Abstract
Recent investigations have emphasized the role of aberrant expression of microRNAs (miRNAs) in progression of almost all types of cancers. Exosomes, membrane-enclosed natural nanovesicles, transport cellular contents, including proteins, mRNAs, and miRNAs, between cells. Unique features of exosomes make them an appropriate carrier for drug delivery. miRNA-381 is one of the downregulated miRNAs in several cancers including triple-negative breast cancer (TNBC) and restoration of its expression in TNBC cells can restrict their migratory ability through targeting several signaling pathways. In current study, we exploited the exosomes isolated from adipose-derived mesenchymal stem cells (ADMSC-exosomes) to deliver miR-381 mimic to MDA-MB-231 cells to elucidate their effects on TNBC cells. The effects of miR-381 loaded ADMSC-exosomes on proliferation, apoptosis, migration, and invasion of MDA-MB-231 cells were analyzed. Our results indicated that ADMSC-exosomes were successfully isolated and internalized by MDA-MB-231 cells. miR-381 mimic was efficiently delivered to MDA-MB-231 cells by ADMSC-exosomes. miR-381 loaded ADMSC-exosomes significantly downregulated the expression of epithelial to mesenchymal transition (EMT) related genes and proteins. Notably, miR-381 loaded ADMSC-exosomes inhibited proliferation, migration, and invasion capacity of MDA-MB-231 and promoted their apoptosis in vitro. Taken together, we showed that ADMSC-exosomes could be used as efficient nanocarriers for RNA-based therapies.

Graphical abstract
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
microRNAs (miRNAs) are a subset of small non-coding RNAs that are involved in post-transcriptional regulation of gene expression. An increasing amount of evidence has shown the involvement of miRNAs in almost all types of cancers. Aberrant expression of miRNAs plays an indispensable role in cancer initiation, progression, and metastasis [1]. Correction of miRNA expression profile can restore normal gene expression profile in cancer cells, which it depends on an efficient delivery system [2].
Exosomes are membrane-enclosed nanovesicles (30–150 nm diameter) produced by various cell types, including stem cells [3, 4]. They play a key role in intercellular communication through the transfer of their content (nucleic acids, proteins, and lipids) between originating cells and recipient cells [3]. High biocompatibility and high stability of exosomes in circulatory system overcome common disadvantages of synthetic nanocarriers, such as liposomes and carbon nanotubes, and cause they attract a great attention as a drug delivery vehicle [5].
Mesenchymal stem cells (MSCs) are multipotent adult stem cells that their unique properties cause their wide-ranging use in disease treatment [6]. Among the cells producing exosomes, MSCs can be an ideal cell source because of their simple isolation, high ex vivo expansion capacity, and high yeild in exosome production [7]. Low toxicity and immunogenicity, high biosafety, and natural and preferential tropism toward target cells make MSC-derived exosomes preferable to exosomes of other cells [8]. Besides, tumor-suppressive potential of MSC-derived exosomes in vivo and in vitro suggest they can also be used as the promising drug delivery vehicles in cancer therapy [9,10,11].
It has been found that miR-381 acts as a tumor suppressor miRNA in different types of cancers including gastric cancer, colon cancer, prostate cancer, and breast cancer [12,13,14,15]. It has been reported that miR-381 is downregulated in breast cancer cell lines and tissues and its downregulation is associated with breast cancer proliferation, metastasis, and drug resistance [14, 16,17,18]. Therefore, miR-381 may be a considerable therapeutic target for breast cancer treatment and finding the appropriate approach to deliver miR-381 to cancerous cells seems essential. The potential risks and biosafety concerns related to conventional transfection methods [19] and lack of experience in exosome-mediated delivery of miR-381 to breast cancer cells caused we perform an in vitro study as a decisive first step toward testing exosomes efficacy in miR-381 delivery.
In our previous study using viral packaging of miR-381 and transduction of MDA-MB-231, we produced stably expressing miR-381 breast cancer cells and showed that miR-381 targets key genes of Wnt signaling pathway and reduces TNBC cells aggressiveness [16]. In the present study, we investigated whether exosomes derived from adipose mesenchymal stem cells (ADMSC-exosomes) can act as a vehicle for delivery of miR-381 to TNBC cells and affect their malignant behaviors.
Material & Methods
ADMSC Isolation & Characterization
Human adipose tissues were obtained from healthy young females (aging from 22 to 35) undergoing liposuction or lipolysis surgery according to procedures approved by the Ethics Committee at the Shahid Beheshti University of Medical Sciences (IR. SBMU.REC.1397.004).
Freshly collected tissues were washed with sterile phosphate-buffered saline (PBS) containing 1% penicillin and streptomycin and then were incubated with 0.1% collagenase I (Sigma, USA) in DMEM/F12 (Gibco, USA) for 30 min at 37 °C and agitated gently. Collagenase was inactivated by adding an equal volume of DMEM/F12 with 10% fetal bovine serum (FBS; Gibco, USA), followed by centrifugation at 1200 RPM for 20 min (Hettich, Germany). Cellular pellet was resuspended in DMEM/F12, seeded in culture flasks and cultured in culture medium (DMEM/F12 containing 15% FBS and 1% penicillin and streptomycin) at 37 °C in a 5% CO2 incubator. The following day, after washing non-adherent cells, the medium was replaced with fresh one. Every 2–3 days, 50% of the medium was replaced with fresh one and cells were trypsinized after growing to optimum confluency.
Flow Cytometry
The expression of mesenchymal stem cell markers was analyzed using flow cytometry according to the manufacturer’s instructions. Briefly, ADMSCs from cells at passage 3 were trypsinized and centrifuged. After suspending of 1 × 106 cells in PBS, 100 μL of the suspension was treated with primary labeled antibodies including CD73-PerCP, CD90-FITC, CD105-PerCP, CD14-FITC, CD34-PE, and CD45-FITC (all from eBioscience, USA), for 30 min, followed by the detection with FACSCalibur flow cytometer (BD Biosciences, USA). Data were analyzed by FlowJo software (Tree Star Inc., Ashland, USA).
In Vitro Adipogenic & Osteogenic Differentiation
The multipotency of ADMSCs was evaluated using osteogenic and adipogenic differentiation medium. Adipogenic differentiation medium is composed of 5 mM insulin (Sigma-Aldrich, St. Louis, MO), 250 nM dexamethasone, 100 mM indomethacin, and 0.5 mM 3-isobutyl-1-methylxanthine (Sigma-Aldrich, St. Louis, MO). Osteogenic differentiation medium is composed of 10 mM β-glycerophosphate (Merck, Feltham, UK), 50 μg/mL ascorbic acid biphosphate (Sigma-Aldrich, St. Louis, MO), and 100 nM dexamethasone (Sigma-Aldrich, St. Louis, MO).
1 × 104 cells/well were seeded onto 24-well plates. Following culture in DMEM/F12 with 10% FBS for 24 h, cells were cultured in differentiation medium at 37 °C. The differentiation medium was changed every three days. After 14 days of adipogenic induction, cells were washed with PBS and fixed with 10% neutral formaldehyde for 30 min at room temperature. Fixed cells were stained with 0.5% Oil Red O dye (Sigma-Aldrich, USA) for 30 min at 37 °C, washed in PBS and observed under the light inverted microscope (Olympus, USA).
Osteogenic differentiation was evaluated after 21 days. Cells were washed and fixed, as mentioned above. Fixed cells were stained 4 min at 37 °C with 0.1% Alizarin red S dye (Sigma-Aldrich, USA) to visualize calcium deposition. The stained plates were air-dried for 2 min and observed under the light inverted microscope.
Cell Lines and Culture Conditions
MDA-MB-231 cells were purchased from the Pasteur Institute of Iran (Tehran, Iran). MDA-MB-231 cells were cultured in DMEM supplemented with 10% FBS and incubated at 37 °C in a 5% CO2 incubator. Culture medium was changed every 2 to 3 days.
Exosome Isolation and Characterization
The ADMSC-conditional medium (ADMSC-CM) was collected from cells passage 3–6. Briefly, ADMSCs were seeded in flasks and incubated at 37 °C and 5% CO2. After reaching 70–75% confluency, the medium of ADMSCs was replaced with the medium containing lower FBS and 1% insulin-transferrin-selenium (ITS; Sigma, USA) every 2 days and MSCs were gradually adapted to FBS-free medium. Subsequently, cells were incubated in serum-free medium for 72 h. After incubation time, CM was collected, filtered by 0.22 μm filter to remove dead cells and debris, and used for exosomes characterization or stored at ˗80 °C. Cell viability was assessed using annexinV/PI staining after 72 h serum starvation.
Exosomes were isolated according to manufacturer’s instructions (EXOCIB; Cibbiotech, Iran). Briefly, the serum-free culture medium was centrifuged at 3000 RPM for 10 min at room temperature to remove debris. Reagent A was added to ADMSC-CM at a 1:5 ratio and mixed by vortexing the tubes for 5 min. The mixture was incubated overnight at 4 °C and centrifuged at 3000 RPM for 40 min at 4 °C. The supernatant was removed completely and exosomes were resuspended with reagent B and stored in ˗80 °C for the following experiments.
Electron Microscopy
Using scanning electron microscopy (SEM) and transmission electron microscopy (TEM), the size and quality of purified exosomes were evaluated. Purified exosomes were diluted in PBS at 1:50 ratio and placed on a sterile glass slides until dried at room temperature. The slides were observed under the scanning electron microscope (KYKY-EM3200, Beijing, China).
For TEM imaging, 20 μL of purified exososmes were placed in 50 μL of 2% agarose gel. The prepared exosomes were fixed in 2.5% glutaraldehyde for 12 h and 2% osmium tetroxide for 30 min, and dehydrated using ascending series of ethanol. The exosomes were impregnated with 812 Resin (TAAB, UK), embedded, and polymerized. The ultrathin sections with a thickness of 60 nm were prepared and stained with 2% uranyl acetate for 3 min and 0.5% lead citrate for 5 min and visualized under electron microscopy (Zeiss EM900).
Dynamic Light Scattering (DLS)
The size distribution of purified exosomes was evaluated by DLS. Isolated exosomes were diluted in PBS at a 1:6 ratio and analyzed using Zetasizer (Malvern, UK). Data were analyzed by the Malvern software (Zetasizer Ver. 7.11).
Exosome Protein Quantification
In order to estimate the protein content of exosomes, BCA Protein Assay Kit (Ariatous, Mashad, Iran) was used. In brief, eight serially diluted standards of bovine serum albumin (BSA) was prepared. 25 μL of BSA standards or samples and 75 μL of BCA working reagent were added to a 96-well ELISA plate and mixed well. The plate was incubated at 60 °C for 60 min before the optical density (OD) measurement at 560 nm using ELISA reader system (BioTek, USA).
Cellular Uptake of ADMSC- Exosomes
To show the internalization of ADMSC-exosomes by MDA-MB-231 cells, PKH67 staining was performed according to the manufacturer’s instructions with some modifications (Sigma, USA). Briefly, purified exosomes (100 μg) and PKH67 (3 μL) were suspended in diluent C (500 μL). Next, an equal volume of each suspension (250 μL) was mixed and incubated at 37 ̊C for 5 min. For inactivation of staining reaction FBS was added and after 1 min, labeled exosomes were re-isolated using EXOCIB kit and incubated at 4 ̊C overnight. Afterwards, cells were treated with labeled exosomes for 24 h. Subsequently, the cells were washed with PBS and fixed using paraformaldehyde and observed under a confocal laser scanning microscope (Leica, Germany).
miRNA Mimic Loading into ADMSC- Exosomes
Electroporation was used to introduce miRNA-381-3p mimics (Exiqon, USA) to ADMSC- exosomes. Resuspended exosomes were diluted in electroporation buffer (composed of 21 mM HEPES, 137 mM NaCl, 5 mM KCl, 0.7 mM Na2HPO4-7H2O, 6 mM Dextrose, and H2O) in 1:1 ratio. miRNA mimics or negative control for miRNA mimic (Exiqon, USA) at a final amount of 100 pmol were added to exosome samples. The mixtures were transferred into cold 0.4 cm electroporation cuvettes and electroporated at 0.400 kV. An Eppendorf AG 22331 electroporation instrument (Eppendorf, Germany) was used for electroporation. Then, MDA-MB-231 cell were treated with 100 μg/ml of ADMSC-exosomes. The relative amount of encapsulated miRNA was determined using real-time PCR.
In this study, all experiments were performed in four groups. Untreated MDA-MB-231 cells, and cells treated with unloaded, scrambled loaded, and miR-381 loaded ADMSC-exosomes.
RNA Extraction and Real-Time PCR
48 h after treatment of MDA-MB-231 cells with ADMSC-exosomes, total RNA, containing miRNAs, was extracted by Hybrid-R™ (GeneAll, Korea) according to the manufacturer’s instructions. After the determination of RNA quantity using NanoDrop (Aosheng, China), 1 μg of total RNA was transcribed to complementary DNA (cDNA) using 1 μL of random hexamer primer. The mixture was incubated at 70 ̊C for 5 min. Following the addition of 5X RT-buffer, dNTP, and RT-enzyme, tubes were incubated at 42 ̊C for 60 min and 70 ̊C for 10 min. RT-Stem loop primers were used for cDNA synthesis of miRNA [20].
Quantitative RT-PCR was performed using the Step One instrument (Applied Biosystems, USA). Relative expression of miR-381 was evaluated in a total volume of 20 μL containing Master Mix probe high ROX (Amplicon, Germany), forward primer, universal reverse primer, TaqMan® probe, and cDNA. Relative expression of target mRNAs was evaluated with SYBR GreenI Master Mix (Amplicon, Germany) in a total volume of 20 μL containing Master Mix, forward and reverse primer, and cDNA. SNORD47 was regarded as the internal reference for miR-381, and β-actin was considered the internal reference of target mRNAs. The relative expression of target genes in experimental and control groups was measured by the 2 –ΔΔCt method. Three independent experiments were performed in duplicate. The sequences of the primers are provided in Table 1.
Western Blot
The total cellular extract and ADMSC-exosomes were mixed with radio immunoprecipitation assay (RIPA) lysis buffer containing phenyl methyl sulfonyl fluoride (PMSF) and boiled at 95 °C for 5 min. Equal amounts of total protein and 20 μg of exosomal proteins were resolved by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Blotting was performed on nitrocellulose membranes followed by blocking in non-fat powdered milk in Tris-buffered saline solution with tween 20 (TBST) for 1 h. Membranes were probed with anti-human CD63 (1 μg/mL, BioLegend), CD81 (1 μg/mL, BioLegend), E-cadherin (1:1000, ab15148; Abcam), N-cadherin (1 μg/mL, ab18203; Abcam), Snail (1:1000, 3879 s; Cellsignal), β-Catenin (1:1000, 8814 s; Cellsignal), and β-Actin (1:1000, sc-47,778; Santa Cruz Biotechnology) as primary antibodies overnight at 4 ̊C. Then the membranes were washed three times with TBST and incubated with horse radish peroxidase (HRP) conjugated anti-rabbit or anti-mouse secondary antibodies for 1 h. Protein bands were visualized using radiographic films and enhanced chemiluminescence (ECL) reagents (Amersham Pharmacia Biotech, Buckinghamshire, UK).
MTT Assay
Approximately 3 × 10 3 MDA-MB-231 cells/ well were seeded in a 96-well plate and incubated in DMEM with 10% FBS. The following day, the medium was replaced with serum-free medium and 100 μg/ml of ADMSC-exosomes were added to each well. After 24, 48, and 72 h, the culture medium was removed, cells were washed with PBS, 100 μL of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; 0.5 mg/mL) was added to each well, and the plate was incubated for 3 h in the dark at 37 ̊C. Then, 150 μL of dimethyl sulfoxide (DMSO) was added to each well, and after 2 h incubation in the dark at 37 ̊C, the absorbance at 570 nm was measured using an ELISA reader (BioTek, USA). Three independent experiments were performed in triplicate.
Annexin V/PI Assay
To detect cell apoptosis, annexin V/PI staining was employed. 5× 104 MDA-MB-231 cells/well were seeded into a 24 well plate. The following day, different groups were treated with 100 μg/ml of ADMSC-exosomes for 48 and 72 h. Then after, collected cells were washed with PBS and annexin V binding buffer. Cells were stained with annexin V/ fluorescein isothiocyanate (FITC) and then with propidium iodide (PI) and analyzed with FACSCalibur flow cytometer (BD Biosciences, USA). All annexin V positive cells were considered as apoptotic cells. The experiments were performed in triplicate.
Scratch Assay
12 × 104 MDA-MB-231 cells/well were seeded into a 24 well plate and were grown until the confluent monolayer was formed. Then, the cells were scratched by a sterile pipette tip to create a wound gap, washed with growth medium to remove cell debris, and the medium was replaced with serum-free medium. Afterwards, 100 μg/ml ADMSC-exosomes were added to each well. Cells migrated from scratch edge were monitored by time lapseimaging for 48 h. Processing and quantification of images was performed using the ImageJ Software (NIH, USA). The experiments were performed in triplicate
Cell Migration Assay
SPL cell culture inserts with 8 μm pore size (SPL, Korea) were used for migration assay. Approximately 5 × 10 4 MDA-MB-231 cells were seeded onto the top chamber of transwell insert in serum-free medium. After 3–4 h, 100 μg/ml of ADMSC-exosomes were added to each chamber. 700 μL medium containing 10% FBS (as a chemoattractant) was added into the bottom chamber in a 24-well plate. After 48 h, top chamber was removed, the cells on upper surface were scraped and washed with PBS. Then, cells were fixed in ethanol 70%, stained with crystal violet, and migrating cells were photographed using the light inverted microscope. The number of migrated cells was counted in five randomly selected fields. Three independent experiments were performed in duplicate.
Cell Invasion Assay
For in vitro invasion assay, we firstly prepared matrigel coated inserts. Matrigel (Corning, USA) was mixed with serum-free DMEM (1:2 v/v) on ice. 100 μL of diluted matrigel was added to each transwell chamber and incubated at 37 ̊C for 2 h to solidify. The next steps were performed like cell migration assay. Three independent experiments were performed in duplicate.
Statistical Analysis
qRT-PCR results were analyzed using REST® 2009. Statistical analysis was performed using GraphPad Prism (GraphPad, San Diego, CA). Results were analyzed using ANOVA for multiple groups and Student’s t.test for two groups.
Results
ADMSC Characterization
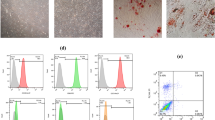
Morphological analysis of ADMSCs using light microscopy indicated homogeneous spindle-shape ADMSCs after three to five passages (Fig. 1a). To evaluate the multipotency of ADMSCs, specific culture mediums were used and differentiation into osteoblast and adipocytewas confirmed by Alizarin red S and Oil Red O staining, respectively (Fig. 1b and c). Analysis of phenotypic markers using flow cytometry showed that ADMSCs were positive for CD-105 (98.5%), CD-90 (99.9%), CD-73 (99.9%) and negative for CD-45 (1.48%), CD-34 (1.4%), and CD-14 (1.61%) (Fig. 1d). The viability of the ADMSCs 72 h after serum starvation has been checked through annexin V/PI assay. The result showed that about 90. 4% of the cells were viable (Fig. 1e).
ADMSC characterization (a) Microscopic images of the first generation of ADMSCs (i) the morphology of the fourth generation of ADMSCs (ii). (b) Alizarin red S staining after 3 weeks of osteogenic induction in ADMSCs. (c) Oil red O staining ofter 2 weeks of adipogenic induction in ADMSCs. (d) Positive markers (CD105, CD90, CD73) and negative markers (CD45, CD34, CD14) of ADMSCs identified by flow cytometry. (e) The viability of the ADMSCs 72 h after serum starvation
Exosome Characterization
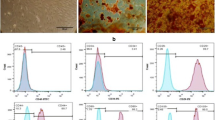
ADMSC-exosomes were characterized by several experiments. Morphological assessment of exosomes was performed by SEM and TEM. The Spherical particles with a diameter of 40–100 nm were displayed in SEM images (Fig. 2a). A complete membrane structure was observed in captured images by TEM (Fig. 2b). To determine the size distribution of ADMSC-exosomes, DLS was used. As demonstrated in Fig. 2c, the average size of purified particles was 83 nm. CD63 and CD81, exosomal surface markers, were detected in ADMSC-exosomes, but not in cell lysate, using western blot (Fig. 2d). The amount of isolated ADMSC-derived exosomes was estimated by exosomal protein concentration. According to BCA assay results, protein content of ADMSC-exosomes was 4000 μg/ml. Also, confocal micrographs indicated that PKH67 labeled ADMSC-exosome were internalized by MDA-MB-231 cells.
ADMSC- exosomes characterization (a) The ultrastructure observed by scanning electron microscopy (SEM) shows ADMSC-exosomes with diameter of 40–100 nm. (b) Transmission electron microscopy (TEM) micrograph of ADMSC-exosomes shows complete membrane structure. Shape and size of exosomes are within the acceptable range. (c) Size distribution of exosomes detected by dynamic light scattering. (d) Expression of exosomal markers (CD63, CD81) detected by western blot analysis
miR-381-Mimic Successfully Loaded to ADMSC- Exosomes
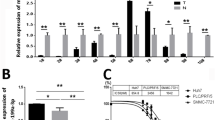
The efficiency of miR-381 loading into ADMSC-exosomes was evaluated by qRT-PCR. MDA-MB-231 cells were treated with unloaded, Scrambled loaded, and miR-381 loaded ADMSC-exosomes and miR-381 level was compared between groups. The expression of miR-381 increased significantly in cells treated with miR-381 loaded exosomes compared to other three groups (p.value < 0.00001) (Fig. 3a).
(a) The relative expression of miR-381 level in MDA-MB-231 cells treated with unloaded, scrambled loaded, and miR-381 loaded ADMSC-exosomes determined by qRT-PCR (****p.value < 0.00001). The relative expression of Twist and Snail (b), LRP6 and CTNNB1 (c) and E-cadherin and N-cadherin (d) mRNA levels in untreated MDA-MB-231 and cells treated with unloaded, scrambled loaded, and miR-381 loaded exosomes determined by qRT-PCR. (e) Protein expression of Snail, CTNNB1, N-cadherin, and E-cadherin in each group was detected using western blot analysis. The data are presented as mean ± SD (*p.value < 0.05, **p value <0.001, and ***p value <0.0001)
miR-381 Loaded ADMSC-Exosomes Decreased the Expression of WNT Pathway Genes and EMT Transcription Factors
It is proved that miR-381 targets 3’UTR of EMT transcription factors (Twist and Snail) and Wnt signaling genes (LRP6 and CTNNB1) [16, 21, 22]. To investigate whether exposure to miR-381 loaded ADMSC-exosomes can effectively alter the expression level of miR-381 target genes, qRT-PCR and western blot analysis were performed. The expression of target genes was evaluated in aforementioned groups. qRT-PCR results showed that the expression of Twist decreased significantly in cells treated with miR-381 loaded exosomes compared with untreated cells (p.value < 0.05) and cells treated with unloaded exosomes (p.value < 0.001). Similarly, the expression of Snail indicated a significant reduction in cells treated with miR-381 loaded exosomes compared with untreated cells (p.value < 0.0001) and cells treated with unloaded (p.value < 0.0001) and scrambled loaded exosomes (p.value < 0.001) (Fig. 3b). The expression of LRP6 in cells treated with miR-381 loaded exosomes was significantly lower than untreated cells (p.value < 0.05) and cells treated with unloaded exosomes (p.value < 0.001). miR-381 loaded exosomes resulted in significant decrease in the expression of CTNNB1 compared to other three groups (p.value < 0.05) (Fig. 3c). As a result of down-regulation of these genes, the expression of E-cadherin as an epithelial marker significantly increased (p.value < 0.0001), while the expression of N-cadherin as a mesenchymal marker significantly decreased (p.value < 0.05) in cells treated with miR-381 loaded exosomes compared to other groups (Fig. 3d). There was no significant difference in expression of Twist, Snail, LRP6, CTNNB1, E-cadherin, and N-cadherin between untreated cells and cells treated with unloaded and scrambled loaded exosomes (p. value > 0.05).
Western blot analysis further validated that after treatment with miR-381 loaded ADMSC-exosomes, the expression of snail, CTNNB1, and N-cadherin decreased and the expression of E-cadherin increased (Fig. 3e).
miR-381 Loaded ADMSC- Exosomes Decrease Viability of MDA-MB-231 Cells
To determine the effects of miR-381 loaded exosomes on viability of MDA-MB-231 cells, MTT assay was performed. Compared with untreated cells, treatment with unloaded exosomes had no significant effect on breast cancer cells viability and proliferation after 24, 48, and 72 h (p.value > 0.05). Similar results were observed in MDA-MB-231 cells treated with miR-381 loaded exosomes after 24 and 48 h (p.value > 0.05), whereas after 72 h, the population of viable cells was significantly decreased in cells treated with miR-381 loaded exosomes compared to other groups (p.value < 0.001; Fig. 4a).
(a) MTT assay for analyzing the effect of miR-381 loaded exosomes on the viability of MDA-MB-231 cells. After 72 h, cell viability significantly decreased in test group compared to control groups (* p < 0.05, **p.value < 0.001). (b) Representative dot plots showing the percentage of apoptotic cells in MDA-MB-231 cells 48 h and 72 h after treatment with miR-381 loaded exosomes and their controls, detected by Annexin V/PI flow cytometry assay. (c) Bar graphs showing the apoptotic index detected by annexin V/PI flow cytometry assay represented in (b), shown as mean ± SD from 3 independent experiments. (*p.value < 0.05, **p.value < 0.001, and *** p.value < 0.0001)
miR-381 Loaded ADMSC- Exosomes Promote Breast Cancer Cells Apoptosis
To evaluate the effect of miR-381 loaded ADMSC-exosomes on apoptosis induction in MDA-MB-231 cells, annexin V/PI assay was performed. There was no significant difference between the apoptotic rate of untreated cells and those treated with unloaded and scrambled loaded exosomes after 48 h (p.value > 0.05). But the apoptotic rate of cells that received miR-381 loaded exosomes showed significant increase compared to unloaded and scrambled loaded exosomes after 48 h (p.value < 0.05). Exposure to miR-381 loaded exosomes significantly increased the apoptosis compared with untreated cells (p.value < 0.001) and cells treated with scrambled loaded (p.value < 0.001) and unloaded exosomes (p.value < 0.0001) after 72 h. Treatment with unloaded exosomes significantly decreased apoptosis percentage compared to untreated cells after 72 h (p.value < 0.0001) (Fig. 4b and c).
Delivery of miR-381- Mimic Using ADMSC- Exosomes Reduced MDA-MB-231 Migration and Invasion
To investigate whether miR-381 loaded ADMSC-exosomes can alter metastatic features of MDA-MB-231 cells, scratch assay and transwell migration and invasion assays were performed. The results of scratch assay indicated that treatment with miR-381 loaded exosomes significantly decrease the motility of MDA-MB-231 cells after 6 h ((p.value < 0.001), 24 h (p.value < 0.0001), and 48 h (p.value < 0.00001)) compared to respective controls and the wound gap remained unfilled until 48 h (Fig. 5a and b). This observation was further confirmed by the results of transwell assays in which MDA-MB-231 cells treated with miR-381 loaded exosomes showed a significant reduction in migration (p.value < 0.001) and invasion (p.value < 0.001) compared to other three groups (Fig. 6a-d). Interestingly exposure to unloaded ADMSC-exosomes have no significant effect in migration and invasion ability of MDA-MB-231 compared to untreated MDA-MB-231(p.value > 0.05).
(a) Migration assay of MDA-MB-231 cells treated with miR-381 loaded exosomes. (b) The results showed a significant decrease in migration of the cells treated with miR-381 loaded exosomes. (c) Invasion assay of MDA-MB-231 cells treated with miR-381 loaded exosomes. (d) The results showed a significant decrease in invasion of the cells treated with miR-381 loaded exosomes (*p value < 0.05 and **p value < 0.001)
Discussion
Numerous reports are indicating that miRNAs are dysregulated in different cancers, and miRNA-based therapy can be a notable therapeutic approach in cancer treatment [23]. Recently, the delivery of concentrated quantities of miRNA mimics or inhibitors by suitable carriers to recipient cells is an issue that has been highlighted by researchers [24]. Unique features of MSC-exosomes make them an appropriate alternative to MSCs. Compared to MSC, MSC-derived exosomes are safe because they lack tumorigenic potential and do not stimulate immune responses. Additionally, similar to their maternal cells, they possess tumor tropism potential and because of their ideal size, they can bypass rapid renal clearance.
Clinical application of MSC-derived exosomes in anti-cancer therapy is in its infancy [8]. It has been shown that exosomes derived from miRNAs-overexpressing bone marrow and Wharton’s jelly mesenchymal stem cells inhibit cancer cell proliferation, migration, and invasion in pancreatic, prostate, oral, and glioblastoma cancer cells [25,26,27,28]. Furthermore, it is reported that direct loading of miR-142-3p inhibitor to exosomes derived from BM-MSC reduces tumorigenicity of breast cancer in the mouse models [29, 30]. Administration of miR-199a- and miR-122-modified ADMSC-exosomes sensitized hepatocellular carcinoma cells to chemotherapeutic agents [31, 32]. So far, there is no report about the application of exosomes derived from human ADMSCs for miRNA delivery to breast cancer cells. Additionally, our miRNA and target genes are completely different from others.
Compared with BM-MSC, ADMSC is the better choice for clinical application. Less invasive isolation procedures, ease of access and harvesting, and their greater amounts make ADMSC a hopeful alternative to BM-MSC in cell-based therapies [33].
According to previously defined criteria (round-shape morphology, size, and specific markers) our results indicated that ADMSC-exosomes were successfully isolated from fresh/frozen CM [34]. It is reported that single freeze-thaw cycle of cell supernatant has no significant effect in EVs’ concentration, size, and morphology [35]. Electroporation was used to direct loading of miR-381 mimic to ADMSC-exosomes. This is an efficient and convenient method to nucleic acid loading into exosomes derived from cells which are hardly transfected such as primary cells [24]. We have previously shown integrity of miRNA electroporated exosomes in our setting [36]. In the current study, effective delivery of miR-381 loaded ADMSC- exosomes to MD-MB-231 indirectly revealed the acceptable condition of electroporated exosomes.
Consistent with our previous study, our results demonstrated that overexpression of miR-381 in TNBC cells using ADMSC-exosomes downregulates CTNNB1 and LRP6, the important genes in Wnt signaling pathway. Wnt is one of the key signaling pathways activated in over 50% of the breast cancer patients and implicated in TNBC tumorigenesis [37,38,39]. Dysregulation of Wnt/β-catenin signaling in the TNBC patient may result in lung and brain metastasis [37]. LRP6 is upregulated in human TNBC cell lines and patients and have a great effect on TNBC migration and invasion [40]. Kong et al. recently proved that miR-381-3p targets LRP6 3’UTR in papillary thyroid carcinoma (PTC) and attenuates proliferation, migration, and invasion of PTC cells [21]. Our results in breast cancer were exactly in line with their findings and this is the first report about miR-381 and LRP6 interaction in breast cancer.
In addition to Wnt signaling, miR-381 targets key transcription factors of EMT process [22, 41]. We showed that miR-381 loaded ADMSC-exosomes significantly downregulate the expression of Twist and Snail in TNBC cells as previously reported in colorectal and lung cancer. It has been reported that the expression of E-cadherin, the most important epithelial marker, significantly decreases in TNBC patients [42]. Twist and Snail are two transcriptional factors cooperate in transcriptional repression of E-cadherin [43]. Twist activates transcription of mesodermal gene expression and Snail suppresses transcription of ectodermal gene expression [44]. Twist and Snail are overexpressed in triple-negative breast cancer [45, 46]. Using computational algorithm (miRanda, miRWalk, TargetScan) Hu et al. showed that only miRNA which targets Snail and Twist 3’UTR is miR-381 [22]. Simultaneous targeting of Wnt signaling pathway and EMT transcription factors by miR-381 loaded ADMSC-exosomes downregulate N-cadherin and upregulate E-cadherin expression.
Of interest, miR-381 mimic delivery by ADMSC-exosomes demonstrated a similar reduction in cell migration and invasion as compared to miR-381 viral transfection into breast cancer cells [16].
Our results showed that unmanipulated ADMSC- exosomes have no considerable effect on the viability, migration, and invasion of MDA-MB-231 cells whereas decrease their apoptosis. Contrary to our results, Lin et al. previously showed that ADMSC-exosomes promote the migration of MCF-7 breast cancer cells, a non- invasive breast cancer cell line, through activation of Wnt signaling pathway [47]. However, Reza et al. reported that ADMSC-exosomes do not exert an identical effects on proliferation and wound healing capacity of different types of ovarian cancer cell lines suggesting that exosomes derived from the same source may have highly variable effects on different types of cancer or different cell lines of same cancer [11]. Additionally, the characteristics of ADMSC are highly donor-specific which can also affect the exosomes’ content and in turn their effects on cancer cells [48].
In conclusion, our data indicate that ADMSC-exosomes can be efficient carriers for miRNA delivery to breast cancer cells. We found that miR-381 loaded ADMSC-exosomes decrease viability, migration, and invasion of TNBC cells through targeting Wnt signaling pathway and EMT transcription factors. This results further confirm therapeutic effects of MSC-derived exosomes in RNA-based therapy in cancer and provide new insights to development of engineered MSC-exosomes for targeted and personalized therapy of cancer. However, our findings need to be supported by further studies. Treatment with miR-381 loaded exosomes should be further optimized in terms of CM preservation and integrity and dose of electroporated exosomes in the upcoming studies. Confirmation of gene expression data by siRNA technology also remain to be studied. Furthermore, additional studies with different cancer cell lines and miRNAs, as well as more advanced in vitro and in vivo tumor models are needed to further confirm these results.
References
Adams, B. D., Kasinski, A. L., & Slack, F. J. (2014). Aberrant regulation and function of microRNAs in cancer. Current biology : CB, 24(16), R762–R776. https://doi.org/10.1016/j.cub.2014.06.043.
Peng, Y., & Croce, C. M. (2016). The role of MicroRNAs in human cancer. Signal Transduction and Targeted Therapy, 1(1), 15004. https://doi.org/10.1038/sigtrans.2015.4.
Marote, A., Teixeira, F. G., Mendes-Pinheiro, B., & Salgado, A. J. (2016). MSCs-derived Exosomes: Cell-secreted Nanovesicles with regenerative potential. Frontiers in Pharmacology, 7, 231. https://doi.org/10.3389/fphar.2016.00231.
Thery, C. (2011). Exosomes: Secreted vesicles and intercellular communications. F1000 biology reports, 3, 15. https://doi.org/10.3410/b3-15.
Batrakova, E. V., & Kim, M. S. (2015). Using exosomes, naturally-equipped nanocarriers, for drug delivery. Journal of controlled release : official journal of the Controlled Release Society, 219, 396–405. https://doi.org/10.1016/j.jconrel.2015.07.030.
Greco, S. J., & Rameshwar, P. (2012). Mesenchymal stem cells in drug/gene delivery: Implications for cell therapy. Therapeutic Delivery, 3(8), 997–1004.
Yeo, R. W., Lai, R. C., Zhang, B., Tan, S. S., Yin, Y., Teh, B. J., et al. (2013). Mesenchymal stem cell: An efficient mass producer of exosomes for drug delivery. Advanced Drug Delivery Reviews, 65(3), 336–341. https://doi.org/10.1016/j.addr.2012.07.001.
Melzer, C., Rehn, V., Yang, Y., Bahre, H., von der Ohe, J., & Hass, R. (2019). Taxol-loaded MSC-derived Exosomes provide a therapeutic vehicle to target metastatic breast Cancer and other carcinoma cells. Cancers, 11(6). https://doi.org/10.3390/cancers11060798.
Wu, S., Ju, G. Q., Du, T., Zhu, Y. J., & Liu, G. H. (2013). Microvesicles derived from human umbilical cord Wharton's jelly mesenchymal stem cells attenuate bladder tumor cell growth in vitro and in vivo. PLoS One, 8(4), e61366. https://doi.org/10.1371/journal.pone.0061366.
Bu, S., Wang, Q., Zhang, Q., Sun, J., He, B., Xiang, C., Liu, Z., & Lai, D. (2016). Human endometrial mesenchymal stem cells exhibit intrinsic anti-tumor properties on human epithelial ovarian cancer cells. Scientific Reports, 6, 37019. https://doi.org/10.1038/srep37019.
Reza, A., Choi, Y. J., Yasuda, H., & Kim, J. H. (2016). Human adipose mesenchymal stem cell-derived exosomal-miRNAs are critical factors for inducing anti-proliferation signalling to A2780 and SKOV-3 ovarian cancer cells. Scientific Reports, 6, 38498. https://doi.org/10.1038/srep38498.
Liang, Y., Zhao, Q., Fan, L., Zhang, Z., Tan, B., Liu, Y., et al. (2015). Down-regulation of MicroRNA-381 promotes cell proliferation and invasion in colon cancer through up-regulation of LRH-1. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie, 75, 137–141. https://doi.org/10.1016/j.biopha.2015.07.020.
Cao, Q., Liu, F., Ji, K., Liu, N., He, Y., Zhang, W., & Wang, L. (2017). MicroRNA-381 inhibits the metastasis of gastric cancer by targeting TMEM16A expression. Journal of experimental & clinical cancer research : CR, 36(1), 29. https://doi.org/10.1186/s13046-017-0499-z.
Xue, Y., Xu, W., Zhao, W., Wang, W., Zhang, D., & Wu, P. (2017). miR-381 inhibited breast cancer cells proliferation, epithelial-to-mesenchymal transition and metastasis by targeting CXCR4. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie, 86, 426–433. https://doi.org/10.1016/j.biopha.2016.12.051.
Rui, X., Gu, T. T., Pan, H. F., Shao, S. L., & Shao, H. X. (2019). MicroRNA-381 suppresses proliferation and invasion of prostate cancer cells through downregulation of the androgen receptor. Oncology Letters, 18(2), 2066–2072. https://doi.org/10.3892/ol.2019.10471.
Mohammadi-Yeganeh, S., Hosseini, V., & Paryan, M. (2019). Wnt pathway targeting reduces triple-negative breast cancer aggressiveness through miRNA regulation in vitro and in vivo. Journal of Cellular Physiology, 234(10), 18317–18328. https://doi.org/10.1002/jcp.28465.
Mi, H., Wang, X., Wang, F., Li, L., Zhu, M., Wang, N., Xiong, Y., & Gu, Y. (2018). miR-381 induces sensitivity of breast cancer cells to doxorubicin by inactivation of MAPK signaling via FYN. European Journal of Pharmacology, 839, 66–75. https://doi.org/10.1016/j.ejphar.2018.09.024.
Yi, D., Xu, L., Wang, R., Lu, X., & Sang, J. (2019). miR-381 overcomes cisplatin resistance in breast cancer by targeting MDR1. Cell Biology International, 43(1), 12–21. https://doi.org/10.1002/cbin.11071.
Schlimgen, R., Howard, J., Wooley, D., Thompson, M., Baden, L. R., Yang, O. O., Christiani, D. C., Mostoslavsky, G., Diamond, D. V., Duane, E. G., Byers, K., Winters, T., Gelfand, J. A., Fujimoto, G., Hudson, T. W., & Vyas, J. M. (2016). Risks associated with Lentiviral vector exposures and prevention strategies. Journal of Occupational and Environmental Medicine, 58(12), 1159–1166. https://doi.org/10.1097/jom.0000000000000879.
Kia, V., Paryan, M., Mortazavi, Y., Biglari, A., & Mohammadi-Yeganeh, S. (2019). Evaluation of exosomal miR-9 and miR-155 targeting PTEN and DUSP14 in highly metastatic breast cancer and their effect on low metastatic cells. Journal of Cellular Biochemistry, 120(4), 5666–5676. https://doi.org/10.1002/jcb.27850.
Kong, W., Yang, L., Li, P. P., Kong, Q. Q., Wang, H. Y., Han, G. X., et al. (2018). MiR-381-3p inhibits proliferation, migration and invasion by targeting LRP6 in papillary thyroid carcinoma. European review for medical and pharmacological sciences, 22(12), 3804–3811. https://doi.org/10.26355/eurrev_201806_15264.
Hu, W. W., Chen, P. C., Chen, J. M., Wu, Y. M., Liu, P. Y., Lu, C. H., et al. (2017). Periostin promotes epithelial-mesenchymal transition via the MAPK/miR-381 axis in lung cancer. Oncotarget, 8(37), 62248–62260. https://doi.org/10.18632/oncotarget.19273.
Bader, A. G., Brown, D., & Winkler, M. (2010). The promise of microRNA replacement therapy. Cancer Research, 70(18), 7027–7030. https://doi.org/10.1158/0008-5472.can-10-2010.
Zhang, D., Lee, H., Zhu, Z., Minhas, J. K., & Jin, Y. (2017). Enrichment of selective miRNAs in exosomes and delivery of exosomal miRNAs in vitro and in vivo. American journal of physiology Lung cellular and molecular physiology, 312(1), L110–Ll21. https://doi.org/10.1152/ajplung.00423.2016.
Shang, S., Wang, J., Chen, S., Tian, R., Zeng, H., Wang, L., Xia, M., Zhu, H., & Zuo, C. (2019). Exosomal miRNA-1231 derived from bone marrow mesenchymal stem cells inhibits the activity of pancreatic cancer. Cancer medicine., 8, 7728–7740. https://doi.org/10.1002/cam4.2633.
Che, Y., Shi, X., Shi, Y., Jiang, X., Ai, Q., Shi, Y., Gong, F., & Jiang, W. (2019). Exosomes derived from miR-143-overexpressing MSCs inhibit cell migration and invasion in human prostate Cancer by Downregulating TFF3. Molecular therapy Nucleic acids, 18, 232–244. https://doi.org/10.1016/j.omtn.2019.08.010.
Xie, C., Du, L. Y., Guo, F., Li, X., & Cheng, B. (2019). Exosomes derived from microRNA-101-3p-overexpressing human bone marrow mesenchymal stem cells suppress oral cancer cell proliferation, invasion, and migration. Molecular and Cellular Biochemistry, 458(1–2), 11–26. https://doi.org/10.1007/s11010-019-03526-7.
Sharif, S., Ghahremani, M. H., & Soleimani, M. (2018). Delivery of exogenous miR-124 to Glioblastoma multiform cells by Wharton's jelly Mesenchymal stem cells decreases cell proliferation and migration, and confers Chemosensitivity. Stem Cell Reviews and Reports, 14(2), 236–246. https://doi.org/10.1007/s12015-017-9788-3.
Naseri, Z., Oskuee, R. K., Jaafari, M. R., & Forouzandeh, M. M. (2018). Exosome-mediated delivery of functionally active miRNA-142-3p inhibitor reduces tumorigenicity of breast cancer in vitro and in vivo. International Journal of Nanomedicine, 13, 7727–7747. https://doi.org/10.2147/ijn.s182384.
Naseri, Z., Oskuee, R. K., Forouzandeh-Moghadam, M., & Jaafari, M. R. (2020). Delivery of LNA-antimiR-142-3p by Mesenchymal stem cells-derived Exosomes to breast Cancer stem cells reduces Tumorigenicity. Stem cell reviews and reports., 16, 541–556. https://doi.org/10.1007/s12015-019-09944-w.
Lou, G., Song, X., Yang, F., Wu, S., Wang, J., Chen, Z., & Liu, Y. (2015). Exosomes derived from miR-122-modified adipose tissue-derived MSCs increase chemosensitivity of hepatocellular carcinoma. Journal of Hematology & Oncology, 8, 122. https://doi.org/10.1186/s13045-015-0220-7.
Lou, G., Chen, L., Xia, C., Wang, W., Qi, J., Li, A., Zhao, L., Chen, Z., Zheng, M., & Liu, Y. (2020). MiR-199a-modified exosomes from adipose tissue-derived mesenchymal stem cells improve hepatocellular carcinoma chemosensitivity through mTOR pathway. Journal of experimental & clinical cancer research : CR, 39(1), 4. https://doi.org/10.1186/s13046-019-1512-5.
Strioga, M., Viswanathan, S., Darinskas, A., Slaby, O., & Michalek, J. (2012). Same or not the same? Comparison of adipose tissue-derived versus bone marrow-derived mesenchymal stem and stromal cells. Stem Cells and Development, 21(14), 2724–2752. https://doi.org/10.1089/scd.2011.0722.
Lotvall, J., Hill, A. F., Hochberg, F., Buzas, E. I., Di Vizio, D., Gardiner, C., et al. (2014). Minimal experimental requirements for definition of extracellular vesicles and their functions: A position statement from the International Society for Extracellular Vesicles. Journal of extracellular vesicles, 3, 26913. https://doi.org/10.3402/jev.v3.26913.
Yuana, Y., Böing, A. N., Grootemaat, A. E., van der Pol, E., Hau, C. M., Cizmar, P., Buhr, E., Sturk, A., & Nieuwland, R. (2015). Handling and storage of human body fluids for analysis of extracellular vesicles. Journal of extracellular vesicles, 4, 29260. https://doi.org/10.3402/jev.v4.29260.
Asadirad, A., Hashemi, S. M., Baghaei, K., Ghanbarian, H., Mortaz, E., Zali, M. R., & Amani, D. (2019). Phenotypical and functional evaluation of dendritic cells after exosomal delivery of miRNA-155. Life Sciences, 219, 152–162. https://doi.org/10.1016/j.lfs.2019.01.005.
Dey, N., Barwick, B. G., Moreno, C. S., Ordanic-Kodani, M., Chen, Z., Oprea-Ilies, G., Tang, W., Catzavelos, C., Kerstann, K. F., Sledge Jr., G. W., Abramovitz, M., Bouzyk, M., de, P., & Leyland-Jones, B. R. (2013). Wnt signaling in triple negative breast cancer is associated with metastasis. BMC Cancer, 13, 537. https://doi.org/10.1186/1471-2407-13-537.
Xu, J., Prosperi, J. R., Choudhury, N., Olopade, O. I., & Goss, K. H. (2015). Beta-catenin is required for the tumorigenic behavior of triple-negative breast cancer cells. PloS one, 10(2), e0117097. https://doi.org/10.1371/journal.pone.0117097.
Zhan, T., Rindtorff, N., & Boutros, M. (2017). Wnt signaling in cancer. Oncogene, 36(11), 1461–1473. https://doi.org/10.1038/onc.2016.304.
Ma, J., Lu, W., Chen, D., Xu, B., & Li, Y. (2017). Role of Wnt co-receptor LRP6 in triple negative breast Cancer cell migration and invasion. Journal of Cellular Biochemistry, 118(9), 2968–2976. https://doi.org/10.1002/jcb.25956.
He, X., Wei, Y., Wang, Y., Liu, L., Wang, W., & Li, N. (2016). MiR-381 functions as a tumor suppressor in colorectal cancer by targeting Twist1. Oncotargets and Therapy, 9, 1231–1239. https://doi.org/10.2147/ott.s99228.
Kashiwagi, S., Yashiro, M., Takashima, T., Nomura, S., Noda, S., Kawajiri, H., Ishikawa, T., Wakasa, K., & Hirakawa, K. (2010). Significance of E-cadherin expression in triple-negative breast cancer. British Journal of Cancer, 103(2), 249–255. https://doi.org/10.1038/sj.bjc.6605735.
Lamouille, S., Xu, J., & Derynck, R. (2014). Molecular mechanisms of epithelial-mesenchymal transition. Nature Reviews Molecular Cell Biology, 15(3), 178–196. https://doi.org/10.1038/nrm3758.
Wang, Y., Liu, J., Ying, X., Lin, P. C., & Zhou, B. P. (2016). Twist-mediated epithelial-mesenchymal transition promotes breast tumor cell invasion via inhibition of hippo pathway. Scientific Reports, 6, 24606. https://doi.org/10.1038/srep24606.
Zhang, Y. Q., Wei, X. L., Liang, Y. K., Chen, W. L., Zhang, F., Bai, J. W., Qiu, S. Q., du, C. W., Huang, W. H., & Zhang, G. J. (2015). Over-expressed twist associates with markers of epithelial Mesenchymal transition and predicts poor prognosis in breast cancers via ERK and Akt activation. PLoS One, 10(8), e0135851. https://doi.org/10.1371/journal.pone.0135851.
Xiao, B., Shi, X., & Bai, J. (2019). miR-30a regulates the proliferation and invasion of breast cancer cells by targeting snail. Oncology Letters, 17(1), 406–413. https://doi.org/10.3892/ol.2018.9552.
Lin, R., Wang, S., & Zhao, R. C. (2013). Exosomes from human adipose-derived mesenchymal stem cells promote migration through Wnt signaling pathway in a breast cancer cell model. Molecular and Cellular Biochemistry, 383(1–2), 13–20. https://doi.org/10.1007/s11010-013-1746-z.
Wegmeyer, H., Broske, A. M., Leddin, M., Kuentzer, K., Nisslbeck, A. K., Hupfeld, J., et al. (2013). Mesenchymal stromal cell characteristics vary depending on their origin. Stem Cells and Development, 22(19), 2606–2618. https://doi.org/10.1089/scd.2013.0016.
Acknowledgements
The authors should thank Cellular and Molecular Biology Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran for providing technical support.
Funding
This project funded by School of Advanced Technologies in Medicine, Shahid Beheshti University of Medical Sciences,Tehran, Iran (Contact grant No: 11985).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
This study was executed under supervision of ethical committee of shahid Beheshti University of Medical Sciences, Tehran, Iran (ethical code: IR.SBMU.REC.1397.004).
Conflict of Interest
Authors declare there are no competing conflict of interests in relation to the work described.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Shojaei, S., Hashemi, S.M., Ghanbarian, H. et al. Delivery of miR-381-3p Mimic by Mesenchymal Stem Cell-Derived Exosomes Inhibits Triple Negative Breast Cancer Aggressiveness; an In Vitro Study. Stem Cell Rev and Rep 17, 1027–1038 (2021). https://doi.org/10.1007/s12015-020-10089-4
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12015-020-10089-4