Abstract
The aim of the present study was to investigate the effects of high doses of copper (Cu) and mercury (Hg) on the cecal microbiota in female mice. Forty-eight Kunming mice were randomly divided into the control group (CCk group), the Cu group (CCu group), the Hg group (CHg group), and the Cu + Hg group (CCH group). At the 90th day, cecal tissues were prepared for histopathological analysis and cecal contents for analysis by 16S rRNA sequencing method. Cecal tissues from treatment groups had histopathological lesions including increased thickness of inner muscularis and outer muscularis, widened submucosa, decreased goblet cells, mild to moderate necrosis of enterocytes, blunting of intestinal villi, and severe atrophy of central lacteal. Furthermore, compared to the CCk group, the abundance of bacteria genera Rikenella, Jeotgailcoccus, and Staphylococcus were significantly decreased, whereas the bacteria genus Corynebacterium was significantly increased in the CCu group. The abundance of bacteria genera of Sporosarcina, Jeotgailcoccus, and Staphylococcus were significantly decreased in the CHg group and CCH group. The bacteria genus Anaeroplasma was significantly increased in the CCH group. The results indicated that high doses of Cu and Hg caused histopathological lesions and changed the diversity of microbiota in the cecum of female mice, which provide a theoretical basis for more accurate assessment of the risk in intestinal diseases caused by Cu and Hg.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
With the rapid industrialization and urbanization in China over the last few decades, pollution of heavy metals has become a serious threat after continuous production, leaching, and emission into the environment. Most heavy metals are difficultly biodegradable, hazardous, and toxic to the environment [1, 2]. Previous studies have shown that certain heavy metals may lead to diarrhea, gastrointestinal disorders, stomatitis, and vomiting [3]. Despite of the toxicity of heavy metals, humans and animals are at the risk of being exposed to heavy metals by accidentally consuming contaminated water and animals. In addition, heavy metals may transfer and accumulate in the food chain. Thus, most living organisms in a given ecosystems are likely or even inevitably to be contaminated due to contaminated air, water, soil, and food chains.
Copper (Cu) and mercury (Hg) are well recognized as pollutants and toxic to the environment. The effects of Cu and Hg depend on quantities. What’s more, Cu is the most widely distributed nonferrous metals in China. Cu in small quantities is an essential chemical element for hemoglobin synthesis in the chicken and a necessary component of various metallo-enzymes [4, 5]. Appropriately, small amounts of Cu can be regarded as nutritional additives, whereas excessive amounts of Cu can lead to liver diseases and severe neurological defects [6]. Furthermore, Cu toxicity can induce damage to intestinal tissues [7]. Hg is regarded as one of the most toxic heavy metals, it is emitted and accumulated in the atmosphere during the gradual process of industrial chemicals or discarded electrical products. All forms of Hg have adverse effects on health at high doses. Hg can abnormally stimulate the organism to cause gingivitis, gastrointestinal disorders, and acute hepatotoxicity [3, 8]. It was reported that inhabitants near the industrial sites of Cu mining and processing areas were exposed to suspended particulate matters (SPM) in air [9]. Minamata disease is the result of Hg toxicity caused by the consumption of Hg-contaminated fish [10]. Cu- and Hg-containing contaminating foods or water are ingested by the digestive system, which inevitably results in direct contact of Hg and Cu with the microorganisms in the gastrointestinal tract.
Gastrointestinal microbiota in the digestive tract plays a pivotal role in nutrition and the body’s health by promoting metabolic functions, preventing pathogen colonization, shaping, and maintaining normal mucosal immunity [11]. Gut bacteria are essential modulators impacting homeostasis. Under normal circumstances, the initial establishment of microbiota can reduce the risk of developing diseases such as allergic disorders, chronic immune-mediated inflammatory diseases, type 1 diabetes, and obesity [12]. On the contrary, deregulation of gut homeostasis by certain bacteria could lead to diseases or even death because of the poisoning of the body caused by bacterial byproducts when the structure of intestinal microbiota have changed [13]. The gut microbiota may be a significant mediator of the bioavailability and toxicity of environmental pollutants including heavy metals [14]. However, non-absorbed heavy metal residuals in the gastrointestinal tract could change the intestinal microbiota, resulting in the changes in the qualitative and quantitative composition of bacteriocenosis [15, 16]. Taken together, the gastrointestinal microbiota and its metabolites could play a key role in the host’s physiology.
However, there is little research on high doses of Cu and Hg causing diseases by changing the gastrointestinal microbiota. Hence, we investigated the effects of high doses of Cu and Hg on the cecal microbiota in mice and analyzed the differences of intestinal microbiota using high-throughput 16S rRNA gene sequencing technology, in order to provide a theoretical basis for more accurate assessment of the risk in intestinal diseases caused by Cu and Hg.
Materials and Methods
Animals and Sample Collection
All animals used in this experiment were approved by the Committee of Animal Welfare. Animal studies and experiments were approved and carried out according to Institutional Animal Care and Use Committee guidelines at College of Animal Science and Technology, Jiangxi Agricultural University.
For the experiments, the treatment of mice were measured by reference to Mitra et al. and Wildemann et al., respectively [17, 18]. After a 7-day adaptation period, 48 Kunming female mice (8-week old) were weighed and randomly divided into four groups: the control group (CCk group, average weight = 27.66 g, 0 mg/kg-bw Cu, 0 mg/kg-bw Hg), the dietary of the Cu group (CCu group, average weight = 27.71 g, 5 mg/kg-bw Cu), the dietary of the Hg group (CHg group, average weight = 27.22 g, 2 mg/kg-bw Hg), and the dietary of the Cu + Hg group (CCH group, average weight = 27.00 g, 2.5 mg/kg-bw Cu, 1 mg/kg-bw Hg). Briefly, mice were housed in cages of n = 12 animals with free access to sunlight and basal diet, purified water with different doses of Cu or/and Hg. Copper chloride (CuCl2) and mercuric chloride (HgCl2) were used as the sources of Cu and Hg in this experiment respectively.
At the 90th day, six mice randomly selected from each group were weighed and euthanized by cervical dislocation after anesthesia. Cecal contents were obtained out of a sterile conditions to 1.5 ml EP tube and stored at − 80 °C for further analysis. Meanwhile, cecal tissues were collected and fixed in 4% paraformaldehyde for histological observation.
Histopathological Examination
Cecal tissues were dissected, rinsed with saline, and then fixed in 4% paraformaldehyde. The tissues were then dehydrated in an ascending gradient of ethanol (70–100%) for dehydration and were made transparent by dipping in xylene three times (for 4 min, 2 min, and 30 s). The paraformaldehyde-fixed samples were embedded in paraffin, and stained with hematoxylin and eosin (H&E). The ultrathin-stained sections were observed using an optical microscope and photographs were taken to evaluate pathology.
Microbial DNA Extraction
Total microbial genomic DNA samples of the intestinal contents of mice was extracted using the DNA extraction kit (Agilent, China) according to the manufacturer’s protocol, and stored at − 20 °C prior to further analysis. The quantity of extracted DNAs were measured using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA,USA) and quality were examined by electrophoresis on 1% agarose and visualized by the GelDoc XR System (Bio-Rad, USA) respectively.
PCR Amplification, Qualification, and Purification
Paired-end sequencing was used to annotate the specific bacterial taxonomic position, since the sequences of the V4 regions of the bacterial 16S rRNA genes include a unique character of a bacterial species. The V4 region flanked by the evolutionary conserved regions were used to design polymerase chain reaction (PCR) primer and it was then amplified using PCR with the primers. The bacterial 16S rRNA universal prime sequences synthesized at Shanghai Personal Biotechnology Co., Ltd. (Shanghai, China) are 563F5′AYTGGGYDTAAAGNG3′ (forward primer) and 802R5′TACNVGGGTATCTAATCC3′ (reverse primer). The PCR components contained 5 μl of Q5 reaction buffer (5 ×), 5 μl of Q5 High-Fidelity GC buffer (5 ×), 0.25 μl of Q5 High-Fidelity DNA Polymerase (5 U/μl), 2 μl (2.5 mM) of dNTPs, 1 μl (10 μM) of each forward and reverse primer, 2 μl of DNA template, and 8.75 μl of ddH2O. The PCR amplification condition was as the follows: initial denaturation at 98 °C for 30 s; denaturation at 98 °C for 15 s, annealing at 50 °C for 30 s, extension at 72 °C for 30 s; final extension 72 °C for 5 min; preservation at 4 °C for 30 min. PCR products were purified with Agencourt AMPure Beads (Beckman Coulter, Indianapolis, IN) and quantified using the PicoGreen dsDNA Assay Kit (Invitrogen, Carlsbad, CA, USA). After the individual quantification step, amplicons were pooled in equal amounts, and paired-end 2 × 300 bp sequencing was performed using the Illlumina MiSeq platform with MiSeq Reagent Kit v3 at Shanghai Personal Biotechnology Co., Ltd. (Shanghai, China).
Sequencing
After the construction of metagenomic libraries following the manufacturer’s instruction (Illumina, USA), we performed mate-pair sequencing on 2 × 300 base pairs (bp) with MiSeq Reagent Kit v3(600-cycles-PE) (Illumina, MS-102-3003) for the librarie on Miseq. The library with fixed adaptors is denatured to single strands and grafted to the flowcell, followed by bridge amplification to form clusters which contain clonal DNA fragments. Before sequencing, the library splices into single strands with the help of linearization enzyme, and then four kinds of nucleotides (ddATP, ddGTP, ddCTP, ddTTP) which contain different cleavable fluorescent dye and a removable blocking group would complement the template one base at a time, and the signal could be captured by a (charge-coupled device) CCD. MiSeq uses two lasers and four filters to detect four types of nucleotide (A, T, G, and C). MiSeq control system (MCS v2.4.1) and real-time analyzer (RTA) is in charge of picture background normalization, signal location correction, cross-talk correction, signals conversion, and sequencing data generation.
Sequence Analysis
Raw sequencing reads with exact matches to the barcodes were assigned to respective samples and identified as valid sequences. Through quality control using Quantitative Insights Into Microbial Ecology (QIIME, v1.8.0) [19]. Briefly, raw sequencing reads with exact matches to the barcodes were assigned to respective samples and identified as valid sequences. The low-quality sequences were filtered through the following criteria [20, 21]: sequences that had a length of < 150 bp, sequences that had average Phred scores of < 20, sequences that contained ambiguous bases, and sequences that contained mononucleotide repeats of > 8 bp. Paired-end reads were assembled using FLASH [22]. After chimera detection, sequences of amplicons of the bacterial 16S rRNA V4 region obtained by high-throughput sequencing were clustered into operational taxonomic units (OTUs) at 97% sequence identity by UCLUST [23]. A representative sequence was selected from each OTU using default parameters [24]. An OTU table was further generated to record the abundance of each OTU in each sample and the taxonomy of these OTUs. OTUs containing less than 0.001% of total sequences across all samples were discarded. To minimize the difference of sequencing depth across samples, an averaged and rounded rarefied OTU table was generated by averaging 100 evenly resampled OTU subsets under the 90% of the minimum sequencing depth for further analysis. Then according to the number of sequence containing each OTU in each sample, the matrix file of OTUs in each sample was constructed. OTUs were labeled with taxonomic information by using the 16S rRNA gene sequence data bank. The sequences were labeled with taxonomic phylum, class, order, family, genus, species, and unclassified [25]. The relative abundance of each bacterial species in the sample were also determined by R software. Additionally, Venn diagrams were plotted to show the shared and unique OTUs in each specified group using the R software (Version 2.11.1) [26].
Bioinformatics and Statistical Analysis
We conducted alpha diversity analysis to estimate whether the identified 16S rRNA sequences could include all the bacteria in the samples. The alpha diversity was analyzed through following programs. Shannon curves were plotted by the QIIME software. Shannon index increases as the number of identified sequences increases in a sample, which indicates that unique bacterial species are increasingly identified in a sample. Saturation of the Shannon curves shows that the number of unique bacterial species does not increase as the number of identified sequences increases, which suggests that the identified 16S rRNA sequences may contain all the bacteria in the samples.
Bacterial relative abundance was determined to identify the most dominant bacterial species, which represented the bacterial profile in the samples. Uparse software package (v7.0.1001) and QIIME software were used to analyze bacterial abundance. LEfSe analysis was used to compare the difference in bacterial abundance between different groups [27]. The complete dataset was submitted to the National Center for Biotechnology Information (NCBI) Short Read Archive database with the accession number PRJNA418396.
Results
Body Weight and Histopathology of Cecal Tissues
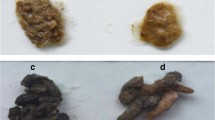
At the 90th day, the weights of mice were analyzed by one-way ANOVA. Data were presented in the format of mean ± SE. Compared to control group (31.49 ± 0.64 g), the weights of mice in the CCu group (29.25 ± 0.87 g), the CHg group (26.13 ± 0.59 g), and the CCH group (27.43 ± 0.40 g) were significantly decreased (P < 0.05). As shown in Fig. 1, compared to the CCk group, cecal tissues in the three treatment groups (CCu group, CHg group, and CCH group) had histopathological lesions including increased thickness of inner muscularis and outer muscularis, widened submucosa, a decreased in the number of goblet cells, mild to moderate necrosis of enterocytes, blunting of intestinal villi and severe atrophy of central lacteal.
Histologic analysis in cecal tissues (HE. 200 ×). The four groups are the control group (CCk group), the Cu group (CCu group), the Hg group (CHg group), and the Cu + Hg group (CCH group). The numbers in the figures indicated (1) inner muscularis, (2) outer muscularis, (3) submucosa, (4) goblet cells, and (5) intestinal villi and central lacteal
Sequence Data and OTUs
There were 21 DNA samples with the concentration above 20 ng/uL out of totally 24 samples, which met the test requirements. The three samples unqualified were removed. A total of 825,808 bacterial 16S rRNA gene sequences were obtained from 21 samples. Six hundred eighty-one thousand and six hundred seventy-three sequences passed the quality control filtering and were regarded as high quality. The 681,673 reads were used for abundance analysis, diversity analysis, and taxonomic comparison; summary information of the sequencing data was presented in Table 1. Additionally, 2912 unique OTUs were constructed by the QIIME software, and on average, there were 2250 unique OTUs in each sample. As shown in Fig. 2a, the OTUs were annotated at the levels of phylum, class, order, family, genus, species, and unclassified by the RDP classifier and the 16S rRNA sequence database, and the GreenGene database. The annotations of all the 2912 were presented in the Venn diagrams Fig. 2b. It showed that four groups shared 1194 OTUs, accounting for 41.0% of the total OTUs. The unique OTUs respectively accounted for 0.76%, 0.93%, 0.96%, and 3.09% in each of the four groups. This procedure showed the uniqueness, similarity, and overlap of the OTUs composition of the samples.
a Statistical results of OTUs identification and classification. The OTU number of phylum, class, order, family, genus, and species. The number of OTUs that could not be classified into any known group were designated as “unclassified.” b Venn diagram showing the unique and shared OTUs in the different groups
Bioinformatics and Statistical Analysis
Shannon curve was used to assess species abundance. It allows the calculation of species richness for a given number of samples based on the construction of rarefaction curves. The Shannon curves (Fig. 3a) of observed species reached a plateau as the number of identified sequences increased, suggesting that the identified sequences could sufficiently cover the bacteria in the samples. The rank abundance curves (Fig. 3b) also became stable, indicating that the species distribution was uniform. This result showed that the 21 samples contained a relatively low proportion of highly abundant bacteria because the end of the curves represented a majority of the reads which belonged to the rare bacteria in different groups.
a Shannon curve. The Shannon curves reached a plateau as the number of identified sequences increased, suggesting that the identified sequences could sufficiently cover the bacteria in samples. b Rank abundance curves. Each curves represents OTU distribution in one sample. The smoother the broken line, the higher the evenness of the community
Microbial Community Structure Analysis
At the phylum level, although the major phyla among the four groups were almost uniform, their relative abundances were different. As shown in Fig. 4a, the total of identified phyla for samples were 11. In the CCk group, the community was mainly composed of Bacteroidetes (45.36%), Firmicutes (30.12%), Proteobacteria (21.52%), and Actinobacteria (2.22%). In the CCu group, the community was mainly composed of Firmicutes (48.08%), Bacteroidetes (34.52%), Actinobacteria (8.70%), Proteobacteria (6.71%), and Spirochaetes (1.20%). In the CHg group, the community was mainly composed of Firmicutes (48.73%), Bacteroidetes (25.00%), Proteobacteria (13.17%), Spirochaetes (6.43%), and Actinobacteria (4.83%). In the CCH group, the community was mainly composed of Firmicutes (60.13%), Bacteroidetes (21.69%), Proteobacteria (6.97%), Actinobacteria (4.83%), and Spirochaetes (4.97%).
The classification was further analyzed and discovered at the genus level. Each group had a variety of diversity at the genus level and the samples showed differences. In addition, a small number of bacteria were not identified. As shown in Fig. 4b, the total of identified genera for the samples were 57. The dominant genera in the CCK group were Acinetobacter (18.94%), Staphylococcus (3.51%), Lactobacillus (3.33%), Odoribacter (2.82%), and Bacteroides (1.79%). The community in the CCu group was mainly represented by Allobaculum (22.94%), Bifidobacterium (4.49%), Oscillospira (4.23%), Bacteroides (4.09%), and Ruminococcus (1.77%). The community in the CHg group was mainly composed of Lactobacillus (6.12%), Treponema (6.43%), Oscillospira (6.19%), Desulfovibrio (6.78%), and Odoribacter (4.18%). The community in the CCH group was mainly composed of Lactobacillus (10.05%), Treponema (4.97%), Bacteroides (4.09%), Oscillospira (3.40%), Allobaculum (3.30%), and Bifidobacterium (2.79%). The dominant genera had obviously different abundances among the four groups.
Analysis of Intestinal Microbiota Among the Different Groups
To explore the composition of intestinal microbiota in different groups, the differences in the relative abundance of the microbiota in samples were detected using LEfSe analysis. As shown in Fig. 5, compared to the CCk group at the phylum level, there was no significant difference in the CCu group, or in the CHg group (P > 0.05) but Tenericutes were significantly increased in the CCH group (P < 0.05). Compared to the CCk group at the genus level, Corynebacterium were significantly increased whereas Rikenella, Odoribacter, Jeotgailcoccus, and Staphylococcus were significantly decreased in the CCu group (P < 0.05). Butyricimonas, Dehalobacterium, Coprococcus, Oscillospira, and Bilophila were significantly increased whereas Sporosarcina, Jeotgailcoccus, Staphylococcus, and Acinetobacter were significantly decreased in the CHg group (P < 0.05). Dehalobacterium, Coprococcus, and Anaeroplasma were significantly increased whereas Sporosarcina, Jeotgailcoccus, Staphylococcus, and Acinetobacter were significantly decreased in the CCH group (P < 0.05).
Cladogram of significant difference between groups, including the comparison between the control group and the CCu group (a), between the control group and the CHg group (b), and between the control group and the CCH group (c). The cladogram shows OTUs of the sample from the phylum to the genus (from the inner to the outer ring in the figures) the hierarchical relations. The average relative abundance of the OUTs was represented by the node size. The yellow node represents no significant difference between the groups (P > 0.05), but other colors (such as green and red) showed that these OTUs showed a significant difference between the groups (P < 0.05). The LDA scores represent differently abundant taxa after treating with Cu or/and Hg (only taxa meeting LDA ≥ 2.5 are shown)
Discussion
Cu at small amounts is an essential nutrient for all animals whereas excessive amounts of Cu may lead to some symptoms including anorexia, vomiting, lethargy, and gastrointestinal bleeding [6]. Hg has strong toxicity to animals and humans in biochemistry and physiology. Inorganic forms of Hg may cause gastrointestinal disorders, such as corrosive esophagitis and hematochezia [3]. Damaged gastrointestinal tract affects the digestion and absorption of nutrient substance, leading to weight loss in animals. In our experiment, we also found that the weight of mice in the Cu group and Hg group decreased significantly compared with the control group. When exposed to excessive Cu, the dysfunction of bile excretion of Cu could lead to the retention of Cu in tissues, individual enterocyte necrosis with enterocytes in the lumen and blunt of the villi [7, 28]. Hg in the form of its water-soluble salts is a highly potent poison, and could lead to extensive corrosive damage to the gastrointestinal tract [29]. Histopathology of the cecum in the Cu group and Hg group showed mild to moderate necrosis of enterocytes, blunt of intestinal villi, and severe atrophy of central lacteal. Hence, the digestion and absorption of substances could be affected by these damaged intestinal tissues. As a result, the weight decreased in the mice. These evidences suggested that the mice were affected by Cu and Hg.
To better understand the composition of the microbial community and diversity of the samples at the phylum and genus level in cecal tissues of female mice, we conducted high-throughput sequencing of the bacterial 16S rRNA. We found that Cu and Hg pollution could induce significant changes in the composition of the gastrointestinal microbiota at both the phylum level and the genus level. LEfSe analysis was used to evaluate the differences in bacterial abundance among different groups.
The Effects of Cu on Intestinal Microbial Community
At the phylum level, we found that there were no significant changes. At the genus level, compared to the CCk group, Corynebacterium were significantly increased whereas Rikenella, Odoribacter, Jeotgailcoccus, and Staphylococcus were significantly decreased in the CCu group (P < 0.05). Corynebacterium glutamicum is the Corynebacterium-Mycobacterium-Nocardia group of actinomycetes. Interestingly, it has been reported that CopRS (previously named as cgtRS9) is a significant regulatory system in Corynebacterium glutamicum for the extracytoplasmic sensing of elevated Cu ion concentrations and for the induction of a set of genes capable of Cu efflux [30]. Therefore, in the Cu-contaminated environment, Corynebacterium can endure Cu damage due to the anti-copper regulatory system CopRS. The number of Corynebacterium goes up relatively than other genera which have no anti-copper regulatory system. It indicated that Corynebacterium can tolerate relatively high levels of Cu and grow in the environment with Cu pollution.
In contrast, Rikenella, Jeotgailcoccus, and Staphylococcus were significantly decreased in the Cu group (P < 0.05) in our study. Up to date, all cultured members of the family Rikenellaceae are described as strictly anaerobic [31]. Nevertheless, copper- and zinc-containing superoxide dismutase (Cu/ZnSOD) is one of the anti-oxidant enzymes found in the periplasm of Gram-negative bacteria [32, 33]. Exposed to excess metal ion, the coordination mode of the Cu (II) ion could mimic the binding of the metal ion in the active center of the CuZnSOD enzyme [34]. Previous studies have demonstrated that the enzyme catalyzed the dismutation of superoxide radicals (O2•−) to molecular oxygen (O2) and hydrogen peroxide (H2O2). The first step reaction (reaction 1) begins with the O2•− substrate binding to Cu (II), and subsequently, Cu (II) is reduced to Cu (I), and O2•− is oxidized to molecular oxygen O2 (reaction 2). The Cu center is cyclically reduced and oxidized by superoxide [35].
Cu exposure promotes the activities of Cu/ZnSOD antioxidant enzymes [36]. Furthermore, this reaction can produce oxygen which can inhibit the growth of the strictly anaerobic bacteria Rikenella. Hence, abundance of this bacteria would decrease in the environment with Cu contamination. Additionally, Jeotgailcoccus and Staphylococcus belong to the family Staphylococcaceae which are Gram-positive coccus bacterium. Few studies have reported that Gram-positive cocci could have been affected in the Cu-contaminated environments. Similar studies reported that Gram-positive Enterococcus hirae is dependent on the activity of two ATPases (CopA and CopB), which are regulated in their expression by the Cu concentration. CopA is probably responsible for Cu uptake and nutrition and CopB (35% similar to CopA) is probably responsible for diminishing Cu stress and detoxification [37]. However, Solioz et al. [38] showed by genetic analysis that wild-type E. hirae can only tolerate up to 8 mM Cu in the media. Hence, excessive Cu is still toxic to E. hirae and do not induce the Cop-operon expression. In our study, Jeotgailcoccus and Staphylococcus could also be inhibited by the presence of high concentrations of Cu.
The Effects of Hg on Intestinal Microbial Community
At the phylum level, we found that the changes in the abundance of predominant bacteria were uniform as well as the CCu group compared with the CCk group, but the changes in the CHg group were significant different at the genus level. Gram-negative bacteria Butyricimonas, Dehalobacterium, Oscillospira, and Bilophila were significantly increased (P < 0.05), whereas Gram-positive bacteria Sporosarcina, Jeotgailcoccus, and Staphylococcus were significantly decreased in the CHg group (P < 0.05). There are specific uptake systems about inorganic Hg on periplasmic proteins in Gram-negative bacteria. In other words, resistance to inorganic Hg is encoded by the genes of the mer operon and can be located on transposons, plasmids, and the bacterial chromosome [39]. Protein MerP probably delivers the toxic cation to the Hg transporter MerT for transport into the cytoplasm to prevent toxic effects in Gram-negative bacteria [37, 40]. This show the effect of Hg contamination on the growth of Gram-negative bacteria was smaller than that of Gram-positive bacteria. Therefore, the abundance of Gram-negative bacteria would increase relatively, while that of Gram-positive bacteria would decrease. These correspond with our experimental results. On the other hand, it is considered that Sporosarcina, Jeotgailcoccus, Staphylococcus, and Acinetobacter were significantly decreased in the CHg group (P < 0.05). It is considered that Sporosarcina and Acinetobacter is aerobic bacteria, Jeotgailcoccus and Staphylococcus are obligate aerobes (oxygen reliant), or facultative anaerobes (having the ability to be aerobic or anaerobic), which are both associated with oxygen. Furthermore, Hg intoxication could lead to disturbed cellular function or biochemical damage which includes induction of free radical formation, and inhibition of activity glutathione peroxidase enzyme (GSH-Px) [41]. Therefore, the presence of Hg reduces oxygen production to a certain extent by inhibiting glutathione peroxidase activity, which may decrease the abundance of aerobic bacteria in our study, such as Sporosarcina, Acinetobacter, Sporosarcina, and Jeotgailcoccus. On the contrary, Coprococcus species are Gram-stain-positive and obligately anaerobic. Hence, the decrease in oxygen contributes to the growth of anaerobic Coprococcus, which may partially explain the increase in abundance of Coprococcus in the CHg group in our study.
We found that there were significant differences in the type and abundance of bacteria between the CCk group and each of the three treatment groups (CCu group, CHg group, CCH group). At the genus level, compared to the CCk group, the changes of intestinal microflora in the CHg group were greater than the changes in the CCu group, which indicated that Hg could have greater effect on the gut microbiota than Cu. In other words, Hg is more toxic than Cu.
The Effects of Cu and Hg on Intestinal Microbial Community
The microbial community structure composition showed that the changes in the abundance of predominant bacteria in the CCH group were similar to the CCu group and CHg group at the phylum level. Furthermore, the abundance of Tenericutes were significantly increased (P < 0.05).
At the genus level, the predominant genus bacteria in the CCH group were consistent with CHg group. In addition, the changes in abundance of Dehalobacterium and Coprococcus in the CCH group were consistent with the changes in the CHg group. Interestingly, the increase was even more pronounced, which may indicate that Hg has a greater effect on these two bacteria than Cu because these two bacteria may have a strong regulatory system to resist Hg damage. In addition, it suggested that the element Cu could have a synergistic relationship with Hg. The abundance of Anaeroplasma were significantly increased (P < 0.05). Anaeroplasma belongs to Gram-positive and is obligately anaerobic. The characteristics of these bacteria are similar to Coprococcus. In this study, the change in abundance of Anaeroplasma was similar to the change in abundance of Coprococcus.
The changes in the abundance of Sporosarcina, Jeotgailcoccus, Staphylococcus, and Acinetobacter in the CCH group were strictly identical with the CHg group. Similarly, it was more suggested that Hg plays a major role in the CCH group.
Conclusion
Taken together, we found that high doses of Cu and Hg might decrease the mice’s body weight and cause histopathological lesions including increased thickness of inner muscularis and outer muscularis, widened submucosa, decreased in the number of goblet cells, mild to moderate necrosis of enterocytes, blunting of intestinal villi, and severe atrophy of central lacteal. Furthermore, high doses of Cu and Hg changed the diversity of microbiota in the cecum of female mice and there may be a synergistic relationship between Cu and Hg.
References
Sun R, Chen L (2016) Assessment of heavy metal pollution in topsoil around Beijing metropolis. PLoS One 11(5):e0155350. https://doi.org/10.1371/journal.pone.0155350
Yang Z, Lu W, Long Y, Bao X, Yang Q (2011) Assessment of heavy metals contamination in urban topsoil from Changchun City, China. J Geochem Explor 108(1):27–38
Duruibe JO, Ogwuegbu MOC (2007) Heavy metal pollution and human biotoxic effects. Int J Phys Sci 2(5):112–118
Huang SH, Weng KP, Lin CC, Wang CC, Lee CT, Ger LP, Wu MT (2017) Maternal and umbilical cord blood levels of mercury, manganese, iron, and copper in southern Taiwan: a cross-sectional study. J Chin Med Assoc 80(7):442–451. https://doi.org/10.1016/j.jcma.2016.06.007
Lee J, Prohaska JR, Thiele DJ (2001) Essential role for mammalian copper transporter Ctr1 in copper homeostasis and embryonic development. Proc Natl Acad Sci U S A 98(12):6842–6847. https://doi.org/10.1073/pnas.111058698
Uriu-Adams JY, Keen CL (2005) Copper, oxidative stress, and human health. Mol Asp Med 26(4–5):268–298. https://doi.org/10.1016/j.mam.2005.07.015
Malinak CM, Hofacre CC, Collett SR, Shivaprasad HL, Williams SM, Sellers HS, Myers E, Wang YT, Franca M (2014) Tribasic copper chloride toxicosis in commercial broiler chicks. Avian Dis 58(4):642–649. https://doi.org/10.1637/10864-051514-Case.1
Wang XY, Lin RC, Dong SF, Guan J, Sun L, Huang JM (2017) Toxicity of mineral Chinese medicines containing mercury element. China J of Chinese Materia Med 42(7):1258–1264. https://doi.org/10.19540/j.cnki.cjcmm.20170224.002
Li Z, Ma Z, van der Kuijp TJ, Yuan Z, Huang L (2014) A review of soil heavy metal pollution from mines in China: pollution and health risk assessment. Sci Total Environ 468–469:843–853. https://doi.org/10.1016/j.scitotenv.2013.08.090
Funabashi H (2010) Minamata disease and environmental governance (environmental governance in Japan). Int J Jpn Sociol 15(1):7–25
De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P (2010) Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A 107(33):14691–14696. https://doi.org/10.1073/pnas.1005963107
Villanueva-Millan MJ, Perez-Matute P, Oteo JA (2015) Gut microbiota: a key player in health and disease. A review focused on obesity. J Physiol Biochem 71(3):509–525. https://doi.org/10.1007/s13105-015-0390-3
Lee WJ, Hase K (2014) Gut microbiota-generated metabolites in animal health and disease. Nat Chem Biol 10(6):416–424. https://doi.org/10.1038/nchembio.1535
Breton J, Daniel C, Dewulf J, Pothion S, Froux N, Sauty M, Thomas P, Pot B, Foligne B (2013) Gut microbiota limits heavy metals burden caused by chronic oral exposure. Toxicol Lett 222(2):132–138. https://doi.org/10.1016/j.toxlet.2013.07.021
Balkrishna AP (2011) The effect of heavy metals on the gastrointestinal microflora (GIM) of metapenaus monoceros and fenneropenaeus indicus. AJMBES Journal 13(1):119–124
Mickėnienė L, Šyvokienė J (1999) The effect of heavy metals on microorganisms of digestive tract of crayfish. Acta Zoologica Lituanica 9(2):37–39
Mitra S, Keswani T, Ghosh N, Goswami S, Datta A, Das S, Maity S, Bhattacharyya A (2013) Copper induced immunotoxicity promote differential apoptotic pathways in spleen and thymus. Toxicology 306:74–84. https://doi.org/10.1016/j.tox.2013.01.001
Wildemann TM, Siciliano SD, Weber LP (2016) The mechanisms associated with the development of hypertension after exposure to lead, mercury species or their mixtures differs with the metal and the mixture ratio. Toxicology 339:1–8. https://doi.org/10.1016/j.tox.2015.11.004
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7(5):335–336. https://doi.org/10.1038/nmeth.f.303
Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE (2006) Metagenomic analysis of the human distal gut microbiome. Science 312(5778):1355–1359. https://doi.org/10.1126/science.1124234
Chen H, Jiang W (2014) Application of high-throughput sequencing in understanding human oral microbiome related with health and disease. Front Microbiol 5:508. https://doi.org/10.3389/fmicb.2014.00508
Magoc T, Salzberg SL (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27(21):2957–2963. https://doi.org/10.1093/bioinformatics/btr507
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26(19):2460–2461
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL (2006) Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72(7):5069–5072. https://doi.org/10.1128/AEM.03006-05
White JR, Nagarajan N, Pop M (2009) Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput Biol 5(4):e1000352
Zaura E, Keijser BJ, Huse SM, Crielaard W (2009) Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol 9(1):259
Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C (2011) Metagenomic biomarker discovery and explanation. Genome Biol 12(6):R60. https://doi.org/10.1186/gb-2011-12-6-r60
Tao TY, Liu F, Klomp L, Wijmenga C, Gitlin JD (2003) The copper toxicosis gene product Murr1 directly interacts with the Wilson disease protein. J Biol Chem 278(43):41593–41596. https://doi.org/10.1074/jbc.C300391200
Clarkson TW, Magos L (2006) The toxicology of mercury and its chemical compounds. Crit Rev Toxicol 36(8):609–662. https://doi.org/10.1080/10408440600845619
Schelder S, Zaade D, Litsanov B, Bott M, Brocker M (2011) The two-component signal transduction system CopRS of Corynebacterium glutamicum is required for adaptation to copper-excess stress. PLoS One 6(7):e22143
Su XL, Tian Q, Zhang J, Yuan XZ, Shi XS, Guo RB, Qiu YL (2014) Acetobacteroides hydrogenigenes gen. nov., sp. nov., an anaerobic hydrogen-producing bacterium in the family Rikenellaceae isolated from a reed swamp. Int J Syst Evol Microbiol 64(Pt 9):2986–2991. https://doi.org/10.1099/ijs.0.063917-0
Perera NCN, Godahewa GI, Lee J (2016) Copper-zinc-superoxide dismutase (CuZnSOD), an antioxidant gene from seahorse (Hippocampus abdominalis ); molecular cloning, sequence characterization, antioxidant activity and potential peroxidation function of its recombinant protein. Fish Shellfish Immu 57:386–399
Hwang CS, Rhie GE, Oh JH, Huh WK, Yim HS, Kang SO (2002) Copper- and zinc-containing superoxide dismutase (Cu/ZnSOD) is required for the protection of Candida albicans against oxidative stresses and the expression of its full virulence. Microbiology 148(11):3705–3713
Csire G, Demjén J, Timári S, Várnagy K (2013) Electrochemical and SOD activity studies of copper(II) complexes of bis(imidazol-2-yl) derivatives. Polyhedron 61(5):202–212
Perry JJ, Shin DS, Getzoff ED, Tainer JA (2010) The structural biochemistry of the superoxide dismutases. Biochim Biophys Acta 1804(2):245–262. https://doi.org/10.1016/j.bbapap.2009.11.004
Martins I, Goulart J, Martins E, Morales-Roman R, Marin S, Riou V, Colaco A, Bettencourt R (2017) Physiological impacts of acute cu exposure on deep-sea vent mussel Bathymodiolus azoricus under a deep-sea mining activity scenario. Aquat Toxicol 193:40–49. https://doi.org/10.1016/j.aquatox.2017.10.004
Nies DH (1999) Microbial heavy-metal resistance. Appl Microbiol Biotechnol 51(6):730–750
Solioz M, Odermatt A (1995) Copper and silver transport by CopB-ATPase in membrane vesicles of enterococcus hirae. J Biol Chem 270(16):9217–9221
Osborn AM, Bruce KD, Strike P, Ritchie DA (1997) Distribution, diversity and evolution of the bacterial mercury resistance (mer) operon. FEMS Microbiol Rev 19(4):239–262
Sahlman L, Skarfstad EG (1993) Mercuric ion binding abilities of MerP variants containing only one cysteine. Biochem Biophys Res Commun 196(2):583–588. https://doi.org/10.1006/bbrc.1993.2289
ElSaeed GSM, Maksoud A, Soheir A, Bassyouni HT, Raafat J, Agybi MH (2016) Mercury toxicity and DNA damage in patients with Down syndrome. Med Res J 15(15):22–2622
Funding
This project was supported by the National Natural Science Foundation of China grant (No. 31492266, Beijing, P. R. China) awarded to PL, the Natural Science Foundation of Jiangxi Province grant (No. 20171ACB21026) awarded to PL and the Technology R&D Program of Jiangxi Province grant (No. GJJ170243, Nanchang, P. R. China) awarded to PL.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
All animals used in this experiment were approved by the Committee of Animal Welfare. Animal studies and experiments were approved and carried out according to Institutional Animal Care and Use Committee guidelines at College of Animal Science and Technology, Jiangxi Agricultural University.
Conflict of Interest
The authors declare that there are no conflicts of interest.
Additional information
All authors have read the manuscript and agreed to submit it in its current form for consideration for publication in the Journal.
Rights and permissions
About this article
Cite this article
Ruan, Y., Wu, C., Guo, X. et al. High Doses of Copper and Mercury Changed Cecal Microbiota in Female Mice. Biol Trace Elem Res 189, 134–144 (2019). https://doi.org/10.1007/s12011-018-1456-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12011-018-1456-1