Abstract
Context
Cyclin-dependent kinase 9 (CDK9) plays a significant role in gene regulation and RNA polymerase II transcription under basal and stimulated conditions. The upregulation of transcriptional homeostasis by CDK9 leads to various malignant tumors and therefore acts as a valuable drug target in addressing cancer incidences. Ongoing drug development endeavors targeting CDK9 have yielded numerous clinical candidate molecules currently undergoing investigation as potential CDK9 modulators, though none have yet received Food and Drug Administration (FDA) approval.
Methods
In this study, we employ in silico approaches including the molecular docking and molecular dynamics simulations for the virtual screening over the natural compounds library to identify novel promising selective CDK9 inhibitors. The compounds derived from the initial virtual screening were subsequently employed for molecular dynamics simulations and binding free energy calculations to study the compound’s stability under virtual physiological conditions. The first-generation CDK inhibitor Flavopiridol was used as a reference to compare with our novel hit compound as a CDK9 antagonist. The 500-ns molecular dynamics simulation and binding free energy calculation showed that two natural compounds showed better binding affinity and interaction mode with CDK9 receptors over the reference Flavopiridol. They also showed reasonable figures in the predicted absorption, distribution, metabolism, excretion, and toxicity (ADMET) calculations as well as in computational cytotoxicity predictions. Therefore, we anticipate that the proposed scaffolds could contribute to developing potential and selective CDK9 inhibitors subjected to further validations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cyclin-dependent kinases (CDKs) belong to the family of serine/threonine kinases, which are involved in regulating the cell cycle and transcriptional control [1]. There are 20 different types of CDKs encoded by the human genome [2]. The CDKs play a significant role in maintaining several biological activities like DNA repair, control and regulation of different cell cycle phases, metabolic regulation, and angiogenesis [3]. CDKs are further classified into two subclasses based on their function, the first class of CDKs includes CDK1, CDK2, CDK4, and CDK6 which regulates different phases of the cell cycle and is known as cell cycle–associated kinases, whereas the second class of CDK directly regulate the gene transcription and called as transcription associated CDKs including CDK7, CDK8, CDK9, CDK12, and CDK13 [4, 5]. Like all kinases, CDKs also contains two-lobed structures the N-terminal lobe (mostly composed of β-sheets) and the C-terminal lobe (predominantly containing α-helix and activation loop) connected by a semi-flexible hinge region (D104–L110) [6, 7]. Each CDK contains a catalytic center which consists of an ATP-binding pocket (Q27-V33) also known as G-loop, a PSTAIRE-like conserved sequence that binds with cyclin, and a T-loop (F174-P196) region that binds to CDK activating kinase [8, 9]. The inactive-active state equilibrium plays a crucial role in the functioning of protein kinases, in which the DFG (aspartate-phenylalanine-glycine) motif in the catalytic domain undergoes conformational changes required for function. The DFG motif (D167-G169) is highly conserved in both sequence and structure among the majority of protein kinases, and the conformation of the DFG motif is primarily known as the “DFG-in” and “DFG-out” conformations. It also determines the shape of the ATP-binding site [10]. The enzymatic activity of each CDK critically depends on the interaction of specific regulatory subunit cyclins such as cyclin A, cyclin D, cyclin K, and cyclin T. CDKs which regulate the cell cycle can bind more than one cyclin subunit while the transcription CDKs only bind with specific regulatory subunits [6]. Cyclin catalyzes the phosphorylation of substrates and participates in the cell cycle, leading to DNA synthesis and mitosis, which promotes cell growth and proliferation [11]. Recent studies have suggested that CDKs are overexpressed in different kinds of cancer and also participate in the progression of cardiovascular and neurodegenerative disorders [7]. CDKs are not only responsible for causing various types of cancer but in several research, it has been noted that CDKs are also involved in the HIV-1 replication cycle [12].
CDK9 is one of the transcriptional CDKs that is a key player in maintaining transcriptional homeostasis by forming heterodimeric complexes with the regulatory subunit cyclin T1, T2a, T2b, and cyclin K [13]. It is also crucial for regulating the transcription, initiation, elongation, and termination of RNA polymerase II [14]. CDK9 is mostly found in two isoforms (short and long) encoded by the same gene, the short form of CDK9 is crucial for regulating overall transcription whereas the long form regulates mainly the DNA repair and apoptosis [6, 14]. There are ample studies that prove that dysregulation of CDK9 signaling results in malignancy and growth of solid tumors; CDK9 is critical for regulating each step of gene transcription and thus can serve as a desired target for inhibiting cancer cell growth and proliferation [5]. In recent years, various potential candidates have been identified as CDK9 inhibitors but the main hurdle while designing CDK9 inhibitors is the selectivity of a molecule. Many of the identified compounds also exhibit inhibitory effects on cell cycle kinases, disrupting the normal cell cycle process and impeding the growth and proliferation of healthy cells. To date, the FDA has not approved any selective CDK9 inhibitors. The only FDA-approved anticancer compound that demonstrates activity against CDK9 in clinical trials is Flavopiridol [15, 16]. Apart from this, Roscovitine and Dinaciclib also displayed inhibition of the CDK9 in clinical studies. Nevertheless, the lack of selectivity exhibited by aforementioned inhibitors sometimes leads to the occurrence of adverse effects, therefore imposing restrictions on their utilization in clinical settings. Several selective inhibitors have been developed to address the clinical difficulties associated with the lack of selectivity and potential toxicity of CDK inhibitors. Notable examples include AZD4573 20, BAY-1143572 21, and BAY-1251152 [17]. Undoubtedly, CDK9 stands as a clinically validated target with significant potential for the development of inhibitors aimed at treating a variety of cancers. Consequently, the pursuit of innovative and distinct chemical scaffolds for targeting CDK9 remains a vital endeavor. The objective of the present study is to find out the novel selective natural CDK9 inhibitor using various computer-aided drug design (CADD) approaches. In this work, we used three different natural compounds (alkaloids, phenol, and flavonoids) databases for virtual screening against the CDK9 receptor. The compounds obtained from the virtual screening were further validated by using molecular docking analysis to find out the binding mode of potential compounds with the CDK9 receptor. The stability of the identified ligand–protein complex was measured by using molecular dynamics (MD) simulations and Molecular Mechanics Poisson-Boltzmann Surface Area (MM-PBSA) calculations.
Materials and methods
Protein preparation
The high-resolution 3D structure of CDK9 protein bound with a well-known FDA-approved drug Flavopiridol was obtained from the Research Collaboratory for Structural Biology (RCSB) Protein Data Bank (PDB) [18]. The non-interacting ions as well as the water molecules were removed and hydrogen atoms were added. The protein was prepared using the “Prepare Protein” protocol available in BIOVIA Discovery Studio (DS) v19 [17, 19]. The minimization of prepared protein was done by using the CHARMm27 force field [20].
Database preparation
A database comprising 1683 natural compounds, including alkaloids, flavonoids, and phenolic compounds was downloaded from MedChemExpess (https://www.medchemexpress.com/). The obtained database was prepared and filtered out using Lipinski’s Rule of Five (Ro5) module embedded in DS [21, 22]. The final obtained total of 1083 compounds containing alkaloids (388), flavonoids (159), and phenols (536) were further used for docking-based virtual screening.
Docking-based virtual screening
Docking-based virtual screening comes out as a promising technique to screen out the active compounds from the small molecule library [23]. The virtual screening was done by using Genetic Optimization of Ligand Docking (GOLDv5.2.2) by incorporating ChemPLP (piecewise linear potential), and ASP (Astex statistical potential) scoring function with 30% efficiency and one conformer for each compound [24].
Molecular docking
The binding pose and key interactions of virtually screened compounds were analyzed by using a well-established protocol known as molecular docking studies [26]. With 100% efficiency Gold PLP (piecewise linear potential) scoring function was used for performing the molecular docking of virtually screened natural compounds [24]. The default scoring functions such as the Gold PLP fitness score and Gold ASP fitness score were used for the selection of potential CDK9 binders [27]. Gold PLP (ChemPLP) is an empirical scoring function implemented in the GOLD program. It combines a range of molecular properties and interaction terms to evaluate the fitness of ligand binding. These properties include van der Waals interactions, hydrogen bonding, and lipophilic contacts. The Gold PLP score attempts to capture the overall complementary and favorable interactions between the ligand and protein. Gold ASP (Astex statistical potential) is another scoring function available in the GOLD program, developed by Astex Pharmaceuticals [28]. It utilizes a statistical potential derived from a large database of protein–ligand complexes to estimate the binding affinity. The Gold ASP fitness score is based on statistical analysis of observed protein–ligand interactions and uses a knowledge-based approach to predict the fitness of protein–ligand binding [29].
Molecular dynamics simulations
MD simulations studies
To examine the protein–ligand complex stability, 500-ns MD simulations were performed for the selected compounds and the conventional inhibitor Flavopiridol [30]. The MD simulation for obtained compounds was performed using the Groningen Machine for Chemical Simulations (GROMACS v5.15) by incorporating the CHARMm27 force field [20, 31]. The parameter files for ligand molecules were generated by using SwissParam [32]. For each selected natural compound associated with the CDK9 receptor, a separate simulation system was built in a dodecahedron box, and the TIP3P water model was employed for hydration [33, 34]. The prepared simulation systems were neutralized by adding sodium ions. Before commencing MD simulations, the Steepest Descent algorithm was applied to perform energy minimization on the prepared systems. This step aimed to alleviate steric hindrance and achieve overall energy minimization. Subsequently, each system was equilibrated by using NVT and NPT ensembles [20]. NVT ensemble was carried out for 1 ns at 300 K by keeping a number of particles (N), volume (V), and temperature (T) constant using a V-rescale thermostat [35]. NPT ensemble was performed at 1 bar at a constant number of particles (N), pressure (P), and temperature (T) by Parrinnello-Rehman barostat under periodic boundary conditions to avoid edge effects [22, 35, 36]. LINC algorithm was employed during simulation to restrain the bond length and particle mesh Ewald (PME) was applied for estimating the long-range electrostatic interactions [31, 37]. Utilizing the DS and GROMACS trajectory analysis tools, the obtained simulation results were analyzed [38].
MD trajectory analysis and hydrogen bond calculation
The protein–ligand complex dynamics on ligand–protein binding was determined by root mean square deviation(RMSD) and root mean square fluctuations (RMSF) calculation additionally the hydrogen bond analysis was performed for each system [22]. The “gmx rmsd,” “gmx rmsf,” and “gmx hbond” commands were implemented for the calculation of RMSD, RMSF, and H-bond respectively [22, 39].
Binding free energy calculation using MM-PBSA
Calculating the binding free energy of a system is a potential measure for estimating the binding affinity of hit compounds for a target protein and has crucial importance in computational drug discovery [40]. The GROMACS plugin tool “g_mmpbsa” was used for the calculation of binding free energy in this study. The MD simulation trajectories were used as an input for binding free energy calculation [41, 42]. The protein–ligand complex binding free energy is calculated as follows:
Pharmacokinetic properties assessment via pkCSM
Pharmacokinetic properties play a crucial role in drug discovery and development. These properties describe how the body affects a drug, including its absorption, distribution, metabolism, and excretion. Understanding and optimizing pharmacokinetic properties are essential for creating safe and effective drugs. Early assessment of pharmacokinetics helps to prioritize compounds with favorable properties for further development. The identified hit compounds were further submitted to the pkCSM webserver (http://structure.bioc.cam.ac.uk/pkcsm) to analyze the pharmacokinetic or ADMET properties [43]. Computational approaches provide a powerful toolbox for analyzing ADME properties and can efficiently predict how drugs are absorbed, distributed, metabolized, and excreted in the body. Experimental ADMET studies can be time-consuming, expensive, and often require a significant amount of resources. Computational methods offer a faster and more cost-effective way to prioritize and screen potential drug candidates before moving to experimental stages [43, 44].
In silico prediction of cytotoxicity with CLC-Pred webserver
In silico screening of cytotoxic effects of drug-like candidates against various cell lines can be a useful step in saving time and cost of the drug development process compared to experimental analysis. In the present work, we have used the freely available webserver the CLC-Pred (cell-line cytotoxicity predictor, https://www.way2drug.com/cell-line/) for the estimation of cytotoxic effects of identified hits FL_72 and PH_435 in cancer cell lines and normal cells [45]. The CLC-Pred uses the PASS (prediction of activity spectra for substances) tool to predict the biological activities of uploaded compounds based on their molecular structures. The SMILES codes of hits were used as input for web application and the results of the prediction include five main characteristics, the probability that the compound will be active (Pa), the probability that the compound will be inactive (Pi), cell lines, tumor type, and the region/tissue. Only activities with Pa > Pi are considered as possible cytotoxic candidates for given cell lines [46, 47].
Results
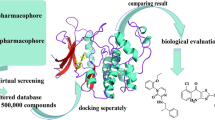
In this study, we used a docking-based virtual screening method followed by MD simulations to identify potential CDK9 inhibitors from a natural drug database. The schematic representation of the work is summarized in (Fig. 1).
Protein preparation
The 3D structure of CDK9 protein co-crystalized with Flavopiridol was downloaded from the RCSB Protein Data Bank (PDB ID: 3BLR) [18]. The DFG motif in 3BLR is typically in the “DFG-in” conformation, where the D167 is positioned in the active site, coordinating with the ATP or inhibitor. The F168 is oriented away from the active site, maintaining the active conformation [48]. The water molecules and non-interacting ions were removed, and hydrogen atoms and missing loops were added to prepare the protein. The “Prepare Protein” protocol available in BIOVIA Discovery Studio (DS) v19 was used to prepare the desired macromolecule [19]. The detailed domain structure of CDK9 is shown in Fig. 2.
The 3D structure of CDK9 (PDB: 3BLR) is shown here with a surface model and the solid ribbon representations. a The important regions of the G-loop or ATP-binding site were shown with green (Q27-V33), hinge region (D104-L110) in blue, orange (cyclin binding domain; P60-E66), yellow (catalytic loop; H148-N154), purple (T loop; F174-P196), and pink color (DFG motif; D167-G169). b The 3D surface model with detailed domains of CDK9
Database preparation
A dataset of 1683 natural compounds, encompassing alkaloids, flavonoids, and phenolic moieties, was assembled. These compounds were then subjected to preparation processes and further organized based on their physiochemical and pharmacokinetic properties. Lipinski’s Rule of Five (Ro5) protocol, available within DS, was utilized for the preparation of a drug-like database (Table S1) [21, 22]. The Ro5 filter yielded a dataset of 1083 chemicals, comprising 536 phenols, 159 flavonoids, and 388 alkaloids. These compounds demonstrated favorable physiochemical properties, indicating their potential suitability as candidates for further investigation (Fig. 3).
Docking-based virtual screening
Molecular docking-based virtual screening was performed using the GOLD program by incorporating ChemPLP, and ASP scoring function with 30% efficiency. During the screening, one conformer for each compound was generated to screen out the potential compounds. The validation of docking parameters was done by using the co-crystalized 3D structure of protein-bound with Flavopiridol (REF). The radius of the binding site was set at 7.43 Å and the XYZ coordinates were set as 53.12, −17.16, and −12.72. The drug was re-docked at the same pocket and the root mean square deviation (RMSD) value was calculated with a crystalized CDK9-Flavopiridol complex. The RMSD value was observed to be in the acceptable range with the value of 1.1 Å (Figure S1). Virtually screened compounds were sorted out based on the Gold PLP score of the REF drug. Flavopiridol displayed a Gold PLP fitness score of 69.81 and a Gold ASP fitness score of 37.43. This score was subsequently utilized for the selection of potential CDK9 inhibitors. Docking results and visual inspection of molecular interactions with the key residues of receptor reveal that only five hit compounds illustrate better interaction and fitness scores when compared with the REF drug. The details of the two-dimensional structure and docking score of hit compounds are shown in (Table 1).
MD simulations studies
MD simulation studies are a powerful tool in the computational drug discovery field that enables the investigation of the dynamic behavior of protein–ligand complex at an atomic level over a set period. These simulations involve the numerical integration of Newton’s equations of motion to simulate the motion and interactions of atoms and molecules in a system [49]. The protein–ligand complexes obtained from molecular docking were further considered as an initial coordinate for MD simulation to check the stability of the ligand–protein binding in an assigned period at given physiological conditions. A total of six systems were prepared including the REF drug and subjected to production run for 500 ns [50]. The CDK9-Flavopiridol complex was used for comparative analysis for identified hit compounds result analysis. The MD simulation results were analyzed through RMSD, RMSF, potential energy, hydrogen bond analysis, and binding mode analysis [25]. The compounds with no important molecular interaction and showing unstable behavior throughout the simulation were discarded from further analysis. Following MD simulations, the obtained systems were ranked based on the calculated binding free energy using the MM-PBSA method. Through MD simulations studies, two natural compounds come out as potential candidates for inhibiting CDK9 with better binding energy and stable interaction with key residues compared to REF and other drug-like compounds and named FL_72 and PH_435.
MD trajectory analysis and hydrogen bond calculation
MD trajectories were used to analyze the system stability throughout the simulation run from backbone RMSD, RMSF, potential energy plots, and hydrogen bond calculation [22, 39]. Compounds that showed unstable behavior and undesirable interaction were discarded from further analysis; detailed MD simulation results have been demonstrated in Table 2 and Fig. 4. The MD results observation suggest that FL_72 and PH_435 showed RMSD values of 3.1 Å and 3.3 Å respectively which is slightly higher than the threshold value of <3 Å, whereas the REF drug-bound CDK9 protein showed RMSD values of 3.0 Å (Fig. 3a).
The root mean square fluctuation (RMSF) values of each simulated system were calculated to analyze the behavior of each protein residue. It measures the average deviation or fluctuation of the atoms in a bimolecular system over a set period. Through RMSF calculation, we observed that both the FL_72 and PH_435 compounds are showing fluctuating results than the REF (Fig. 4b). Further, the potential energy plots suggested that FL_72 and PH_435 were showing stable behavior throughout the 500-ns MD simulation run compared to REF (Fig. 4c). The average number of hydrogen bonds present in each system during the 500-ns simulation run was calculated by using simulation trajectories. The hydrogen bond analysis was performed along with other MD simulation calculations to obtain a more comprehensive understanding of each system’s dynamics and function. The hydrogen bond analysis results revealed that both FL_72 and PH_435 form a more prominent and greater number of hydrogen bonds compared to REF (Fig. 4d and Table 2).
Binding free energy calculation using MM-PBSA
The binding affinity of the identified drug-like candidates towards the CDK9 receptor was inferred by calculating the binding free energy (ΔG) using the MM-PBSA method [40]. The last 50-ns trajectory data was used for calculating the ΔG values. The observed average binding free energy value was for FL_72 − 86.99 kJ/mol, PH_435 − 64.58 kJ/mol, and REF − 63.75 kJ/mol (Fig. 5a, Table 3). The ΔG values emphasized that both the identified compounds FL_72 and PH_435 show a greater binding affinity for CDK9 compared to REF, interestingly PH_435 displayed significantly better affinity towards CDK9 in MM-PBSA calculations.
The per-residues contribution obtained from free energy calculation can provide more details about protein inhibitor interactions. It can be noticed from Fig. 5 that the known inhibitor Flavopiridol (REF) and selected hits (FL_72 and PH_435) target similar residues with different energetics. In particular, L25, F30, V33, V79, F103, L156, and A166 significantly contribute to binding via various hydrophobic interactions. The residues shown on the upper side of the graph, such as K35, K48, E66, D104, D109, A153, and D167, may contribute to polar interactions (Fig. 5b).
Binding mode and intermolecular interaction analysis
Flavopiridol is a small molecule inhibitor that has been extensively studied as a CDK9 inhibitor. It binds to the ATP-binding site of CDK9 and competes with ATP for binding, thereby inhibiting the kinase activity of CDK9. The binding site of Flavopiridol on CDK9 is located in the catalytic domain of the protein. Specifically, it occupies the ATP-binding pocket of CDK9, which is a conserved region responsible for binding and transferring phosphate groups during kinase activity. Flavopiridol forms multiple interactions with the residues lining the ATP-binding site, including hydrogen bonds, hydrophobic interactions, and pi-stacking interactions.
It is been already notified that Cys106 and Asp167 are important residues in CDK9 protein that have been implicated in the regulation of its activity and inhibition [2]. The role of Cys106 present in the hinge region in CDK9 inhibition can vary depending on the specific context and inhibitor being studied. Cys106 is located in the N-terminal lobe of the CDK9 kinase domain, away from the ATP-binding pocket. In certain cases, binding of an inhibitor or ligand to Cys106 can induce allosteric effects that modulate CDK9 activity. Asp167 is located in the active site of CDK9, specifically within the catalytic domain. It plays a significant role in the catalytic activity of CDK9 and can also influence the binding of inhibitors and their inhibitory potency [2]. Allosteric regulation mediated by Cys106 can influence the conformation and stability of CDK9, which in turn affects its susceptibility to inhibition [2]. The crystal structure of CDK9 bound with Flavopiridol (PDB: 3BLR) also shows hydrogen bond interaction with Cys106 which further confirms the role of Cys106 in CDK9 inhibition [18, 51].
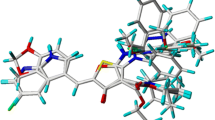
After a simulation run of 500 ns, we analyzed the molecular interaction pattern of each system and it was observed that with Flavopiridol-CDK9 complex displayed van der Waals interactions with active site residues Lys48, Val79, Phe103, Glu107, His108, Asp109, Lys151, and Ala153. The drug displayed hydrogen bond interactions with Asp167. Moreover, residues such as Ile25, Val33, Lys35, Ala46, Cys106, Leu156, and Ala166 are observed to form hydrophobic interactions. In the present work, we observed that all the key residues responsible for π-π interaction and hydrogen bond interaction in the crystal structure of the Flavopiridol-CDK9 complex were forming stable interactions throughout the 500-ns simulations. In the case of FL_72 residues, Gly26, Gln27, Lys48 Val79, Phe103, Phe105, His108, Asp109, Val155, and Asn154 are forming van der Waals interactions and the hinge region residues, Asp104, Cys106, Ala153 of the catalytic loop, and Asp167 from the DFG motif are stabilizing the protein–ligand complex by forming hydrogen bond interactions. On other hand Ile25, Val33, Ala46, Leu157, and Ala166 forms hydrophobic interactions. Additionally, Phe30 is forming a π-π-stacked interaction. By analyzing the molecular interaction of PH_435, we found out that residues responsible for van der Waals interactions were Ile25, Gly26, Val33, Lys48, Phe103, Phe105, Gly112, and Asn154. Hinge region residues Asp104, Cys106, Asp109, and Asp167 in the DFG motif are key formers of hydrogen bond interaction with PH_435 and CDK9, while residues Ala46, Val79, Ala111, Ala153, Leu156, and Ala166 are making hydrophobic interactions. Additionally, Asp167 is also participating in the formation of π-anion bond. Overall molecular interaction analysis suggests that with FL_72 and PH_435 all the previously mentioned key residues Cys106, Asp104, and Asp167 are making stable hydrogen bonds making protein–ligand complex stable (Fig. 6, Table 4).
Binding mode of a REF, b FL_72, and c PH_435 with the active site residues of CDK9. REF, FL_72, and PH_435 are shown in brown, pink, and yellow respectively represented in the stick model. The lower panel represents the 2D molecular interaction of d REF, e FL_72, and f PH_435 with active site residues. The hydrogen bonds are shown in green dash lines while π-π, π-alkyl, π-cation π-sulfur, and π-σ interactions are shown as pink, orange, yellow, and purple dash lines respectively
Pharmacokinetic properties assessment via pkCSM
Computational tools allow researchers to predict ADME properties of compounds in the early stages of drug discovery, helping to select the most promising candidates for further development. This reduces the risk of investing resources in compounds with unfavorable properties.
The obtained results suggested that FL_72 and PH_435 have intermediate levels of water solubility which is compatible with REF. The caco-2 permeability prediction is a useful parameter for oral absorption of a drug, which indicates the acceptable value of 1.45 and 1.12 for FL_72 and REF respectively, but unfortunately PH_435 shows a value of 0.51 which does not fall under the acceptable cut-off criterion for caco-2 permeability according to pkCSM reference values. The intestinal absorbance level of a compound below 30% is considered as poorly soluble and less absorbed; in this study, REF as well as both FL_72 and PH_435 showed good absorption values of 83.01%, 92.02%, and 80.79% respectively. The skin permeability potential score is higher than the acceptable range for REF, FL_72, and PH_435. Both FL_72 and PH_435 and REF were investigated for being p-glycoprotein substrates and inhibitors; we observed that all three compounds were predicted as p-glycoprotein substrates. Interestingly, FL_72 was observed as not an inhibitor for p-glycoprotein whereas REF and PH_435 came out as an inhibitor of p-glycoprotein. A drug that is considered a substrate of p-glycoprotein can potentially act as an inhibitor or inducer of its function, p-glycoprotein functions as a biological barrier by removing xenobiotics and toxins from the cell. The volume of distribution (VD) is an essential pharmacokinetic parameter that needs to be estimated during the drug discovery process. VD explains the relationship between the dose of an administered drug and the amount of drug present in plasma to tissue, the higher the VD more of a drug is distributed in tissue. The REF, FL_72, and PH_435 were observed as fairly distributed in tissue. The pharmacokinetic parameters, blood–brain barrier permeability (BBBP), and central nervous system permeability (CNSP) for REF, FL_72, and PH_435 were observed to be very low which indicates that all three compounds have a very rare chance of causing CNS-related toxicity. Metabolism-related pharmacokinetic parameter analysis includes cytochrome P450 analysis, which is an important enzyme in the human body for drug detoxification. The cytochrome P450 enzymes play a crucial role in drug metabolism by oxidizing a large variety of xenobiotic substances. While predicting the pharmacokinetic properties we considered all the isoforms of cytochrome P450, overall analysis suggested that FL_72 and PH_435 both showed acceptable results compared to REF. Drug clearance is measured as a combination of hepatic clearance and renal clearance. Transport of cationic substrates is mainly mediated by organic cation transporter 2 (OCT2) which is a renal uptake transporter unambiguously expressed on tubular epithelia of the kidney and plays a significant role in drug disposition and drug renal clearance. The renal OCT2 substrate and total clearance excretion properties mentioned in pkCSM were also predicted during the pharmacokinetic properties assessment. The total clearance was observed as a value of 0.46 ml/min/kg, 0.17 ml/min/kg, and 0.36 ml/min/kg for REF, FL_72, and PH_435 respectively. On the other side, none of the compounds were predicted as a substrate of renal OCT2. We observed that REF, FL_72, and PH_435 come out as not an inhibitor for hERG I (the human ether-à-go-go-related gene) which indicates that compounds are not cardiotoxic. On the other hand, similar to REF both FL_72 and PH_435, it inhibits hERG II. The oral rat acute toxicity (LD50) and oral rat chronic toxicity (LOAEL) were also predicted for FL_72, PH_435, and REF. Other important toxicity parameters such as hepatotoxicity, skin sensitization, T. pyriformis toxicity, AMES toxicity, and Minnow toxicity were also predicted for REF, FL_72, and PH_435 (Table S3). The overall analysis of pharmacokinetic or ADMET properties suggested that both the identified natural compounds FL_72 and PH_435 are showing acceptable predicted values compared to REF, interestingly in some pharmacokinetic parameters Fl_72 is showing better results than REF (Table S2).
In silico prediction of cytotoxicity with CLC-Pred webserver
The CLC-Pred tool is used to predict the cytotoxicity of tumor cell lines using the PASS-based CLC-Pred database to develop potential anti-cancer agents. Cytotoxicity prediction was performed for both the identified hits. As per the interpretation of CLC-Pred, if the Pa value is > 0.5, the probability of action is considerably high, whereas the Pi value indicates the inactivity of compounds or activities with Pa > Pi is also considered as possible potential candidates. The results obtained after the prediction suggest that FL_72 and PH_435 have a higher probability of being active mainly in leukemia and lung carcinoma (Table 5).
Discussion
The cell cycle progression is critically controlled by proline directed serine/threonine kinases known as cyclin-dependent kinases (CDK) at each step [6]. CDK9 regulates the transcription elongation of RNA polymerase II; it also modulates the expression and activity of different oncogenes [14, 52]. It has been experimentally proved by researchers that CDK9 plays key role in tumor progression and pathogenesis; hence, it can be a promising pharmacological target for a variety of cancer, specifically tumor associated with transcriptional dysregulation [53, 54]. The only FDA-approved drug available as CDK inhibitor in market is Flavopiridol which does not show selectivity towards CDK9; hence, there is an absolute need for developing new candidate which can selectively binds to CDK9 and inhibit the cancer cell progression [55]. In this work, we have used natural compounds databases to design the potential selective CDK9 inhibitor using computer-aided drug-designing techniques in order to save time efforts required for experimental analysis. Present study is primarily focused on identifying natural compounds as CDK9 antagonist by using docking-based virtual screening followed by molecular docking, MD simulation, and binding free energy calculation for identified hits as well as the reference drug (Fig. 2). The co-crystalized 3D structure of CDK9 protein with Flavopiridol was obtained from the RCSB Protein Data Bank (PDB ID: 3BLR) [18]. The “Prepare Protein” protocol available in DS was used to prepare the protein [19]. A dataset of 1683 natural compounds, encompassing alkaloids, flavonoids, and phenolic moieties, was assembled and used for virtual screening. Molecular docking-based virtual screening was performed with 30% efficiency and one conformer for each compound using GOLD program by incorporating ChemPLP and ASP scoring functions [26]. Virtually screened compounds were further filtered out based on the Gold PLP score of REF drug. The Flavopiridol displayed a Gold PLP fitness score of 69.81 and Gold ASP fitness score of 37.43. Docking results and visual inspection of molecular interactions with the key residues of receptor suggests that only five hit compounds shows better interaction and fitness score than the REF drug which can be used for performing further computational calculations. The docking score of hit compounds and detailed of two-dimensional structure are shown in (Table 1). In order to check the stability of the complex upon ligand binding the protein–ligand complexes obtained from molecular docking were further considered as initial coordinates for MD simulation. The RMSD, RMSF, potential energy, hydrogen bond analysis, and binding mode analysis were performed after MD simulation calculation (Table 2 and Fig. 3). The overall MD analysis suggest that FL_72 and PH_435 showed stable RMSD value of 3.1 Å and 3.3 Å respectively which is quite higher than the threshold value of < 3 Å. The REF drug bound to CDK9 protein showed RMSD values of 3.0 Å (Fig. 3). RMSF calculation also pointed out that the FL_72 and PH_435 compounds are showing comparatively acceptable results with the REF (Fig. 3). To analyze the stable behavior of the complex throughout the 500-ns MD simulation run, the potential energy was calculated for REF, FL_72, and PH_435 (Fig. 3). The average number of hydrogen bonds present in each system during the 500-ns simulation run was calculated by using simulation trajectories (Fig. 3 and Table 2). The molecular interaction pattern of each system was analyzed and it was observed that with Flavopiridol-CDK9 complex displayed van der Waals interactions with active site residues Lys48, Val79, Phe103, Glu107, His108, Asp109, Lys151, and Ala153. The drug displayed hydrogen bond interactions with Asp167. On the other hand, residues such as Ile25, Val33, Lys35, Ala46, Cys106, Leu156, and Ala166 are observed to form π-alkyl interactions. In the case of FL_72 residues, Gly26, Gln27, Lys48 Val79, Phe103, Phe105, His108, Asp109, Val155, and Asn154 are forming van der Waals interactions, and Asp104, Cys106, Ala153, and Asp167 are stabilizing the protein–ligand complex by forming hydrogen bond interactions. On other hand Ile25, Val33, Ala46, Leu157 and Ala166 forms π-alkyl bonding. Additionally, Phe30 forms π-alkyl bonding. Additionally, Phe30 forms a π-π-stacked interaction. By analyzing the molecular interaction of PH_435, we found out that residues responsible for van der Waals interactions were Ile25, Gly26, Val33, Lys48, Phe103, Phe105, Gly112, and Asn154. Residues Asp104, Cys106, Asp109, and Asp167 are key formers of hydrogen bond interaction with PH_435 and CDK9. Moreover Ala46, Val79, Ala111, Ala153, Leu156, and Ala166 are making π-alkyl bonding. Additionally, π-anion bond was also observed with PH_435 and Asp167. Overall, molecular interaction analysis suggests that FL_72 and PH_435 are making stable hydrogen bonds with all the previously mentioned key residues Cys106, Asp104, and Asp167 (Fig. 4, Table 3). Computational tools allow researchers to predict ADME properties of compounds in the early stages of drug discovery, helping to select the most promising candidates for further development [43, 56]. The overall analysis of pharmacokinetic or ADMET properties was performed by using pkCSM online server [43]. The ADMET results suggested that both the identified natural compounds FL_72 and PH_435 are showing acceptable predicted values for each pharmacokinetic property compared to REF; interestingly, in some pharmacokinetic parameters, FL_72 is showing better results than REF (Table S3). The cytotoxicity of tumor cell lines using the PASS-based CLC-Pred database predicted that identified hits, FL_72 and PH_435, have a great potency mainly in leukemia and lung carcinoma which further theoretically promotes these compounds as potential anti-cancer agents.
Limitation of the study
The computational drug design approach used in the present work is able to provide valuable insights, but has several limitations that need to be considered. Firstly, we agree that the accuracy of our molecular dynamics simulations and docking studies is constrained by the used force fields and scoring functions, which may not fully justify the complexity of protein–ligand interactions. Additionally, the conformational sampling in our MD simulations might not explore the complete conformational space, potentially overlooking important binding modes. The other factor which limit our research could be the use of implicit solvent models, while computationally efficient, may not accurately represent all solvent interactions in real time. The physiological environment required for a drug, including factors such as ionic strength and pH, was tried to keep as same as the biological environment but it could affect the predicted binding affinities and stabilities.
In this work, we tried to incorporate a long MD simulation run of 500 ns to get more reliable and stable results. Despite these limitations, our study provides a robust framework for understanding the initial stages of drug design, and future work will focus on addressing these limitations through improved models, enhanced sampling techniques, and extensive experimental validation.
Conclusion
CDK9 plays a crucial role in cancer progression and has emerged as an important target for cancer therapy. Dysregulation of CDK9 can lead to abnormal expression of genes, disrupting the balance between cell growth and cell death which can positively contribute to cancer formation. In the present work, we have used computational methods to identify novel selective CDK9 inhibitors obtained from natural compounds databases. In the first step, we performed the docking base virtual screening of natural compounds, and then the screened compounds were further subjected to molecular docking studies. The docking results conferred the total five potential compounds having better docking scores and interaction with key residues compared to reference drug Flavopiridol were subjected to the 500-ns production. The detailed analysis of MD simulation results theoretically suggests two potential natural compounds (FL_72 and PH_435) as novel selective CDK9 inhibitors based on stable RMSD behavior, binding free energy value, and detailed interaction pattern with the important residues.
Data availability
No datasets were generated or analysed during the current study.
References
Fang C, Xiao Z, Guo Z (2011) Generation and validation of the first predictive pharmacophore model for cyclin-dependent kinase 9 inhibitors. J Mol Graph Model 29:800–808. https://doi.org/10.1016/j.jmgm.2011.01.003
Anshabo AT, Milne R, Wang S, Albrecht H (2021) CDK9: a comprehensive review of its biology, and its role as a potential target for anti-cancer agents. Front Oncol 11:678559. https://doi.org/10.3389/fonc.2021.678559
Ghosh A, Mukherjee S, Jha PC, Manhas A (2023) Identifying natural product inhibitors against CDK9 enzyme via combined multicomplex-based pharmacophore modeling, interaction studies and molecular dynamics simulations. Comput Biol Med 161:107055. https://doi.org/10.1016/j.compbiomed.2023.107055
Chou J, Quigley DA, Robinson TM, Feng FY, Ashworth A (2020) Transcription-associated cyclin-dependent kinases as targets and biomarkers for cancer therapy. Cancer Discov 10:351–370. https://doi.org/10.1158/2159-8290.CD-19-0528
Wu T, Qin Z, Tian Y, Wang J, Xu C, Li Z, Bian J (2020) Recent developments in the biology and medicinal chemistry of CDK9 inhibitors: an update. J Med Chem 63:13228–13257. https://doi.org/10.1021/acs.jmedchem.0c00744
Malumbres M (2014) Cyclin-dependent kinases. Genome Biol 15:122. https://doi.org/10.1186/gb4184
De Vivo M, Bottegoni G, Berteotti A, Recanatini M, Gervasio FL, Cavalli A (2011) Cyclin-dependent kinases: bridging their structure and function through computations. Future Med Chem 3:1551–1559. https://doi.org/10.4155/fmc.11.113
Wu M, Han J, Liu Z, Zhang Y, Huang C, Li J, Li Z (2020) Identification of novel CDK 9 inhibitors based on virtual screening, molecular dynamics simulation, and biological evaluation. Life Sci 258:118228. https://doi.org/10.1016/j.lfs.2020.118228
Whittaker SR, Mallinger A, Workman P, Clarke PA (2017) Inhibitors of cyclin-dependent kinases as cancer therapeutics. Pharmacol Ther 173:83–105. https://doi.org/10.1016/j.pharmthera.2017.02.008
Ung PM-U, Schlessinger A (2015) DFGmodel: predicting protein kinase structures in inactive states for structure-based discovery of type-ii inhibitors. ACS Chem Biol 10:269–278. https://doi.org/10.1021/cb500696t
Kciuk M, Gielecińska A, Mujwar S, Mojzych M, Kontek R (2022) Cyclin-dependent kinases in DNA damage response. Biochim Biophys Acta - Rev Cancer 1877:188716. https://doi.org/10.1016/j.bbcan.2022.188716
Rice AP (2016) Cyclin-dependent kinases as therapeutic targets for HIV-1 infection. Expert Opin Ther Targets 20:1453–1461. https://doi.org/10.1080/14728222.2016.1254619
Sonawane YA, Taylor MA, Napoleon JV, Rana S, Contreras JI, Natarajan A (2016) Cyclin dependent kinase 9 inhibitors for cancer therapy. J Med Chem 59:8667–8684. https://doi.org/10.1021/acs.jmedchem.6b00150
Bacon CW, D’Orso I (2019) CDK9: a signaling hub for transcriptional control. Transcription 10:57–75. https://doi.org/10.1080/21541264.2018.1523668
Morales F, Giordano A (2016) Overview of CDK9 as a target in cancer research. Cell Cycle 15:519–527. https://doi.org/10.1080/15384101.2016.1138186
Montagnoli A, Valsasina B, Croci V, Menichincheri M, Rainoldi S, Marchesi V, Tibolla M, Tenca P, Brotherton D, Albanese C et al (2008) A Cdc7 kinase inhibitor restricts initiation of DNA replication and has antitumor activity. Nat Chem Biol 4:357–365. https://doi.org/10.1038/nchembio.90
Zhang H, Huang J, Chen R, Cai H, Chen Y, He S, Xu J, Zhang J, Wang L (2022) Ligand- and structure-based identification of novel CDK9 Inhibitors for the potential treatment of leukemia. Bioorg Med Chem 72:116994. https://doi.org/10.1016/j.bmc.2022.116994
Baumli S, Lolli G, Lowe ED, Troiani S, Rusconi L, Bullock AN, Debreczeni JE, Knapp S, Johnson LN (2008) The structure of P-TEFb (CDK9/cyclin T1), its complex with flavopiridol and regulation by phosphorylation. EMBO J 27:1907–1918. https://doi.org/10.1038/emboj.2008.121
Studio D (2015) Dassault systemes BIOVIA, discovery studio modelling environment, release 4.5. Accelrys Softw. Inc. https://doi.org/10.11436/mssj.17.98
Sapay N, Tieleman DP (2011) Combination of the CHARMM27 force field with united-atom lipid force fields. J Comput Chem 32:1400–1410. https://doi.org/10.1002/jcc.21726
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings1PII of original article: S0169-409X(96)00423-1. The article was originally published in advanced drug delivery reviews 23 (1997) 3. Adv Drug Deliv Rev 46:3–26. https://doi.org/10.1016/S0169-409X(00)00129-0
Singh P, Kumar V, Lee G Jung TS, Ha MW Hong JC, Lee KW (2022) Pharmacophore-oriented identification of potential leads as CCR5 inhibitors to block HIV cellular entry. Int J Mol Sci 23. https://doi.org/10.3390/ijms232416122
Zhang B, Li H, Yu K, Jin Z (2022) Molecular docking-based computational platform for high-throughput virtual screening. CCF Trans High Perform Comput 4:63–74. https://doi.org/10.1007/s42514-021-00086-5
Jones G, Willett P, Glen RC, Leach AR, Taylor R (1997) Development and validation of a genetic algorithm for flexible docking. 11Edited by F. E. Cohen. J Mol Biol 267:727–748. https://doi.org/10.1006/jmbi.1996.0897
Kumar V, Kumar R, Parate S, Yoon S, Lee G, Kim D, Lee KW (2021) Identification of ACK1 inhibitors as anticancer agents by using computer-aided drug designing. J Mol Struct 1235:130200. https://doi.org/10.1016/j.molstruc.2021.130200
Meng X-Y, Zhang H-X, Mezei M, Cui M (2011) Molecular docking: a powerful approach for structure-based drug discovery. Curr Comput Aided Drug Des 7:146–157. https://doi.org/10.2174/157340911795677602
Yadav MB, Singh P, Jeong YT (2023) Regioselective Synthesis and molecular docking studies of functionalized imidazo [1,2-a]pyridine derivatives through MCRs. Mol Divers. https://doi.org/10.1007/s11030-023-10669-9
Mooij WTM, Verdonk ML (2005) General and targeted statistical potentials for protein-ligand interactions. Proteins 61:272–287. https://doi.org/10.1002/prot.20588
Liu J, Su M, Liu Z, Li J, Li Y, Wang R (2017) Enhance the performance of current scoring functions with the aid of 3D protein-ligand interaction fingerprints. BMC Bioinformatics 18:343. https://doi.org/10.1186/s12859-017-1750-5
Salsbury FR (2010) Molecular dynamics simulations of protein dynamics and their relevance to drug discovery. Curr Opin Pharmacol 10:738–744. https://doi.org/10.1016/j.coph.2010.09.016
Hess B, Kutzner C, Van Der Spoel D, Lindahl E (2008) GRGMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput 4:435–447. https://doi.org/10.1021/ct700301q
Zoete V, Cuendet MA, Grosdidier A, Michielin O (2011) SwissParam: a fast force field generation tool for small organic molecules. J Comput Chem 32:2359–2368. https://doi.org/10.1002/jcc.21816
Mark P, Nilsson L (2001) Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J Phys Chem A 105:9954–9960. https://doi.org/10.1021/jp003020w
Boonstra S, Onck PR, van der Giessen E (2016) CHARMM TIP3P water model suppresses peptide folding by solvating the unfolded state. J Phys Chem B 120:3692–3698. https://doi.org/10.1021/acs.jpcb.6b01316
Bussi G, Donadio D, Parrinello M (2007) Canonical sampling through velocity rescaling. J Chem Phys 126. https://doi.org/10.1063/1.2408420
Parrinello M, Rahman A (1981) Polymorphic transitions in single crystals: a new molecular dynamics method. J Appl Phys 52:7182–7190. https://doi.org/10.1063/1.328693
Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG (1995) A smooth particle mesh Ewald method. J Chem Phys 103:8577–8593. https://doi.org/10.1063/1.470117
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14:33–38. https://doi.org/10.1016/0263-7855(96)00018-5
Kulkarni AM, Kumar V, Parate S, Lee G, Yoon S, Lee KW (2022) Identification of new KRAS G12D inhibitors through computer-aided drug discovery methods. Int J Mol Sci 23, https://doi.org/10.3390/ijms23031309
Wang C, Greene D, Xiao L, Qi R, Luo R (2018) Recent developments and applications of the MMPBSA method. Front Mol Biosci 4. https://doi.org/10.3389/fmolb.2017.00087
Kumari R, Kumar R, Lynn A (2014) G-Mmpbsa -a GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model 54:1951–1962. https://doi.org/10.1021/ci500020m
Paissoni C, Spiliotopoulos D, Musco G, Spitaleri A (2015) GMXPBSA 2.1: a GROMACS tool to perform MM/PBSA and computational alanine scanning. Comput Phys Commun 186:105–107. https://doi.org/10.1016/j.cpc.2014.09.010
Pires DEV, Blundell TL, Ascher DB (2015) PkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J Med Chem 58:4066–4072. https://doi.org/10.1021/acs.jmedchem.5b00104
Veber DF, Johnson SR, Cheng H-Y, Smith BR, Ward KW, Kopple KD (2002) Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem 45:2615–2623. https://doi.org/10.1021/jm020017n
Lagunin AA, Rudik AV, Pogodin PV, Savosina PI, Tarasova OA, Dmitriev AV, Ivanov SM, Biziukova NY, Druzhilovskiy DS, Filimonov DA et al (2023) CLC-Pred 2.0: a freely available web application for in silico prediction of human cell line cytotoxicity and molecular mechanisms of action for druglike compounds. Int J Mol Sci 24:1689
Chinnasamy P, Arumugam R (2018) In silico prediction of anticarcinogenic bioactivities of traditional anti-inflammatory plants used by tribal healers in Sathyamangalam wildlife Sanctuary. India Egypt J Basic Appl Sci 5:265–279. https://doi.org/10.1016/j.ejbas.2018.10.002
Bendjedid S, Benouchenne D (2023) In silico studies for assessing physicochemical, pharmacokinetic and cytotoxic properties of bioactive molecules identified by LC-MS in aloe vera leaf extracts. South African J Bot 157:75–81. https://doi.org/10.1016/j.sajb.2023.03.052
Martin MP, Endicott JA, Noble MEM (2017) Structure-based discovery of cyclin-dependent protein kinase inhibitors. Essays Biochem 61:439–452. https://doi.org/10.1042/EBC20170040
Liu X, Shi D, Zhou S, Liu H, Liu H, Yao X (2018) Molecular dynamics simulations and novel drug discovery. Expert Opin Drug Discov 13:23–37. https://doi.org/10.1080/17460441.2018.1403419
Sakano T, Mahamood MI, Yamashita T, Fujitani H (2016) Molecular dynamics analysis to evaluate docking pose prediction. Biophys Physicobiology 13:181–194. https://doi.org/10.2142/biophysico.13.0_181
Cassandri M, Fioravanti R, Pomella S, Valente S, Rotili D, Del Baldo G, De Angelis B, Rota R, Mai A (2020) CDK9 as a valuable target in cancer: from natural compounds inhibitors to current treatment in pediatric soft tissue sarcomas. Front Pharmacol 11.https://doi.org/10.3389/fphar.2020.01230
Łukasik P, Załuski M, Gutowska I (2021) Cyclin-dependent kinases (CDK) and their role in diseases development-review. Int J Mol Sci 22. https://doi.org/10.3390/ijms22062935
Shen Y, Wang Y, Zhang Y, Ma S, Yang L, Zhao C, Huang X (2022) Targeting cyclin-dependent kinase 9 in cancer therapy. Acta Pharmacol Sin 43:1633–1645. https://doi.org/10.1038/s41401-021-00796-0
Karati D, Mahadik SRK, Trivedi P, Kumar D (2023) Molecular Insights on selective and specific inhibitors of cyclin dependent kinase 9 enzyme (CDK9) for the purpose of cancer therapy. Anticancer Agents Med Chem 23:383–403
Zhang XH, Hsiang J, Rosen ST (2021) Flavopiridol (Alvocidib), a cyclin-dependent kinases (CDKs) inhibitor, found synergy effects with niclosamide in cutaneous t-cell lymphoma. J Clin Haematol 2:48–61. https://doi.org/10.33696/haematology.2.028
van de Waterbeemd H, Gifford E (2003) ADMET in silico modelling: towards prediction paradise? Nat Rev Drug Discov 2:192–204. https://doi.org/10.1038/nrd1032
Funding
This research was supported by funding from the Basic Science Research Program and LAMP program through the National Research Foundation of Korea (NRF) grant funded by the Korea government(MSIT) (grant #: 2020R1A6A1A03044344 and RS-2023-00301974 to J.C.H).
Author information
Authors and Affiliations
Contributions
Conceptualization, P.S. and V.K.; methodology, P.S.; software, K.W.L.; validation, P.S. and V.K.; formal analysis, J.C.H., J.S.L., and K.W.L.; resources, J.C.H. and K.W.L.; data curation, V.K.; writing—original draft preparation, P.S.; writing review and editing, V.K., P.S., supervision, J.C.H., T.S.J and K.W.L.; funding acquisition, J.C.H. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Singh, P., Kumar, V., Jung, T.S. et al. Uncovering potential CDK9 inhibitors from natural compound databases through docking-based virtual screening and MD simulations. J Mol Model 30, 267 (2024). https://doi.org/10.1007/s00894-024-06067-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-024-06067-z