) were considered a statistically significant difference
Methylation of CGI was further confirmed by analyzing its expression in association with demethylation after treatment with a demethylating agent, 5-Aza-CdR, in gastric cancer cell lines (SGC-7901, BGC-823 and MKN-45). The result demonstrated that after the treatment of 5-Aza-CdR, the expression of methylation of CGI in GC cell strains decreased to that in GES, which is a normal gastric mucosa cell line (Fig. 3a–c, P = 0.005 (SGC-7901), 0.002 (BGC-823), 0.028 (MKN-45), respectively). The relative expression level of miRNA-212 increased significantly after the 5-Aza-CdR treatment (Fig. 3d, P = 0.016 (SGC-7901), 0.000 (BGC-823), 0.015 (MKN-45), respectively). This further suggested that miR-212 was methylation-silenced in gastric cancer cells.
Identification of miR-212 targets
We hypothesis that MYC might be miR-212 target, which will be helpful to verify the function of miR-212 in carcinogenesis. Anti-microRNA antisense oligodeoxyribonucleotides (AMOs) of miR-212 were transfected into normal gastric mucosa cell line GES, MYC mRNA, and protein were detected using real-time RT–PCR and Western blot. The result showed that miR-212 can be silenced effectively by AMO-212 (Fig. 4b, P = 0.042). After AMO-212 transfection, the level of MYC mRNA increased significantly compared to the groups transfected with mutant AMO-212 and blank control (Fig. 4b, P = 0.015, 0.013, respectively). Moreover, the expression of MYC protein increased twofold compared to the negative control. (P = 0.019, Fig. 4c). These data proved that MYC might be one of the target genes of miR-212. At the same time, the result of Western blot showed that the expression level of MECP2 protein, a recognized target of miRNA-212, was also increased about twofold compared to the negative control. (AMO-212 vs. Mutant, P = 0.000; AMO-212 vs. blank control, P = 0.001, Mutant AMO-212 vs. blank control P = 0.248, Figs. 4d, 5).
Expression of MYC mRNA and protein on normal gastric mucosa cell line GES by AMO-212 transfection. a Expression of MYC and mir-212 mRNA in GES by AMO-212 vector transfection. The level of MYC and mir-212 mRNA in GES was evaluated by real-time RT–PCR, and the results were quantified by determining the intensities of the bands compared to that of ß-actin. Every experiment was repeated three times; the data are presented as mean ± SD. The relative expression levels of MYC and mir-212 mRNA were compared to those of GES, Significances were calculated using ANOVA. P < 0.05 (*), P < 0.01 (**) were considered a statistically significant difference. b–c Down-regulation of MYC after AMO-212 transfection. GES cells were examined for MYC expression by Western blot after transfected with the AMO-212, with the cells transfected with mutant AMO-212, no AMO-212 as the negative control, blank control, respectively. The results were quantified by determining the intensities of the bands compared to that of ß-actin. Relative protein levels are shown. c–d The expression of MYC and MECP2 in GES cells after AMO-212 transfection. A representive result of Western blot from three experiments was shown. MYC or/and MECP2 expression after transfected with the AMO-212, and with mutant AMO-212 was compared to that in the blank control. The data are presented as mean ± SD. Significances were calculated using ANOVA. P < 0.05 (*), P < 0.01 (**) were considered a statistically significant difference
Verification of the potential target genes of miR-212. BGC-823 cells were co-transfected with MYC pGL-3-3′-UTR, pGL-3 control, and negative control. 24 h later, luciferase activity was measured. Values are presented as relative luciferase activity after normalization to Renilla control luciferase activity. The ratio of luciferase activity of each construct was calculated by luminometer. Data (mean ± SD) are from three independent experiments. Significances were calculated using the ANOVA. P < 0.05 (*), P < 0.01 (**) were considered a statistically significant difference
MYC is a target of miR-212
Although the data above partly supported the hypothesis that MYC is a target genes of the miR-212, but the undirected relationship could not be ruled out. So miR-212 mimics was synthesized, and dual luciferase reporter assay was applied to prove the hypothesis. As expected, luciferase activity of the MYC pGL3-3′-UTR was significantly inhibited compared to the blank control and the random control (pGL3-3′-UTR mutant) after the introduction of miR-212 mimics into BGC-823 cells (P = 0.005, 0.001). And there was no significant difference between the random control and the blank control (P > 0.05). These data suggested that MYC is a potential target of miR-212.
Discussion
In this study, we addressed for the first time that miR-212 is significantly reduced in gastric cancer cells compared to normal gastric mucosa cell. It is tempting to speculate that this decrease may be the result of its putative promoter hypermethylation. The evidence presented in this study shows that gastric cancer cells were hypermethylated, and methylation inhibitor 5-Aza-CdR restored the expression of miR-212. To explore the possible role of miR-212 in gastric cancer cells, we transfected AMO-212 into gastric cancer cell lines to detect its possible target genes.
Studies have confirmed that in certain regions of chromosome, DNA molecules by epigenetic regulation were able to induce tumor production. Interestingly, researchers found that some miRNAs are located in or around CpG islands and the expression of these miRNAs are susceptible to epigenetic regulation, while some of the miRNAs as tumor suppressor genes are usually induced silencing by over-methylation, leading to tumorigenesis [10]. Recent reports demonstrated that, however, the expression of some miRNAs, such as miR-34b and Let-7a-3, is regulated by DNA methylation epigenetic mechanisms [11, 12], while some miRNAs locating in CpG island or in which, at the same time found that some miRNAs with tumor suppression function are usually silenced due to DNA hypermethylation leading to tumorigenesis [11]. In addition, recent studies demonstrated that expression of some miRNAs is regulated by epigenetic mechanisms in human cancers, indicating methylation of miRNAs and tumorigenesis are closely linked [13]. At present, there were many researches for expression profile and epigenetic of miRNAs in gastric cancer, Rosa et al. had a review for miRNAs expression profile researches in stomach diseases, and listed some abnormal expression of miRNAs [1]. Takayuki et al. discovered that DNA methylation of certain miRNA genes was associated with H.pylori infection [14]. In the present study, we got the similar result that the lower expression of miR-212 is partly due to the hypermethylation of the CpG island of miR-212. With 5-Aza-CdR, a demethylating agent, the expression of miR-212 in GC cell strains recovered to that of the normal cells.
With three softwares, it was predicted that MYC may be one of the targets. In order to further verify this speculation, specific antisense oligodeoxyribonucleotides of miR-212 was transfected into normal gastric mucosa cell line GES, luciferase reporter assay was used and MECP2, one of the known targets of miR-212 were analyzed in this paper. All of the study supported the hypothesis that MYC is the target of miR-212. Of course, many questions are still unknown, such as whether the miR-212 is directly combined with the MYC mRNA transcription and translation, whether the level of miR-212 methylation has a tangible impact on the clinical progress of gastric cancer? To answer these questions, we will further collect clinical specimens to clarify miR-212 methylation and the clinical relationship between the risks of gastric cancer.
The c-Myc oncogene is a master regulator of genes involved in diverse cellular processes and plays a significant role in cell proliferation, differentiation, transformation, and apoptosis. The c-Myc protein located upstream of signaling pathways regulating cellular replication/growth as well as apoptosis/growth arrest, c-Myc may be helpful to integrate processes determining cell numbers and tissue size in physiology and disease [15]. The abnormalities of c-myc gene occur frequently in tumor tissue, such as amplification, abnormal expression of mRNA or protein could promote the excessive proliferation of tumor cells. But once tumor cells were induced differentiation, c-myc mRNA levels are correspondingly reduced in tumor tissues [16, 17], so this ‘dual potential’ allows c-Myc to act as its own tumor suppressor.
In a word, miR-212 is significantly reduced in gastric cancer cells, and the change of epigenetic such as methylation of the putative promoter regions or modifications of histone, to certain extent, plays an important role in the regulation of miRNAs expression and tumorigenesis.
References
Visone R, Petrocca F, Croce CM. Micro-RNAs in gastrointestinal and liver disease. Gastroenterology. 2008;135(6):1866–9.
Garzon R, Fabbri M, Cimmino A, et al. MicroRNA expression and function in cancer. Trends Mol Med. 2006;12(12):580–7.
Chivukula RR, Mendell JT. Circular reasoning: microRNAs and cell-cycle control. Trends Biochem Sci. 2008;33(10):474–81.
Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–66.
Esquela-Kerscher A, Slack FJ. Oncomirs- microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–69.
Yang L. Incidence and mortality of gastric cancer in China. World J Gastroentrol. 2006;12(1):17–20.
Chen CF, Ridzon DA, Broomer AJ. Real- time quantification of microRNAs by stem-loop RT- PCR. Nucleic Acids Res. 2005;33(20):e179.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using Real-Time quantitative PCR and the 2−∆∆Ct method. Methods. 2001;25(4):402–8.
Kaneda A, Kaminishi M, Sugimura T, et al. Decreased expression of the seven ARP2/3 complex genes in human gastric cancers. Cancer Lett. 2004;212(2):203–10.
Lehmann U, Hasemeier B, Römermann D, et al. Epigenetic inactivation of microRNA genes in mammary carcinoma. Verh Dtsch Ges Pathol. 2007;91:214–20.
Weber B, Stresemann C, Brueckner B, et al. Methylation of human microRNA genes in normal and neoplastic cells. Cell Cycle. 2007;6(9):1001–5.
Kozaki K, Imoto I, Moqi S, et al. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res. 2008;68(7):2094–105.
Yang N, Coukos G, Zhang L. MicroRNA epigenetic alterations in human cancer: one step forward in diagnosis and treatment. Int J Cancer. 2008;122(5):963–8.
Ando T, Yoshida T, Enomoto S, et al. DNA methylation of microRNA genes in gastric mucosae of gastric cancer patients: its possible involvement in the formation of epigenetic field defect. Int J Cancer. 2009;124(10):2367–74.
Vita M, Henriksson M. The Myc oncoprotein as a therapeutic target for human cancer. Semin Cancer Biol. 2006;16(4):318–30.
van Waardenburg RC, Meijer C, Pinto-Sietsma SJ, et al. Effects of c-myc oncogene modulation on differentiation of human small cell lung carcinoma cell lines. Anticancer Res. 1998;18(1A):91–5.
Robson S, Pelengaris S, Khan M. c-Myc and downstream targets in the pathogenesis and treatment of cancer. Recent Pat Anticancer Drug Discov. 2006;1(3):305–26.
Acknowledgments
We thank Miss Yong Hua for excellent advice and critical reading of this paper.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Xu, L., Wang, F., Xu, XF. et al. Down-regulation of miR-212 expression by DNA hypermethylation in human gastric cancer cells. Med Oncol 28 (Suppl 1), 189–196 (2011). https://doi.org/10.1007/s12032-010-9691-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12032-010-9691-0