Abstract
Cyclic-dependent kinase 2 (CDK2) is one of the primary protein kinases involved in the regulation of cell cycle progression. Flavopiridol is a flavonoid derived from an indigenous plant act as a potent antitumor drug showing increased inhibitory activity toward CDK2. The presence of deleterious variations in CDK2 may produce different effects in drug-binding adaptability. Studies on nsSNPs of CDK2 gene will provide information on the most likely variants associated with the disease. Furthermore, investigating the relationship between deleterious variants and its ripple effect in the inhibitory action with drug will provide fundamental information for the development of personalized therapies. In this study, we predicted four variants Y15S, V18L, P45L, and V69A of CDK2 as highly deleterious. Occurrence of these variations seriously affected the normal binding capacity of flavopiridol with CDK2. Analysis of 10-ns molecular dynamics (MD) simulation trajectories indicated that the predicted deleterious variants altered the CDK2 stability, flexibility, and surface area. Notably, we noticed the decrease in number of hydrogen bonds between CDK2 and flavopiridol mutant complexes in the whole dynamic period. Overall, this study explores the possible relationship between the CDK2 deleterious variants and the drug-binding ability with the help of molecular docking and MD approaches.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cyclin-dependent kinase 2 (CDK2) is a member of highly conserved family of protein kinases that govern the eukaryotic cell cycle. Human CDK2 is highly parallel to the gene products of S. cerevisiae cdc28, and S. pombe cdc2 [1]. CDK2 is a catalytic subunit of the cyclin-dependent protein kinase complex, whose activity limited to the G1-S phase, and is essential for cell cycle G1/S phase transition [2]. The CDK2 is regulated by the regulatory subunits of the complex including cyclin A or E [3, 4] and also by protein phosphorylation. In addition, CDK2 is treated as a potent therapeutic target to inhibit the activity of cell cycle in cancerous cells. Thus, research works were started to develop small molecule to inhibit CDK2 which can act as a potential therapy for various cancers [5]. Flavopiridol is a potential antitumor drug, currently used in phase II trials and showed an inhibitory activity toward CDK2 [6]. Genetic variations in the CDK2 gene may affect its regular biological activity such as cell cycle, cell division, DNA damage repair, meiosis, and mitosis. In annexation with these normal processes, genetic variation has the potent to alter the binding adaptability of drugs and also leads to failure of inhibitory action. Understanding the impact of deleterious genetic variations promises to have a greatest impact on our ability to identify the basis of individual variation in response to therapeutics.
Recent progress of the high throughput human genome analysis has provided a wealth of information detailing tens of millions of human genetic variations between individuals [7, 8]. Single-nucleotide variations are commonly observed in all the human genome occurs at the rate of greater than one percentage are referred as “Single Nucleotide Polymorphisms” (SNPs). It was estimated that SNPs occur in just about every 3,000 bp in human genome [7] and frequency of occurrence of the different alleles differs in different populations. Several studies tried to explain how SNPs can cause deleterious effect on the stability and function of a protein [9–13]. Given the large number of SNPs, a comprehensive experimental study of effects on the biological function is a daunting task. A powerful replacement will be the use of in silico methods to predict the change in molecular phenotype allowing prioritization to the experimental studies.
Recently, more sophisticated bioinformatics algorithms are available to predict the effect of amino acid substitutions on protein structure and function. Some of the variation tolerance methods follow the same kind of procedure, in which the missense variant is marked with its property related to the damage it may cause to the protein structure [14]. In other computational methods, predictions were based on the difference in the free energy of unfolding between a native-type protein and its mutant [15]. The ultimate goal of all these approaches is to determine the deleterious nsSNPs from the neutral ones. To understand the atomistic level changes and the dynamic behavior of protein with respect to the potential mutations, we conducted molecular dynamics (MD) simulations analysis. Dynamic motions of proteins were studied intensively by the structural biologists. The motions are known to play a vital role in protein folding, enzyme catalysis, recognition processes, and signaling [16–25]. MD simulations study helps us to explore and to understand how an amino acid substitution can create rigorous changes in the protein structure and its function. Offman et al. [26, 27] found strong correlation between MD analysis and experimental work on the molecular basis of the most common protein upon N370S mutation in causing Gauche’s disease. Based on these studies, we believe that MD simulation could provide more reliable structural information on CDK2 mutations.
The major objective of this in silico analysis is to determine the highly deleterious variants in CDK2 gene. Figure 1 displays the work flow adopted for SNP analysis in CDK2 gene. This was achieved using the publicly available bioinformatics tools like Sorting Intolerant From Tolerant (SIFT) [28], Polymorphism Phenotyping (PolyPhen) version 2 [29], PANTHER [30], and I-mutant 3.0 [31]. Next, molecular docking studies were carried out on the CDK2 protein with its potential inhibitor flavopiridol. We performed docking analysis for both native and mutant CDK2 proteins using the molecular docking suite AutoDock 4.0 [32]. Finally, we conducted MD simulation study for the native and mutant complexes of CDK2-flavopiridol using GROMACS 4.5.3 package [33, 34]. MD simulations explain the structural conformational changes occurred by the incorporation of deleterious mutations in CDK2 protein with respective to the time-scale level.
Materials and Methodology
Computational Methods for Finding Deleterious Variants
The ability to distinguish between deleterious and benign variants by computationally could significantly boost the prediction of disease-related mutations by helping in the identification of deleterious candidate from a pool of data. Recently, different computational algorithms were developed to predict the impact of disease-associated variants. Some of the methods classify deleterious variants according to the predicted pathogenicity and other methods predict the deleterious variants based on protein stability changes upon mutation. We used both these approaches to identify deleterious variants in the CDK2 gene. Sequence evolutionary information-based methods (SIFT, PANTHER) and the combination of protein structural and functional parameter-based methods (PolyPhen 2, I-Mutant 3) are some of the most reliable tools used for deleterious nsSNPs prediction. SIFT, PANTHER, and I-Mutant 3 make their prediction in two categories either tolerated or deleterious, while PolyPhen 2 predicts in three categories benign, possibly damaging, and probably damaging. SIFT prediction is based on the sequence homology and the physicochemical properties of amino acids which dictated by the substituted amino acid. SIFT score ≤0.05 indicates the amino acid substitution is intolerant or deleterious, whereas the score ≥0.05 predicted as tolerant [35, 36]. PANTHER estimates the likelihood of a particular variant causing a functional impact on the protein [30]. PANTHER uses multiple sequence alignment (MSA) and hidden markov model (HMM)-based statistical modeling methods to perform evolutionary analysis on the coding nsSNPs. PANTHER subPSEC scores vary from zero (neutral) to about −10 (more deleterious). Query sequences which are obtained subPSEC value ≤−3 categorized to be deleterious. PolyPhen 2.0 utilizes sequence phylogenetic and structural information in identifying the deleterious substitution. A mutation is classified as “probably damaging” if its probabilistic score is above 0.85–1, “possibly damaging” if its probabilistic score is above 0.15–0.84 and the remaining mutations classified as “benign” [29]. I-Mutant 3 [31] is a support vector machine (SVM)-based algorithm for the prediction of protein stability changes in the occurrence of single point mutation. The output is the predicted free energy change which is calculated from the unfolding Gibbs free energy change of the mutated protein minus the unfolding Gibbs free energy value of the native protein (Kcal/mol). DDG <0 means that the mutated protein has less stability and vice versa.
Protein–Ligand Docking Study
For docking analysis, mutant structures were generated based on the crystal structure of CDK2 protein using Swiss Protein Data Bank (PDB) Viewer [37]. Protein–ligand interaction studies were performed between native and the four mutated models of CDK2 protein with its inhibiting compound, flavopiridol. In order to carry out the docking analysis, we used the AutoDock4 suite as a molecular docking tool [32]. AutoDock4 is a suite of programs making it possible to predict how ligand binds to large macromolecules. In this docking simulation, we used semi-flexible docking protocols. Throughout the docking simulation, the target protein was kept rigid. The ligand being docked is usually flexible and, therefore, explores an arbitrary count of torsional degrees of freedom with addition to the six spatial degrees of freedom spanned by the rotational and parameters. AutoDock4 provides different optimization algorithms to search the space of possible protein–ligand combinations, such as simulated annealing (SA), genetic algorithm (GA), and a hybrid evolutionary algorithms (EA) termed Lamarckian GA (LGA) combining the GA with a local search strategy [38, 39]. The energy functions used in docking simulations attempt to account for the intermolecular energies between ligand and protein as well as the intermolecular energies arising from the ligand conformation itself. AutoDock4 uses a grid-based approach to approximate the energy calculations used by the energy function. During the evaluation of a candidate conformation, the grids are used as lookup tables storing the values used in the calculation, thus making the overall docking simulation remarkably fast. The graphical user interface program AutoDock Tools (ADT) was used to prepare, run, and analyze the docking simulations. Polar hydrogens, solvation parameters, and kollman united atom charges were added into the receptor PDB file for the preparation of protein in docking simulation. Gasteiger charges were added on the ligand PDB file. The geometry of CDK2 structures were optimized through steepest descent method with 1,000 steps each of GROMACS 4.5.3 package. Each minimization was carried out with GRO-MOS-96 [40] 43a1 force field.
MD Simulation Procedure
MD simulations for native and mutant complexes were done with the aid of GROMACS 4.5.3, that adopts GROMOS96 43a1 force field parameter for energy minimizations. Energy-minimized structures of the native CDK2 and four mutant models were used as a starting point for MD simulations. All the proteins were solvated in a cubic box with wall extending at least 0.9 nm from all atoms and filled with simple point charge (SPC) [41] water molecules. Periodic boundary conditions were applied on the system to maintain the number of particle, constant pressure, and temperature (NPT). In order to obtain electrically neutralized system, we utilized GENION procedure from the GROMACS package to replace random water molecule with Na+ or Cl− ions. The temperature was kept constant using a Berendsen algorithm [42] with a coupling time of 0.2. In order to soak the macromolecules into the water molecules both the native and the mutant-minimized systems were equilibrated for 10,000 ps by position restrained MD simulation at 300 K. Then the equilibrated systems were subjected to 10-ns MD simulations. To treat long-range coulombic interactions, particle mesh Ewald method [43] was used and to perform simulations SANDER module [44] was used. To constrain bond lengths involving in hydrogen bond formations, the SHAKE algorithm was used at a time step of 2 fs. The coordinates were saved at regular time intervals of 1 ps. The coulomb interactions were truncated at 0.9 nm and the van der Waals force was maintained at 1.4 nm.
Analysis of MD Trajectories
Structural properties of the native and mutant models of CDK2 protein were calculated from the trajectory files with the built-in functions of GROMACS 4.5.3. The trajectory files were analyzed through the use of g_rmsd and g_rmsf GRO-MACS algorithms to get the root mean square deviation (RMSD) and root mean square fluctuations (RMSF) graph. Total number of hydrogen bonds formed between protein and ligand during the simulation was calculated using g_hbond utility. Number of hydrogen bonds determined based on the donor acceptor angle larger than 90 nm and donor acceptor distance smaller than 3.9 nm [45]. Distance formed between CDK2 protein and ligand was analyzed using g_dist GROMACS utility. Further solvent accessible surface area (SASA) calculated by g_sas utility. To generate the three-dimensional backbone RMSD, RMSF of c-alpha atom, hydrogen bond, distance between molecules, and SASA analysis motion projection of the molecules in phase space of the system were plotted for all five simulations using graphing, advanced computation and exploration (GRACE) program.
Results
Sources of CDK2 Information
Dataset for the evaluation of potential variants in CDK2 gene was retrieved from dbSNP [46] and Swiss-Prot [47] database. We selected the 11 nsSNPs for investigation, and its related information was retrieved from OMIM [48], PubMed, and Swiss-Prot database. Related experimental data about the CDK2 protein and PDB structural file with PDB ID 1HCL [49] were obtained from Swiss-Prot database and PDB [50], respectively. For docking analysis, the ligand molecule, flavopiridol, was retrieved from Drug Bank database [51].
Deleterious Variants in CDK2 by In Silico Tools
Identifying the deleterious variants has become more feasible with the aid of improved in silico tools. In this study, we analyzed 11 variants of CDK2 gene using four different in silico tools namely SIFT, PolyPhen 2, PANTHER, and I-Mutant 3 to determine their pathological significance. Table 1 displays the distribution of the deleterious and neutral variants of CDK2 gene with the corresponding amino acid substitution. SIFT makes inferences from sequence similarity using mathematical operations. SIFT constructs a MSA and considers the position of the missense variants. SIFT calculates the probability based on the amino acids present at each position in the MSAs, and classifies a missense variant “tolerant” or “intolerant”. SIFT can be applied to not only naturally occurring variations but also artificial missense mutations. Among the 11 variants analyzed in SIFT tool, 6 (54.5 %) were identified as deleterious which obtained score ≤0.05. PolyPhen 2 utilizes both the sequence- and the structure-based information to describe the effect of an amino acid substitution, and the effect of mutation is predicted by a Bayesian classifier. The sequence-based features are position-specific independent count (PSIC) scores, MSA properties, and position of mutation in relation to domain boundaries as defined by Pfam [52]. The structure-derived features include changes in solvent accessibility for buried residues, solvent accessibility, and crystallographic B-factor. By PolyPhen 2, 7 nsSNPs (63.63 %) were predicted to be probably damaging the effect on the protein structure and function of CDK2 protein and the remaining four variants were classified as benign which obtained score <0.15. In order to verify the prediction of evolutionary-based SIFT results, we used PANTHER, a HMM-based evolutionary approach to verify its effect on protein function upon a single point mutation. PANTHER predicts all the 11 variants as highly deleterious. In order to improve overall prediction accuracy, we used I-Mutant 3 a SVM-based stability prediction tool. A score <0 means the variant decreases the stability of protein. Conversely, a score >0 means variant increase the stability of protein. Among the 11 variants of the CDK2 gene, 9 variants (81.81 %) showed the negative DDG values, were considered to be less stable and deleterious. Remaining two nsSNP, N3K and G16R, showed positive DDG value and classified as non-deleterious. By comparing the results of all the four in silico tools used in this study, four variants of CDK2 gene specifically Y15S, V18L, P45L, and V69A were filtered as highly deleterious. Among these variants, P45L variant was found in genes of 210 diverse human cancers patients [53].
CDK2–Flavopiridol Interaction Analysis in the Presence of CDK2 Deleterious Variant
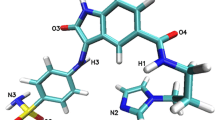
Experimental study of flavopiridol shows considerable inhibitory effect toward CDK2 protein [6, 54]. Amino acid variants in CDK2 protein may affect the binding efficacy of protein with flavopiridol. This has to be analyzed to improve the potentiality of drugs to inhibit CDK2 protein actions. Hence, we analyzed the binding ability of flavopiridol with native and mutant models of CDK2 protein using in silico docking tool-Autodock4. Before entering into docking analysis, we evaluated the binding sites of native CDK2 protein. De Azevedo et al. [54] and Kim et al. [6] observed that flavopiridol directly binds at the ATP-binding pocket between the larger C-terminal and smaller N-terminal domains. Kim et al. [6], in their crystallography study, demonstrated the role of surrounding residues Ile10, Lys33, Glu81, Leu83, His84, Gln85, Asp86, Lys89, and Asp145 in the inhibitory effect of flavopiridol toward CDK2. Our in silico docking analysis showed the similar results as of experimental studies, i.e., flavopiridol binds at the very similar position of ATP-binding site to the native CDK2 protein (Fig. 2a). In contrast to the experimental studies, flavopiridol binds at different positions in all the four mutant models Y15S, V18L, P45L, and V69A (Fig. 2b–e). Change in the binding residues will certainly affect the complementarities between the mutant models and flavopiridol. Shape complementarity and non-covalent interactions were believed to drive protein–ligand interaction. Non-covalent bonds such as hydrogen bonds, van der Waals contacts, and electrostatic forces are the major forces involved in protein–ligand interactions. Calculating the interaction energies of non-covalent bonds is extremely important to understand the binding ability of ligand molecule. The number of hydrogen bonds formed between the protein and the ligand, van der Waals interacting energies, and electrostatic interacting energies were computed using Autodock4. The binding energy and the non-covalent bonds interaction energy between CDK2 protein (native and mutant) and flavopiridol molecule were calculated and displayed in Table 2. In the native complex, the significant contribution of van der Waals and electrostatic energy was observed as −7.38 and 0.48 kcal/mol, respectively. On the contrary, mutant models Y15S, V18L, P45L, and V69A interacting with flavopiridol showed an increased van der waals and electrostatics energies as −7.06, −6.93, −6.49, −7.35 and 0.29, 0.43, 0.35, 0.36 kcal/mol, respectively. The total ligand receptor binding energy exhibited by the native, Y15S, V18L, P45L, and V69A complexes were −6.47, −6.17, −5.9, −5.54, and −6.04 kcal/mol, respectively. Lower binding energy of the native complex shows the better interaction and complete agreement with flavopiridol compound. This docking analysis gives a “theoretical quantitative” assessment on the binding efficiency of CDK2 mutant proteins with flavopiridol drug.
Structural Stability and Flexibility Analysis in CDK2–Flavopiridol Complex
MD simulations provide detailed information about the motions and dynamics of the system studied. Analyses of the trajectories from MD simulations allow us to calculate the stability and flexibility changes for the time averaged atomic motions. The overall protein stability changes upon mutation were evaluated by RMSDs values. We calculated the backbone RMSD for all the atoms from the initial structure, and considered to be a primary criterion to determine the convergence of the protein structure concerned. The backbone RMSD was calculated for both the native and the mutant models of CDK2 protein from the appropriate trajectory files (Fig. 3). We observed a substantial structural deviation in mutant models Y15S, V18L, P45L, and V69A when compared to native CDK2 protein structure. The native and the mutant P45L and V69A structure obtained a mean deviation range of ~0.1 to ~0.25 nm in the entire 10-ns simulation period. But the mutant model, Y15S and V18L, obtained a high RMSD range of ~0.1 to ~0.3 nm and ~0.1 to ~0.3 nm, respectively. This difference in the deviation range in mutant model explains the stability changes and which reflects the impact of substituted amino acid in CDK2 protein. MD provides valuable information about long-range motions and correlated motions, and it allows thermal fluctuations, and long period motions to be distinguished [55, 56]. In order to evaluate the structural flexibility, we calculated the RMSFs values from the 10-ns simulation trajectory data. The RMSFs values of native and mutant models are shown in Fig. 4. The native CDK2 protein residues 13–100 showed a higher fluctuation which belongs to the glycine-rich loop (G-loop) and identified as an inhibitory segment of CDK2 roofing the ATP-binding site [57]. Decrease in fluctuation was observed for all the four mutant models in the residue position of 13–100 indicate the reduction in the flexibility at the inhibitory site. Including these functional residue regions, RMSFs of all the mutant models were remarkably deviated from the native structure in the entire residue portions. Differences in the RMSF value indicate the mode of flexibility change in the mutant models and also reflect the impact of substituted deleterious variants in CDK2 protein.
Hydrogen Bonding and Minimum Distance Change Analysis
Hydrogen bonds are the most significant weak interactions in chemistry and biology [58–60]. A hydrogen bond arises when a hydrogen atom covalently bound to a molecule interacts with an electronegative atom in the same or another molecule. As they are responsible for maintaining the stability of protein structure, determining the hydrogen bonds significantly revels about the stability of protein [61]. Therefore, to assess the stability between protein and ligand, it is essential to gain knowledge of the hydrogen bond strength in protein–ligand interactions. Figure 5 depicts the number of hydrogen bonds formed between CDK2 and flavopiridol complex in both native and mutant state. Native complex of CDK2–flavopiridol exhibits a maximum of five hydrogen bonds throughout the 10-ns simulation period. Mutant protein models Y15S, V18L, P45L, and V69A complexes with flavopiridol obtained maximum of three hydrogen bonds in the entire simulation period. Notably, four mutant protein–ligand complexes obtained less number of hydrogen bonds when compared with the native CDK2–flavopiridol complex. Decrease in the number of hydrogen bond formation between mutant proteins–flavopiridol complexes explains the impact of deleterious amino acid substitution and their ability to destroy the hydrogen bond formations between mutant proteins and ligand.
Furthermore, the minimum distance between the CDK2 protein and ligand was computed for both the native and the mutant complexes (Fig. 6). The minimum distance between native CDK2 and flavopiridol was observed as ~0.1 to ~0.2 nm in the whole 10-ns simulation period. But for the four mutant complexes Y15S, V18L, P45L, and V69A increase in the distance range was observed as ~0.15 to ~0.3 nm. From this minimum distance analysis, we infer that the distance between mutant protein and ligand was increased in compared native complex which may due to the substitution of deleterious amino acid variants.
Effect of CDK2 Deleterious Variants in SASA of Protein Structure
The SASA of a bimolecular is that, accessible to a solvent. Solvation effect plays an important role in maintaining protein stability and folding. Likewise, the solvation effects accompanied the protein–ligand binding process and the rearrangement of solvent molecules. These solvation effects can be calculated by explicit solvent models, such as MD simulations, using a sphere of water molecules [62]. Solvent accessibility was generally divided into exposed and buried regions, indicating the high accessibility and least accessibility of the amino acid residues to the solvent molecules [63]. SASA was calculated for both the native and the mutant trajectory values of CDK2 protein. From Fig. 7, it was observed that native CDK2 protein obtained SASA of ~88 to ~96 nm2 in the whole 10-ns simulation period, while the mutant models Y15S, V18L, P45L, and V69A obtained less SASA of ~78 to ~88 nm2, ~82 to ~94 nm2, ~80 to ~92 nm2, and ~78 to ~88 nm2, respectively. In compare with native protein, all the four mutant proteins obtained less SASA. Reduced in the SASA of mutant proteins indicates that there may be a shift in amino acid residues from accessible area to buried state, which may be the cause of ligand binding other than the active site in mutant proteins.
Discussion
Substitution of deleterious nsSNPs in the coding region may lead to alteration in the protein structure and function which account for susceptibility to disease and altered drug-binding site. Identification of such a deleterious nsSNPs from neutral one is the main task in detecting pathogenesis of disease, individual susceptibility to disease, identifying molecular targets for drug treatment, and for conducting individualized medicine. Several experimental studies were carried out to analyze the relationship between nsSNPs and drug response in cancer treatments. Garcia-Campelo et al. [64] reported the involvement of polymorphism in repair genes such as ERCC1, RRM1, XPD, and BRCA1 resistance to anticancer drug cisplatin. Giovannetti et al. [65] demonstrated the role of nsSNPs in the DNA repair protein to be candidate biomarkers of primary resistance to gemcitabine-/cisplatin-based polychemotherapeutic agent in the treatment of pancreatic cancer. In another analysis, Wang and Moult [66] reported the role of nsSNPs in individuals by inducing or influencing the disease by affecting protein–protein interactions (PPI), protein expression, alternative splicing, stability, folding and ligand binding, or catalysis. These mounting studies on nsSNPs insist their role in better understanding the phenotypic changes occurred among the individuals which enhance toward new drug design and development. The exponential increase in the number of SNPs makes impossible by wet laboratory experiments to determine the biological significance of each nsSNP. Alternatively, bioinformatics tools can be used to investigate the potentially deleterious nsSNPs that might affect important drug targets before further consideration by wet laboratory techniques. Previously, our group had identified and analyzed the effects of deleterious nsSNPs in several proteins at structural and functional levels, and drug-binding capability using various in silico tools [67–69]. In this study, we performed a systematic in silico study to determine the potential deleterious and their structural and functional significance with molecular docking and MD approach.
Sequence-based methods (based on homology and evolutionary conservation) such as SIFT and PANTHER have more advantage over the structure-based methods (PolyPhen 2.0 and I-Mutant 3.0), as they include all types of effect at the protein sequence level, and can be used on any human protein with knowledge relative protein sequence. They are unable to reveal the underlying mechanisms of how SNPs result in changed protein phenotypes. On the other hand, structure-based methods have limited applicability due to unavailability of known 3D structures. Analyzing deleterious nsSNPs by both sequence and structure level have the added benefit of being able to determine the reliability of the predicted results by cross verifying the outputs obtained from both the approaches. Tools that combine sequence- and structure-based approaches use different algorithms and methodologies in their prediction, thereby having a wider coverage of the different aspects of SNP analysis. Both the methods have disadvantages and advantages in their predictions. So, user only must decide which tool is most optimal to the specific objectives of their analysis to gain the optimum knowledge. To determine the possible effects of nsSNPs in CDK2 gene, we employed four widely used in silico tools SIFT, PolyPhen 2, I-Mutant 3, and PANTHER. SIFT predicted 50 % of nsSNPs as deleterious, whereas PolyPhen 2, I-Mutant 3, and PANTHER predicted 87.5 % of nsSNPs to be deleterious. Basically all the four algorithms follow different strategies for predicting the impact of deleterious variant and, we would expect that the results to be in some ways, dissimilar. However, the positive predictions that overlap all these four in silico tools should give high reliability to behave similarly. The variation in their predictions might be due to the difference in features utilized by the methods or the training dataset. Comparing the prediction of all the four methods, four amino acid variants Y15S, V18L, P45L, and V69A were identified as highly deleterious and filtered for further structure and functional investigations. In docking analysis, several factors enhancing the protein–ligand interactions were analyzed. All the parameters elucidated the less binding capability of mutant models with flavopiridol drug. Particularly, the number of hydrogen bonds and its forming residues differed in the mutant proteins in comparison with native CDK2 protein. Mutant proteins Y15, V18L, and P45L and their residues Val251, Thr218, and Arg214, respectively, form single hydrogen bond with the ligand oxygen atoms. Native protein residue Thr14 formed two hydrogen bonds by interacting with two oxygen atoms of ligand, and the another mutant protein V69A residues Lys250 and Gln256 obtained two hydrogen bonds with the flavopiridol molecule by interacting with two different oxygen atoms of the flavopiridol. Even though the mutant model V69A obtained two hydrogen bonds with flavopiridol, but the ligand fails to binds on the ATP-binding site of V69A and other three (Y15S, V18L, P45L) mutant proteins. It actually interprets the structural changes in the CDK2 protein due to the inclusion of deleterious amino acids. Furthermore, we performed a serious of MD simulation analysis on the native and the mutant models of CDK2–flavopiridol complexes which will provide insight knowledge about the atomic level motion in time-scale level. Five different analyses such as RMSD, RMSF, hydrogen bond, minimum distances, and SASA were performed on the 10-ns simulation trajectories. Molecular stability and flexibility changes were observed by RMSD and RMSF analyses. Stability is the fundamental property enhancing bimolecular function, activity, and regulation. CDK2 protein stability analysis results inferred that all the four mutant proteins Y15S, V18L, P45L, and V69A obtained averagely high RMSD values than the native protein. Higher the deviation increases the stability of a protein. Higher the stability increase in the rigidity of a protein and decrease in the stability increases the flexibility of a protein. Conformational changes are required for many protein functions [70–72], but the conformational flexibility and rigidity must be finely balanced [73]. Hence, from the stability analysis of native and mutant CDK2 proteins it was observed that mutant proteins obtained high rigidity due to the incorporation of deleterious amino acids and these changes may affect the binding adaptability of flavopiridol. From the RMSF analysis, we observed decrease in flexibility for all the four mutant proteins in the first half of the residues ranging from 0 to ~100. But native protein obtained high flexibility in the same residue range. Protein active sites always have high flexibility, which facilitates good interaction with binding drug molecules. In docking analysis, we observed that flavopiridol binds at the highly flexible region (0 to ~100) of the native protein. But the substitution of deleterious amino acids in mutant protein affected the original flexibility of CDK2 protein and obtained new flexible region range from ~200 to ~250. In all the four mutant proteins, flavopiridol binds within the newly formed flexible residues region. Thus, in consistence with the RMSD, RMSF analysis also confirmed that the substitution of amino acids adversely affected the structure of CDK2 protein. Besides from the different electrostatic forces, the hydrogen bonds across the protein–ligand interactions serve as a main contributor in maintaining the stable contact between the molecules. Furthermore, the incorporation of deleterious nsSNP may change the possible electrostatic formation between the molecules. Consequently, in all the four mutant complexes Y15S, V18L, P45L, and V69A less number of hydrogen bonds were observed between protein and ligand molecule. In the whole simulation period, mutant complexes obtained an average of one hydrogen bond and maximum of three hydrogen bonds. Whereas, the native CDK2–falvopiridol complex obtained an average of two hydrogen bonds and maximum of five hydrogen bonds in the entire 10-ns simulation period. Decrease in the number of hydrogen bonds reveals the change in binding stability in CDK2–flavopiridol mutant complexes. The path traced by hydrogen bonds is an essential characteristic that may be used to infer how a perturbation caused by a mutation [74]. Minimum distances between protein–ligand complexes were analyzed in both native and mutant condition. Distance between the CDK2 protein and flavopiridol were increased in all the four mutant complexes when compared to the native complex. Increased in the distance may reduce the binding affinity between the CDK2–flavopiridol mutant complexes. Further in SASA analysis, it was found that with respect to the native, less area of solvent accessible surface observed in all the four mutant proteins. Lesser the accessible area, decrease in probability of regular interaction with ligand molecules and other biomolecules. For example, residues with high solvent accessible area are significantly having higher probability to interact with DNA [75]. Thus, SASA analysis indicates that the presence of deleterious mutations in CDK2 protein results in change in the hydrophilic area of the mutant proteins. In the last few decades, molecular science has made many advances in benefit of medicine, including the international hapmap project, human genome project, and genome wide association studies [76]. SNPs are now recognized as the important source of human genetic variability and considered as a valuable resource for mapping complex genetic traits [77]. So for thousands of DNA, variants were identified that are associated with diseases and phenotypic changes [78]. With the knowledge of genetic relationship between phenotypes and the drug response, personalized medicine will tailor to treat patients with specific genotype. Based on this context, we explored the relationship between deleterious variants of CDK2 protein and its impact on the drug (flavopiridol)-binding ability with the help of in silico molecular docking and MD approaches. This analysis could pay a way for the development of new drugs with respect to the acquired structural changes in CDK2 protein. The proposed methodology demonstrates the convincing links between genetic variation and drug responses via computational methods to experimental biologists. This information can be later translated into useful pharmacogenomic assessments.
References
Ninomiya-Tsuji, J., Nomoto, S., Yasuda, H., Reed, S. I., & Matsumoto, K. (1991). Cloning of a human cDNA encoding a CDC2-related kinase by complementation of a budding yeast cdc28 mutation. Proceedings of the National Academy of Sciences, 88, 9006–9010.
Morgan, D. O. (2007). The cell cycle: principles of control. London: New Science Press.
Barrett, J. F., Lewis, B. C., Hoang, A. T., Alvarez, R. J, Jr, & Dang, C. V. (1995). Cyclin A links c-Myc to adhesion-independent cell proliferation. Journal of Biological Chemistry, 270, 15923–15925.
Haas, K., Johannes, C., Geisen, C., Schmidt, T., Karsunky, H., Blass-Kampmann, S., et al. (1997). Malignant transformation by cyclin E and Ha-Ras correlates with lower sensitivity towards induction of cell death but requires functional Myc and CDK4. Oncogene, 15, 2615–2623.
Malumbres, M., Pevarello, P., Barbacid, M., & Bischoff, J. R. (2008). CDK inhibitors in cancer therapy: what is next? Trends in Pharmacological Sciences, 29, 16–21.
Kim, K. S., Sack, J. S., Tokarski, J. S., Qian, L., Chao, S. T., Leith, L., et al. (2000). Thio and oxoflavopiridols, cyclin-dependent kinase 1-selective inhibitors: synthesis and biological effects. Journal of Medicinal Chemistry, 43, 4126–4134.
Cargill, M., Altshuler, D., Ireland, J., Sklar, P., Ardlie, K., Patil, N., et al. (1999). Characterization of single nucleotide polymorphisms in coding regions of human genes. Nature Genetics, 22, 231–238.
Hinds, D. A., Stuve, L. L., Nilsen, G. B., Halperin, E., Eskin, E., Ballinger, D. G., et al. (2005). Whole-genome patterns of common DNA variation in three human populations. Science, 307, 1072–1079.
Reumers, J., Maurer-Stroh, S., Schymkowitz, J., & Rousseau, F. (2006). SNPeffect v2.0: a new step in investigating the molecular phenotypic effects of human non-synonymous SNPs. Bioinformatics, 22, 2183–2185.
Reumers, J., Schymkowitz, J., Ferkinghoff-Borg, J., Stricher, F., Serrano, L., & Rousseau, F. (2005). SNPeffect: a database mapping molecular phenotypic effects of human non synonymous coding SNPs. Nucleic Acids Research, 33, D527–D532.
Packer, B. R., Yeager, M., Burdett, L., Welch, R., Beerman, M., Qi, L., et al. (2006). SNP500 cancer: a public resource for sequence validation, assay development, and frequency analysis for genetic variation in candidate genes. Nucleic Acids Research, 34, D617–D621.
Jegga, A. G., Gowrisankar, S., Chen, J., & Aronow, B. J. (2007). Poly Doms: a whole genome database for the identification of non-synonymous coding SNPs with the potential to impact disease. Nucleic Acids Research, 35, D700–D706.
Yang, J. O., Hwang, S., Oh, J., Bhak, J., & Sohn, T. K. (2008). An integrated database-pipeline system for studying single nucleotide polymorphisms and diseases. BMC Bioinformatics, Suppl, 9(Suppl 12), S19.
Thusberg, J., Olatubosun, A., & Vihinen, M. (2001). Performance of mutation pathogenicity prediction methods on missense variants. Human Mutation, 32, 358–368.
Capriotti, E., Calabrese, R., & Casadio, R. (2006). Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information. Bioinformatics, 22, 2729–2734.
Iakoucheva, L. M., Brown, C. J., Lawson, J. D., Obradovic, Z., & Dunker, A. K. (2002). Intrinsic disorder in cell signaling and cancer associated proteins. Journal of Molecular Biology, 323, 573–584.
Daidone, I., Amadei, A., Roccatano, D., & Di Nola, A. (2003). Molecular dynamics simulation of protein folding by essential dynamics sampling: folding landscape of horse heart cytochrome c. Biophysical Journal, 85, 2865–2871.
Daniel, R. M., Dunn, R. V., Finney, J. L., & Smith, J. C. (2003). The role of dynamics in enzyme activity. Annual Review of Biophysics and Biomolecular Structure, 32, 69–92.
Teague, S. J. (2003). Implications of protein flexibility for drug discovery. Nature Reviews Drug Discovery, 2, 527–541.
Yuan, Z., Zhao, J., & Wang, Z. X. (2003). Flexibility analysis of enzyme active sites by crystallographic temperature factors. Protein Engineering, 16, 109–114.
Chen, C. J., Xiao, Y., & Zhang, L. S. (2005). A directed essential dynamics simulation of peptide folding. Biophysical Journal, 88, 3276–3285.
Eisenmesser, E. Z., Millet, O., Labeikovsky, W., Korzhnev, D. M., Wolf-Watz, M., Bosco, D. A., et al. (2005). Intrinsic dynamics of an enzyme underlies catalysis. Nature, 438, 117–121.
Hung, A., Tai, K., & Sansom, M. S. P. (2005). Molecular dynamics simulation of the M2 helices within the nicotinic acetylcholine receptor transmembrane domain: Structure and collective motions. Biophysical Journal, 88, 3321–3333.
Schlessinger, A., & Rost, B. (2005). Protein flexibility and rigidity predicted from sequence. Proteins, 61, 115–126.
Olsson, M. H. M., Parson, W. W., & Warshel, A. (2006). Dynamical contributions to enzyme catalysis: critical tests of a popular hypothesis. Chemical Reviews, 106, 1737–1756.
Offman, M. N., Krol, M., Silman, I., Sussman, J. L., & Futerman, A. H. (2010). Molecular basis of reduced glucosylceramidase activity in the most common Gaucher disease mutant, N370S. Journal of Biological Chemistry, 285, 42105–42114.
Offman, M. N., Krol, M., Rost, B., Silman, I., & Sussman, J. L. (2011). Comparison of a molecular dynamics model with the X-ray structure of the N370S acid-b-glucosidase mutant that causes Gaucher disease. Protein Engineering, Design & Selection, 24, 773–775.
Kumar, P., Henikoff, S., & Ng, P. C. (2009). SIFT: predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nature Protocols, 4, 1073–1081.
Ramensky, V., Bork, P., & Sunyaev, S. (2002). Human non-synonymous SNPs: server and survey. Nucleic Acids Research, 30, 3894–3900.
Mi, H., Guo, N., Kejariwal, A., & Thomas, P. D. (2007). PANTHER version 6: protein sequence and function evolution data with expanded representation of biological pathways. Nucleic Acids Research, 35, 247–252.
Capriotti, E., Fariselli, P., Rossi, I., & Casadio, R. (2008). A three-state prediction of single point mutations on protein stability changes. BMC Bioinformatics, 9(Suppl 2), S6.
Morris, G. M., Huey, R., Lindstrom, W., Sanner, M. F., & Belew, R. K. (2009). AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. Journal of Computational Chemistry, 30, 2785–2791.
Hess, B., Kutzner, C., van der Spoel, D., & Lindahl, E. (2008). GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. Journal of Chemical Theory and Computation, 4, 435–447.
Kutzner, C., van der Spoel, D., Fechner, M., Lindahl, E., & Schmittn, U. W. (2007). Speeding up parallel GROMACS on high latency networks. Journal of Computational Chemistry, 28, 2075–2084.
Ng, P. C., & Henikoff, S. (2003). SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Research, 13, 3812–3814.
Ng, P. C., & Henikoff, S. (2001). SIFT: predicting deleterious amino acid changes that affect protein function. Genome Research, 11, 863–874.
Guex, N., & Peitsch, M. C. (1997). SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis, 18, 2714–2723.
Rosin, C. D., Scott H. R., Hart, W. E., & Belew, R. K. (1997). A comparison of global and local search methods in drug docking. In proceedings of the 7th intmiariorial conference 011 geiieric algorithms (ICGA-Y7), 221–228.
Morris, G. M., Goodsell, D. S., Halliday, R. S., Huey, R., & Hart, W. E. (1998). Automated docking using a Lamarckian genetic algorithm and empirical binding free energy function. Journal of Computational Chemistry, 19, 1639–1662.
Van Gunsteren, W. F., Billeter, S. R., Eising, A. A., Hunenberger, P. H., & Kruger, P. (1996). Biomolecular simulation: the GROMOS96 manual and user guide; vdf Hochschulverlag AG an der ETH Zurich and BIOMOS b.v: Zurich, Groningen.
Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W., & Klein, M. L. (1983). Comparison of simple potential functions for simulating liquid water. Journal of Chemical Physics, 79, 926.
Berendsen, H. J. C., Postma, J. P. M., van Gunsteren, W. F., DiNola, A., & Haak, J. R. (1984). Molecular dynamics with coupling to an external bath. Journal of Chemical Physics, 81, 3684–3690.
Essmann, U., Perera, L., Berkowitz, M. L., Darden, T., & Lee, H. (1995). A smootparticle meshes Ewald method. Journal of Chemical Physics, 103, 8577–8593.
Case, D. A., Pearlman, D. A., Caldwell, J. W., Wang, J., & Ross, W. S. (2002). AMBER simulation software package. San Francisco: University of California.
Baker, E. N., & Hubbard, R. E. (1984). Hydrogen bonding in globular proteins. Progress in Biophysics and Molecular Biology, 44, 97–179.
Sherry, S. T., Ward, M. H., Kholodov, M., Baker, J., Phan, L., Smigielski, E. M., et al. (2001). dbSNP: the NCBI database of genetic variation. Nucleic Acids Research, 29, 308–311.
Amos, B., & Rolf, A. (1996). The SWISS-PROT protein sequence data bank and its new supplement TREMBL. Nucleic Acids Research, 24, 21–25.
Amberger, J., Bocchini, C. A., Scott, A. F., & Hamosh, A. (2009). McKusick’s online Mendelian inheritance in man (OMIM). Nucleic Acids Research, 37, 793–796.
Schulze-Gahmen, U., De Bondt, H. L., & Kim, S. H. (1996). High resolution crystal structures of human cyclin-dependent kinase 2 with and without ATP: bound waters and natural ligand as guides for inhibitor design. Journal of Medicinal Chemistry, 3, 4540–4546.
Kouranov, A., Xie, L., de la Cruz, J., Chen, L., & Westbrook, J. (2006). The RCSB PDB information portal for structural genomics. Nucleic Acids Research, 34, D302–D305.
Knox, C., Law, V., Jewison, T., Liu, P., & Ly, S. (2011). DrugBank 3.0: a comprehensive resource for ‘Omics’ research on drugs. Nucleic Acids Research, 39, D1035–D1041.
Finn, R. D., Mistry, J., Tate, J., Coggill, P., & Heger, A. (2010). The Pfam protein families’ database. Nucleic Acids Research, 3, D211–D222.
Greenman, C., Stephens, P., & Smith, R. (2007). Patterns of somatic mutation in human cancer genomes. Nature, 446, 153–158.
De Azevedo, W. F, Jr, Mueller-Dieckmann, H. J., Schulze-Gahmen, U., Worland, P. J., Sausville, E., et al. (1996). Structural basis for specificity and potency of a flavonoid inhibitor of human CDK2, a cell cycle kinase. Proceedings of the National Academy of Sciences of the United States of America, 93, 2735–2740.
Garcia, A. E. (1992). Large-amplitude nonlinear motions in proteins. Physical Review Letters, 68, 2696–2699.
Cui, Q., & Bahar, I. (2006). Normal mode analysis: Theory and applications to biological and chemical systems. Boca Raton: Chapman & Hall.
Bossemeyer, D. (1994). The glycine-rich sequence of protein kinases: a multifunctional element. Trends in Biochemical Sciences, 19, 201–205.
Pimentel, G. C., & McClellan, A. L. (1960). The hydrogen bond. San Francisco: Freeman.
Desiraju, G., & Steiner, T. (1999). The weak hydrogen bond-in structural chemistry and biology. Oxford: Oxford University Press.
Schuster, P., Zundel, G., & Sandorfy, C. (1976). The hydrogen bond-recent developments in theory and experiments (Vol. I–III). Amsterdam: Elsevier Science Publishing.
Gerlt, J. A., Kreevoy, M. M., Cleland, W. W., & Frey, P. A. (1997). Understanding enzymic catalysis: the importance of short, strong hydrogen bonds. Chemistry & Biology, 4, 259–267.
Becker, O., Mackerell, A. Jr., Roux, B., & Watanabe, M., (Eds.). (2001). Computer simulation of biomolecular system: theoretical and experimental. Computational biochemistry & biophysics. New York: Marcel Dekker.
Gilis, D., & Rooman, M. (1997). Predicting protein stability changes upon mutation using database-derived potentials: solvent accessibility determines the importance of local versus non-local interactions along the sequence. Journal of Molecular Biology, 272, 276–290.
Garcia-Campelo, R., Alonso, Curbera G., Anton Aparicio, L. M., & Rosell, R. (2005). Pharmacogenomics in lung cancer: an analysis of DNA repair gene expression in patients treated with platinum-based chemotherapy. Expert Opinion on Pharmacotherapy, 6, 2015–2026.
Giovannetti, E., Pacetti, P., Reni, M., Leon, L. G., & Mambrini, A. (2011). Association between DNA-repair polymorphisms and survival in pancreatic cancer patients treated with combination chemotherapy. Pharmacogenomics, 12, 1641–1652.
Wang, Z., & Moult, J. (2001). SNPs, protein structure, and disease. Human Mutation, 17, 263–270.
Rajasekaran, R., George Priya Doss, C., Sudandiradoss, C., Ramanathan, K., Purohit, R., & Sethumadhavan, R. (2008). Effect of deleterious nsSNP on the HER2 receptor based on stability and binding affinity with herceptin: a computational approach. Comptes Rendus Biologies, 331, 409–417.
Rajasekaran, R., George Priya Doss, C., Arun Prasad, G., & Sethumadhavan, R. (2011). In silico identification and analysis of drug resistant mutants of ABL tyrosine kinase based on detrimental missense mutations. Current Signal Transduction Therapy, 6, 396–404.
George Priya Doss, C., Sudandiradoss, C., Rajasekaran, R., Purohit, R., Ramanathan, K., & Sethumadhavan, R. (2008). Identification and structural comparison of deleterious mutations in nsSNPs of ABL1 gene in chronic myeloid leukemia a bio-informatics study. Journal of Biomedical Informatics, 41, 607–612.
Hsu, Y. H., Johnson, D. A., & Traugh, J. A. (2008). Analysis of conformational changes during activation of protein kinase Pak2 by amide hydrogen/deuterium exchange. Journal of Biological Chemistry, 283, 36397–36405.
Mohamed, A. J., Yu, L., Backesjo, C. M., Vargas, L., & Faryal, R. (2009). Bruton’s tyrosine kinase (Btk): function, regulation, and transformation with special emphasis on the PH domain. Immunological Reviews, 228, 58–73.
Muller, C. W., Schlauderer, G. J., Reinstein, J., & Schulz, G. E. (1996). Adenylate kinase motions during catalysis: an energetic counterweight balancing substrate binding. Structure, 4, 147–156.
Vihinen, M. (1987). Relationship of protein flexibility to thermostability. Protein Engineering, 1, 477–480.
Bikadi., Z., Demko, L., & Hazai, E. (2007). Functional and structural characterization of a protein based on analysis of its hydrogen bonding network by hydrogen bonding plot. Archives of Biochemistry and Biophysics, 461, 225–234.
Ahmad, S., Gromiha, M. M., & Sarai, A. (2004). Analysis and Prediction of DNA-binding proteins and their binding residues based on composition, sequence and structural information. Bioinformatics, 20, 477–486.
International HapMap Consortium. (2005). A haplotype map of the human genome. Nature, 437(7063), 1241–1242.
Collins, F. S., Guyer, M. S., & Charkravarti, A. (1997). Variations on a theme: cataloging human DNA sequence variation. Science, 278, 1580–1581.
Hindorff, L. A., Sethupathy, P., Junkins, H. A., Ramos, E. M., Mehta, J. P., Collins, F. S., et al. (2009). Potential etiologic and functional implications of genome wide association loci for human diseases and traits. Proceedings of the National Academy of Sciences, 106, 9362–9367.
Acknowledgments
The authors take this opportunity to thank the management of Vellore Institute of Technology University for providing the facilities and encouragement to carry out this work.
Conflict of interest
All authors have no potential conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nagasundaram, N., George Priya Doss, C. Predicting the Impact of Single-Nucleotide Polymorphisms in CDK2–Flavopiridol Complex by Molecular Dynamics Analysis. Cell Biochem Biophys 66, 681–695 (2013). https://doi.org/10.1007/s12013-012-9512-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12013-012-9512-5