Abstract
Streptomyces badius DB-1 produces α-amylase extracellularly, and its production was enhanced 5.1-fold (from 9.47 ± 0.51 to 48.23 ± 1.45 U mL−1) due to optimization by one-variable-at-a-time and statistical approaches. Soluble starch emerged as the most influential factor that strongly affected enzyme production. The purified enzyme is a monomer with a molecular mass of ~57 kDa and optimally active at 50 °C and pH 6.0. The enzyme hydrolyzes soluble as well as raw starches into simpler sugars with a high proportion (>40.0 %) of maltotetraose. It is optimally active at moderate temperature and generates maltooligosaccharides from starch, thus, useful as an antistale in bread making. It also plays a role in increasing the formation of maltooligosaccharides due to transglycosylation activity, thus, finds application in functional foods. This is the first report on the production of raw starch-digesting α-amylase by S. badius with transglycosylation activity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Amylases are glycoside hydrolases (GHs), which act on α-(1,4)- and/or α-(1,6)-glycosidic linkages between adjacent glucose units of starch polymer and liberate diverse products. The maltooligosaccharides are being increasingly used as biopreservatives and in functional foods in the recent years [1]. They have characteristic features that modify the flavor and physiochemical properties of foods. Maltooligosaccharides are non-digestible in the upper gastrointestinal tract and thus considered prebiotics which have beneficial effects on human health [2, 3], thus, used as major ingredients of various food products such as beverages, infant milk powders, confectionery, bakery products, yoghurts, and dairy desserts [4]. As chemical synthesis of maltooligosaccharides larger than maltotriose is not economically feasible, there is an immense interest in the use of maltooligosaccharide-forming amylases in food industry [2]. The application of amylases is not only limited to food industry but also in pharmaceuticals, textile, and paper industries [5]. Amylases account for approximately 25 % of total enzyme sales worldwide and have almost replaced chemical hydrolysis of starch in starch saccharification [6]. Starch-hydrolyzing enzymes are classified into three major GH families: α-amylases (GH13 family), β-amylases (GH14), and glucoamylases (GH15). α-Amylases (1,4-α-d-glucan glucanohydrolase, EC 3.2.1.1) are endo-amylases hydrolyzing α-(1,4)-glycosidic bonds of starch polymers randomly to generate shorter oligosaccharides and different α-limit dextrins [7]. α-Amylases are widely distributed throughout microbial, animal, and plant kingdoms. Over the past few decades, microbes are preferred over other sources to produce amylases, since they are easier to cultivate and manipulate for producing enzymes of the desired characteristics [8]. There are several reports on amylase production by microorganisms including fungi [5], actinobacteria [6], and bacteria [7]. Actinobacteria are well known to produce bioactive compounds including antibiotics and enzymes. Among them, Streptomyces species have been a prominent source of extracellular hydrolytic enzymes. Streptomyces species such as Streptomyces lonarensis [9], Streptomyces sp. MSC702 [10], Streptomyces gulbargensis [11], and Streptomyces erumpens MTCC 7317 [12] have been reported to secrete extracellular amylase.
To maximize enzyme yield without increasing the cost of production is of prime importance in industrial enzymology. High enzyme production can be attained by either genetic manipulation of microbes or process optimization. Optimization of fermentation parameters has been traditionally used in the development of economically feasible bioprocesses to meet the industrial demands [12]. Formulation of an appropriate fermentation medium is, therefore, a crucial factor for enhancing the product yield. Submerged fermentation is more ideal for the production of industrially important enzymes because of the ease of controlling fermentation variables such as pH, temperature, and aeration [13]. Medium optimization by the conventional methods involves varying one factor at a time while keeping others constant, which is based on the assumption that effect of every variable is mutually independent and may be effective in some situations. This is quite time consuming and requires a lot of experimentation to achieve optimal enzyme yield [9]. Despite its limitations, it is still useful in standardizing various physical and nutritional parameters. Plackett-Burman design (PB) and central composite design (CCD) in response surface methodology (RSM) are most popular statistical tools [14]; these not only allow rapid screening of a large number of factors and to understand interactions but also reflect the role of each component [15]. Statistical approaches have been used to optimize various fermentation processes for the production of enzymes [16], metabolite and biomass.
Raw starch-degrading α-amylases act on the starch granules without requiring gelatinization step, thereby reducing the energy cost of starch processing. Two Streptomyces spp. [17, 18] and Thermobifida fusca [19] have also been reported to secrete raw starch hydrolyzing amylases. Raw starch-hydrolyzing enzymes are receiving immense attention in the starch saccharification. Furthermore, some α-amylases display transglycosylation activity, thus, transferring specific sugar moieties to the hydroxyl group of other saccharides [20], therefore, are useful in synthesizing maltooligosaccharide glycosides. Amylases with transglycosylation activity have been used in the modification of bioactive natural compounds to enhance their stability and solubility [21].
Bread and bakery goods are mostly consumed food products around the world. Microbial enzymes such as α-amylases and xylanases have been used as additives in bread making to improve overall texture and taste of bread. Amylases hydrolyze wheat starch into fermentable sugars for the yeast to act upon [7]. α-Amylases with intermediate thermostability are useful as antistale to reduce crumb firmness during storage, thereby increasing the shelf life of bread [22].
In this investigation, an attempt has been made to formulate a cost effective medium for maximizing amylase production by using both conventional and statistical approaches. The enzyme was purified and confirmed its applicability in raw starch saccharification, maltooligosaccharide synthesis and bread making. This is the first report on the production of raw starch-digesting α-amylase by Streptomyces badius DB-1with transglycosylation activity.
Materials and Methods
Microorganisms and Other Materials
The actinobacterial strains used in this investigation were procured from Dr. Debananda Ningthoujam, Department of Biochemistry, Manipur University, Imphal (India). The primers used in this investigation were purchased from Sigma-Aldrich (USA). pGEM-T easy vector kit was purchased from Promega, USA. Glucose, maltose, maltotriose, maltotetraose, maltopentaose, soluble potato starch, cyclodextrins, amylose, amylopectin, and pullulan were purchased from Sigma-Aldrich (USA). All other chemicals were of analytical grade. While raw starches were procured from the local market.
Selection of Amylase-Producing Actinobacteria Strains
Thirty actinobacterial cultures were screened for the production of amylase by the qualitative halo-assay method. Cells were grown on starch peptone agar (g L−1: starch (10.0), peptone (5.0), Na2HPO4 (3.0), NaH2PO4 (1.0), MgCl2 (0.15), pH 7.0). After 3 days of incubation, amylase-producing strains were identified by flooding with Lugol’s iodine solution (containing 3.0 % KI and 0.3 % I2). The actinobacterial strains forming starch clearing zones against dark blue background were selected and further screened on the basis of quantitative amylase assay [23] and the end products formed by the enzyme action. The formation of end products was analyzed by thin-layer chromatography (TLC) technique. The strain DB-1 exhibited high amylolytic activity and liberated a relative higher quantity of maltooligosaccharides.
Amylase Assay and Protein Estimation
The amylase activity was determined by quantitating reducing sugars liberated during enzyme-substrate reaction using dinitrosalicylic acid (DNSA) reagent [23]. The enzyme reaction was performed at 50 °C for 15 min using soluble starch (0.5 %, w/v) as substrate in 0.1 M phosphate buffer (pH 6.0). One unit of amylase is defined as the amount of crude enzyme that liberates 1 μmol of reducing sugar (as maltose)/min under the assay conditions. The total protein was quantitated by the Bradford method using bovine serum albumin (BSA) as the standard [24].
Identification of Strain DB-1
The strain was identified on the basis of scanning electron micrograph, fatty acid methyl ester (FAME) profile, 16S ribosomal DNA (rDNA) gene sequence analysis, and spectra of MALDI-TOF-MS of ribosomal proteins. Scanning electron microscopy of cells of strain DB-1 was carried out at Advanced Instrumentation Research Facility (AIRF), Jawaharlal Nehru University, New Delhi, India. The analysis of cellular fatty acid methyl ester profile by gas chromatography was performed at the Institute of Microbial Technology (CSIR-IMTECH), Chandigarh, India. Genomic DNA was extracted from the actinobacterial biomass according to Tripathi and Rawal [25]. The 16S rDNA gene was amplified by using universal 16S rDNA primers (27F and 1492R), and amplified gene product was cloned in pGEM-T easy vector (Promega, USA) and sequenced at central instrumentation facility (CIF), University of Delhi South Campus, New Delhi, India. The 16S rDNA gene sequence was deposited at the National Centre for Biotechnology Information (NCBI). Phylogenetic tree of 16S rDNA gene sequence of strain DB-1 was constructed using the neighbor-joining method and MEGA 5.2 software. Analysis of MALDI-TOF-MS of ribosomal proteins was carried out at National Centre for Cell Science, Pune, India.
Inoculum Preparation and Cultivation Media
The inoculum for amylase production was prepared in starch peptone broth. One percent 40-h-old inoculum containing 2.0 × 107 cfu mL−1 was used for inoculating amylase production medium. In order to select a suitable medium that supports good enzyme production, the actinobacterial isolate was cultivated in 14 different media of varied compositions (pH 7.0) at 30 °C and 200 rpm for 72 h in an incubator shaker. Among these, medium 5 (g L−1: starch (20.0), peptone (10.0), yeast extract (1.0), Na2HPO4 (3.0), NaH2PO4 (1.0), and MgSO4 (0.2)) supported luxurious growth and high enzyme titer. The medium composition was further modified by using classical and statistical optimization methods.
Experimental Design for Optimization of Medium Component
The effect of various physiological and nutritional parameters of the production medium were investigated first by one-variable-at-a-time (OVAT) approach, followed by identifying significant parameters that affect amylase production by PB design. The interactions among significantly important parameters were further investigated by RSM.
One-Variable-at-a-Time Approach
Selection of Physiological and Nutritional Parameters
The effect of incubation period on enzyme production was assessed by incubating the culture flasks at 30 °C for 7 days. The flasks were harvested at an interval of 24 h, and the culture supernatants were used in amylase assays. The effect of pH (5.0–9.0) was assessed using 50 mM acetate (5.0), phosphate (6.0–8.0), and glycine-NaOH (9.0) buffers in the medium 5. The effect of temperature (15–40 °C) and agitation (100–300 rpm) on amylase production were also evaluated.
To select a suitable carbon source for amylase production, 2.0 % (w/v) starch of production medium 5 was substituted with equimolar proportions of different carbon sources such as maltodextrin, disaccharides (maltose and lactose), monosaccharides (glucose, fructose, and galactose), and sugar alcohols (glycerol and sorbitol). Likewise, peptone (1.0 %) as nitrogen source was substituted with equimolar nitrogen concentration of both organic (casein hydrolysate, tryptone, soybean meal, beef extract, triammonium citrate, and urea) and inorganic nitrogen sources (ammonium chloride, ammonium nitrate, ammonium sulfate, and potassium nitrate). After cultivation and harvesting, culture supernatants were used in amylase assays.
Plackett-Burman Design
After selecting the favorable physicochemical and nutritional factors for amylase production by S. badius DB-1, various variables such as soluble starch, NH4Cl, yeast extract, phosphate strength, MgSO4, and inoculum size were evaluated for their effect on enzyme production by PB design (Table 1). PB design is a two-factorial design, which identifies the critical physicochemical parameters required for elevated amylase production by screening n variables in n + 1 experiments. All variables were examined at two widely spaced levels, minimum (−) and maximum (+) to generate a set of 12 experiments using Design expert® 6.0 (Stat Ease, Inc., Minneapolis, USA).
The response of parameters on the production was calculated by the following equation
where, E is the effect of individual parameter and M+ and M− are responses (amylase activities) of trials at which each factor was at its higher and lower levels, respectively. N represents the total number of trials.
Response Surface Methodology
To determine the interactions among critical variables and their optimum levels to enhance enzyme yield is the next step in the formulation of production medium. The determination of interactions among the significant variables selected on the basis of p values, were studied by RSM design using CCD. The critical parameters examined are shown in Table 2. Each of which was evaluated at five coded levels (−2, −1, 0, +1, +2) and a total of 20 experimental runs with three factors and six repetitions at the center were generated using Design expert® 6.0.
ANOVA Analysis, Statistical Modeling, and Model Validation
The data recorded in RSM experiments were subjected to analysis of variance and further expressed according to the following quadratic model equation to predict the optimal points.
where Y was the predicted response, β0 was an intercept, β1, β2, and β3 were linear coefficients, β11, β22, and β33 were squared coefficient, and β12, β13, and β23 were interaction coefficients. In this study, A, B, C, A2, B2, C2, AB, AC, and BC were the independent variables. The ANOVA study, 3D curves, contour, and one-factor plots were also employed in order to illustrate the relationships among significant parameters and to find the optimum concentration of each factor for amylase production. The statistical significance of the model equation and the model terms are expressed via the coefficient of determination (R 2) and the adjusted R 2. The significance of quadric model was checked by Fisher’s test value (F value). The point optimization method was employed in order to check accuracy of the model. A set of experiments by taking closed values of optimized process parameters predicted by model were set up and assessed for practical validation of theoretical observations.
Enzyme Production in a Laboratory Fermenter
S. badius was cultivated in a 7-L laboratory fermenter (Applikon Biotechnology, the Netherlands) containing 4 L medium at constant aeration (0.1 vvm) under optimized cultural conditions. The samples were drawn at the desired intervals and assayed for amylase activity. The dry cell weight (dcw) and amylase production in terms of dcw were calculated. The spectrum of extracellular enzyme production during cultivation was checked on 10 % SDS-PAGE gel [26].
Amylase Purification
The cell biomass was removed by centrifugation at 8000 rpm for 10 min. One liter of enzyme supernatant was passed through 10 kDa molecular weight cutoff membrane. The retentate with most of the amylase activity was subjected to fractional acetone precipitation (30, 50, and 80 %). The 80 % acetone precipitated fraction was dissolved in phosphate buffer (0.05 M and pH 6.0) containing 1.7 M ammonium sulfate and further loaded onto pre-equilibrated phenyl sepharose column. The proteins were eluted by using 0.05 M phosphate buffer containing reverse gradient of ammonium sulfate (1.7 to 0.0 M). Each fraction was dialyzed against 0.05 M phosphate buffer to remove the salt and assayed for amylase. For purified enzyme, 1 unit of amylase is defined as the amount of enzyme that liberates 1 μmol of reducing sugar (as glucose)/min under the assay conditions.
Determination of Molecular Mass of the Enzyme
A fraction showing amylase activity was run on 10 % SDS-polyacrylamide gel electrophoresis [26] for checking the purity of amylase. The activity of pure protein was confirmed by zymogram analysis on native PAGE as described by Nisha and Satyanarayana [27]. The molecular mass of the purified protein was estimated from its relative position to those of a set of protein markers with known molecular weight standards according to Nisha and Satyanarayana [27]. Different molecular weight standards such as cytochrome c (12.4 kDa), carbonic anhydrous (29 kDa), bovine serum albumin (66 kDa), yeast alcohol dehydrogenase (150 kDa), and sweet potato β-amylase (200 kDa) were used to determine the molecular mass of the enzyme on a semi-log graph plotted by taking V e/V o values on x-axis and molecular weight on y-axis.
Effect of pH and Temperature on the Activity and Stability
Effect of pH on the activity of amylase was determined at different pH values (5.0–10.0) using 0.1 M acetate buffer (4.0–5.0), phosphate buffer (6.0–8.0), and glycine-NaOH (9.0–10.0). The pH stability of the enzyme was studied by incubating the enzyme in buffers of pH 6.0, 7.0, 8.0, and 9.0 at room temperature for 6 h. The residual activity was then determined. The effect of temperature was studied in the range 20–70 °C. The thermostability of the enzyme was studied by pre-incubating the enzyme at temperature ranging from 40 to 60 °C in the presence and absence of 10 mM CaCl2 and determining the residual activity.
Substrate Specificity and Enzyme Kinetics
To determine the substrate specificity of α-amylase, it was incubated in the presence of (0.5 % (w/v)) soluble starch, wheat flour, rice flour, water chestnut, corn and sago starch powder (raw starches), pullulan, amylose, amylopectin, α-cyclodextrin, β-cyclodextrin, and γ-cyclodextrin, and α-amylase activity was determined. The K m, V max, and K cat values were graphically determined from the Lineweaver-Burk plot.
Effect of Metal Ions and Other Modulators
The effect of different cations in the form of chloride/sulfate salts (CuSO4, CoCl2, CaCl2, MgSO4, HgCl2, BaCl2, PbCl2, MnCl2, ZnCl2, and FeSO4) and various modulators like ethylenediaminetetraacetic acid (EDTA), ethylene glycol-bis(2-aminoethylether)-N,N,N′,N′-tetraacetic acid (EGTA), Woodward’s reagent K (WRK), dithiothreitol (DTT), phenylmethylsulfonyl fluoride (PMSF), iodoacetate (IAA), and N-bromosuccinimide (NBS) was studied at 1 and 5 mM. The effect of various detergents such as SDS (0.1 and 1.0 %) and Triton-X-100 (0.1–0.2 %) were incorporated into the reaction mixtures and incubated for 30 min in 0.1 M sodium phosphate buffer at room temperature. The residual α-amylase was determined under optimal conditions.
Analysis of Enzyme Action Pattern on Soluble Starch and Raw Starches
The types of sugars formed were analyzed on pre-activated silica gel plates (TLC Silica gel 60F254, Merck, Germany). The reaction mixture comprising 0.5 mL soluble starch solution or wheat starch or sago starch powder (0.5 % (w/v) prepared in phosphate buffer (0.1 M, pH 6.0)) and 0.5 mL of a suitably diluted enzyme sample was incubated in a water bath at 50 °C for 20 and 60 min. One microliter from the 10-mg mL−1 stocks of standard sugars (glucose, maltose, maltotriose, maltotetraose, and maltopentaose) and samples drawn at different time intervals from amylase-starch reaction mixtures were spotted on TLC plates. Chromatograms were developed using butanol/ethanol/water (5:3:2) as the solvent phase. The plates were dried at 37 °C for 2 h and sprayed with aniline-diphenylamine reagent [28]. The plates were kept in an oven at 100 °C for an hour and observed for individual sugars appeared as dark blue spots on a white background.
Analysis of End Products Formed on the Hydrolysis of Soluble Starch and Raw Starches
The hydrolysis products of the enzyme-substrate reaction containing suitably diluted amylase and 0.5 % substrate were assessed by high-performance liquid chromatography (HPLC; Shimadzu) using an Aminex HPX-87P column (Biorad, USA) with acetonitrile/water (70:30, v/v) as the mobile phase. Elution was conducted at 0.6 mL min−1 flow rate and sugars were detected by a refractive index detector (RID).
Transglycosylation Activity of α-Amylase
Maltooligosaccharides such as maltose, maltotriose, maltotetraose, and maltopentaose were used as substrate and incubated with pure enzyme for 2 h. The samples were drawn at 20 min, 1 h and 2 h and analyzed by TLC.
Application of α-Amylase in Starch Saccharification
The efficacy of purified enzyme in starch saccharification was assessed as described by Nisha and Satyanarayana [27]. Different starches (raw wheat starch and sago starch powder (20 %, w/v) prepared in sodium phosphate buffer (0.1 M, pH 6.0) were subjected to gelatinization at 105 °C for 10 min. Thereafter, the starch slurry was cooled to 50 °C and incubated with the amylase for 4 h. Aliquots were drawn at intervals of 2 h, and the reducing sugars liberated were determined. The percent saccharification of starch was computed using the formula:
The factor 0.95 normalizes the conversion for the weight gain caused by addition of water molecule to glycosyl moiety on hydrolysis [29].
Application of α-Amylase in Bread Making
Wheat flour (300 g) was mixed with dry yeast (10 g), NaCl (4.0 g), and 60 % (w/v) water. α-Amylase (5.0 U g−1) was added and mixed mechanically for 30 min to produce the dough, which was kept undisturbed for proofing followed by fermentation for 1 h and baked at 240 °C for 20 min. Bread making was performed in the presence of α-amylase of S. badius DB-1 (test) and with the commercial α-amylase (control). The amount of reducing sugars of test and control bread were determined by the following the method of Miller [23]. The protein concentration was quantified according to Bradford [24]. The moisture content percentage was calculated from wet and dry weight of the bread. The shelf life of both test and control was checked by keeping them at room temperature till they become dry and hard. The bread made were then checked for moisture contents, softness, reducing sugars, protein, and shelf life. The test and control bread were suspended in deionized water (1 mg mL−1) and the supernatants after centrifugation were analyzed for sugars formed by the action of S. badius DB-1 α-amylase on wheat flour in the bread by thin layer chromatography (TLC).
Results and Discussions
Screening of Amylase-Producing Actinobacteria and Identification of Strain DB-1
On the basis of halo-amylase assay, 21 among 30 actinobacterial cultures displayed zones of clearance surrounding the growing colonies on starch peptone medium. Among the tested 21 amylolytic actinobacterial isolates, a strain DB-1 produced a high amylolytic activity (9.47 U mL−1) under unoptimized cultural conditions (Table S1) and formed a relatively higher amount of reducing sugars (data not shown). It grew as rough velvety colonies on the agar plate (Fig. S1a). Microscopic morphological observation of a 3-day culture of strain DB-1 grown on malt extract yeast extract agar (International Streptomyces Project medium 2) revealed well-developed aerial mycelium without fragmentation specifying that strain DB-1 has a typical characteristic of the genus Streptomyces (Fig. S1b). In general, cellular fatty acid profile cannot be used to define Streptomyces species [30], since the genus Streptomyces encompasses a large number of species with varied cellular fatty acid profile, but are still being used for the rapid characterization of large numbers of Streptomycetes isolated from the environment [31]. The major cellular fatty acids of the strain DB-1 are shown in Table S2. Like other species of Streptomyces, the strain DB-1 contained 15:0 anteiso fatty acids in higher proportion [32]. The 16S rDNA gene analysis revealed that the strain DB-1 shows 99.0 % homology to that of S. badius strain G4-3 (Fig. S2). The 16S rDNA gene sequence was submitted to NCBI (accession no. KU886090). In MALDI-TOF-MS-based identification method, the strain shows ≥1.7 log value in database, which confirms that strain belongs to the particular genus, and the strain that shows ≥2.0 log value confirms to particular species. Based on the protein spectra obtained for the isolate DB-1, a dendrogram was constructed using MALDI biotyper software 3.0 (Briker Daltonik). It showed 2.023 score with S. badius and 1.74 score with Streptomyces griseus.
Process Optimization Using OVAT and Statistical Methods (PB and RSM) and Batch Fermentation
Among 14 media tested, medium 5 supported luxurious growth of the actinobacterium and high amylase titer. The strain DB-1 exhibited high amylase activity after 3 days of incubation. Actinobacterial cultures have been reported to secrete extracellular amylase in 48 to 96 h [10, 33]. Streptomyces cheonanensis VUK-A has been reported to produce high enzyme titer in 72 h [34]. Among physiological parameters, the pH plays a critical role in morphological changes of microorganisms and enzyme production. Likewise, the effect of temperature is directly associated with microbial cell growth, therefore, influences the enzyme yield [12]. The pH 7.0 was found to be suitable for amylase production by actinobacterial species [34]. It has been reported earlier that mesophilic actinobacteria produce good amylase titres at 30 °C [33, 34]. Agitation and aeration plays a critical role in supporting the growth of aerobes, therefore, directly affecting growth and enzyme production [35]. In this investigation, maximum amylase production of the strain DB-1 was recorded at 200 rpm.
Among carbon sources, soluble starch (17.50 U mL−1), maltodextrin (17.02 U mL−1), and maltose (16.31 U mL−1) supported a high amylase production. This is consistent with the earlier report, where the use of soluble starch was shown to enhance amylase production by Streptomyces albidoflavus [36]. In another report, 2.0 % maltose enhanced enzyme yield [6]. d-inositol and sorbitol have been reported to induce a high α-amylase production in a Streptomyces sp. [10]. In some reports, monosaccharides such as glucose [33] and mannose [37] had been found to be a suitable carbon source for amylase production in actinobacteria. The use of soluble starch as the carbon source is more economical than using maltodextrin and maltose in terms of low cost, therefore, soluble starch was chosen for further optimization process. The amylase synthesis by S. badius DB-1 is higher in the presence of starch, maltose, and maltodextrin, suggesting that the enzyme synthesis is inducible. Among nitrogen sources tested, peptone (17.03 U mL−1), casein hydrolysate (16.901 U mL−1), and ammonium chloride (17.19 U mL−1) supported comparatively higher enzyme titres than in others.
The results of PB design for amylase production by S. badius are presented along with predicted responses in Table S3. Analysis of data obtained from Plackett and Burman design revealed that three variables (starch, yeast extract, and MgSO4) significantly affect enzyme production. The F values of starch, yeast extract, and MgSO4 were 205.6, 13.29, and 17.08, respectively. The most influential factor was starch (p value, 0.0001) while less effect of phosphate strength (p value, 0.9936) was observed, therefore, low level of phosphate strength was further used. Increased concentration of yeast extract (p value, 0.0219) showed positive response on amylase production as it has trace minerals and ions that enhance the enzyme yield. Narayana and Vijayalaxmi [36] observed the inclusion of 0.5 % yeast extract in the medium exerted positive effect. Metal ions often influence the activity of amylases [38]. In our study, MgSO4 (p value, 0.0145) was showing positive response to improve enzyme yield. NH4Cl (p value, 0.1471) and inoculum size (p value, 0.2471) exerted negative effect on amylase production at higher concentration. The size of inoculum played an important role in the amylase production by Aspergillus oryzae [39]. Gangadharan et al. [40] reported that a high inoculum size did not show any significant effect on amylase production. In PB data analysis, the F value of 35.08 and the values of “Prob > F” are less than 0.05 indicated that the model terms are significant. The dummy variable included in the design had no impact on α-amylase production, implying that the model is significant. Based on regression analysis of the model, the coefficient of determination (R 2) of the model was 0.9840 which suggested that up to 98.40 % variability in the data can be explained by the model. The predicted R 2 of 0.8557 and adjusted R 2 of 0.9559 are in good agreement with each other.
RSM is a collection of statistical techniques used to design experiments, build models, and investigate the effect of main process parameters. It searches optimum condition of each factor for desirable response. The optimum levels of significant variables and effect of their interactions on amylase production were further studied by RSM using CCD design (Table S4). The second-order regression equation for the amylase production as a function of starch (%; A), yeast extract (%; B), and MgSO4 (%; C) is given below:
Amylase production = + 45.55 + 2.36 * A + 0.35 * B − 0.098 * C − 9.09 * A 2 − 9.20 * B 2 − 9.50 * C 2 − 0.39 * A * B − 0.41 * A * C − 0.83 * B * CThe F value of the model was 49.92 and the Prob > F value was 0.0001, indicating that the model is highly significant. Coefficients A of the three linear A, B, and C squared model terms A2, B2, and C2 are significant. The coefficient of determination (R 2) was 0.9782 for amylase production. The predicted R 2 (0.8559) and adjusted R 2 (0.9586) are in reasonable agreement between experimental and predicted values for amylase production. The adjusted R 2 corrects the R 2 value for the sample size and for the number of terms in the model. Adequate precision is a measure of the signal to noise ratio and a value greater than 4.0 is desirable. The adequate precision value of 17.023 indicated an adequate signal and suggested that the model can be used to navigate the design space. ANOVA study of amylase production showed a satisfactory adjustment of the statistical model with the experimental data (Table S5).
The response surface is used essentially to find out the optimum values of the variables for which the response is maximized and to find a desirable location in the design space [41]. The effect of interaction of various physiochemical parameters on amylase production (z-axis) was studied by plotting three-dimensional response surface curves against any two independent variables. Amylase production was maximum when mid-values (0) of three components (starch, yeast extract, and MgSO4) were taken, while decreasing or increasing concentration led to a decline in enzyme production as indicated by the response surface curve (Fig. S3). Starch (2.25 %), yeast extract (0.55 %), and MgSO4 (0.26 %) supported a high enzyme titer (48,230 U L−1).
The enzyme production enhanced up to ~5.1-fold after optimization using classical and statistical approaches (Table 3). The experimental model and regression equation were validated by quantitating amylase production in a set of experiments predicted by the software. The enzyme production predicted by the quadratic model and that recorded experimentally are in good agreement, thus proving the validity. A peak in enzyme production was attained in 39 h in the fermenter as compared with that of 72 h in shake flasks (Fig. S4). The production of amylase and biomass of S. badius DB-1 at different time intervals during fermentation were represented in Table S6. A reduction in fermentation time for sustained enzyme production was due to better control of process conditions as reported in xylanase production by Bacillus halodurans TSEV1 [42]. The profile of extracellular enzyme production during fermentation was shown in Fig. S5.
Purification and Molecular Mass Determination of α-Amylase
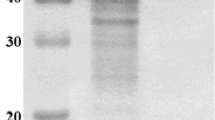
When grown on starch-containing medium (M5), S. badius DB-1 secreted amylases into the culture medium (approximately 48.23 U mL−1). The crude enzyme of the strain DB-1 showed multiple starch hydrolysis bands on the native gel (zymogram, not shown), indicating the presence of more than one starch-hydrolyzing enzymes. Streptomyces sp. IMD 2679 produced three α-amylases with similar starch hydrolysis pattern [43]. The α-amylase of strain DB-1was purified to apparent homogeneity by using ultrafiltration, fraction acetone precipitation, and hydrophobic interaction chromatography. A summary of the alpha amylase purification is presented in Table 4. Overall, the specific activity increased to 246.9 U mg−1, this corresponds to 8.83-fold purification. SDS-PAGE analysis showed a single band of approximately 57 kDa and native PAGE zymogram revealed a single band of starch hydrolysis (Fig. 1). In gel filtration chromatography, molecular mass of purified protein was approximately 57 kDa, confirming monomeric nature of amylase (Fig. S6); this is similar to that produced by Kitasatospora sp. MK-1785 (57 kDa) [20].
Effect of pH and Temperature on the Activity and Stability
The activity of α-amylase was stable in the pH range of 6.0–9.0 with optimum at pH 6.0 (Fig. 2a) and retained more than 85 % activity after 4 h of incubation (Fig. 2b). The enzyme was optimally active at 50 °C (Fig. 3a) and t 1/2 has increased from 17 to 40 min in the presence of CaCl2 indicating that amylase is calcium dependent (Fig. 3b, c). Goldberg and Edward [44] reported that 50 % of amylase activity of Streptomyces thermoviolaceus subsp. apingens was lost in the absence of calcium ions at 60 °C even after 13 min of incubation, while only 28 % of its activity was lost after 30 min incubation under similar conditions in the presence of 10 mM calcium chloride. Amylases of actinobacteria require calcium ions for their activity and stability [11, 20], while a calcium-independent amylase has also been reported from marine Streptomyces sp. [6]. Maltotriose forming exo-α-amylase of Kitasatospora sp. MK-1785 was optimally active at pH 6.5 and 55 °C [20]. A maltotriose-forming amylase of Microbacterium imperiale showed optimum activity at pH 6.5–7.0 and 50 °C [45].
Enzyme Kinetics and Substrate Specificity
The enzyme exhibited K m (1.12 mg mL−1), V max (454 μmol mg−1 min−1), and K cat (1472 s−1). Maltotriose-forming amylase of T. fusca exhibited K m value (0.88 mg mL−1) [19]. When different types of starches were used as substrates, the highest enzyme activity was observed on soluble starch. A comparable enzyme activity was also observed on wheat and sago starch powders and amylose (Table 5). There was no activity on α,β,γ-cyclodextrin and pullulan. Maltogenic amylases with transglycosylation activity have been reported to efficiently hydrolyze cyclodextrins [46]. α-Amylase of an actinobacterium, Kitasatospora sp. MK-1785 with transglycosylation showed very low activity on α- and β-cylodextrin, but a little higher activity (14.4 U mg−1) on γ-cyclodextrin [20].
Effect of Metal Ions and Modulators on the α-Amylase Activity
Calcium ion enhanced the enzyme activity (Table 6). The amylase activity was moderately stimulated by Mg2+, while Cu2+, Hg2+, and Zn2+ ions strongly inhibited α-amylase, as reported earlier [20]. The chelating agents (EDTA and EGTA) affected α-amylase activity suggesting that the enzyme requires Ca2+ and some other cations for its activity. Exo-α-amylase activity of Kitasatospora sp. MK-1785 was also inhibited by EDTA [20]. NBS strongly inhibited the enzyme activity indicating the role of tryptophan residue(s) on amylase activity. The sulfhydryl inhibitors like DDT, β-mercaptoethanol, N-ethylmaleimide, and iodoacetate did not inhibit the enzyme activity suggesting that thiol group is not involved in the enzyme catalysis. Woodward’s reagent K inhibited enzyme activity, suggesting the role of carboxyl group(s) in the catalytic activity [47].
Analysis of Hydrolysis Products
TLC analysis of the end products of hydrolysis of soluble starch, wheat starch, and sago starch powders were glucose, maltose, maltotriose, and maltotetraose, confirming that the amylase produced by S. badius is an α-amylase (Fig. 4). The amounts of maltotriose and maltotetraose increased with increase in reaction time (Table 7). Kaneko et al. [18] observed the formation of all sugars with higher amount of maltose from the raw starches. Whereas, Primarini and Ohta [17] reported all sugars except glucose from raw starch hydrolysis by the amylase-1 of Streptomyces sp. no. 4.
TLC analysis of end products formed on hydrolysis of soluble starch, wheat starch, and sago starch powder. Lane M represents a standard mixture of glucose (G1), maltose (G2) and maltooligosaccharides, maltotriose (G3), maltotetraose (G4), and maltopentaose (G5). Lanes 1, 2, and 3 represent end products of hydrolysis of soluble starch, wheat flour, and sago starch powder, respectively. The samples were taken after 10 min. Lanes 4, 5, and 6, samples drawn after 60 min of reaction on soluble starch, wheat flour, and sago starch powder, respectively
Transglycosylation Activity
The enzyme hydrolyzed all maltooligosaccharides tested and formed glucose, maltose, maltotriose, and maltotetraose within 20 min (Fig. 5a, b). The formation of low level of higher maltooligosaccharides such as maltopentaose and maltohexaose were observed, indicating that α-amylase exhibits transglycosylation activity. Transglycosylation activity is useful in improving the solubility and stability of many compounds [21]. Glycosylation is also used in the synthesis of maltooligosyl glycosides, which have been used in functional foods.
TLC analysis of transglycosylation activity of amylase. a Lane M represents a standard mixture of glucose (G1), maltose (G2) and maltooligosaccharides, maltotriose (G3), maltotetraose (G4), and maltopentaose (G5). Lanes 1, 2, 3, and 4 represent end products of hydrolysis of maltose, maltotriose, maltotetraose, and maltopentaose, respectively. The samples were drawn after 20 min incubation. Lanes 5, 6, 7, and 8 represent end products of hydrolysis of maltose, maltotriose, maltotetraose, and maltopentaose after 60 min of incubation. b Lane M represents a standard mixture of glucose (G1), maltose (G2) and maltooligosaccharides maltotriose (G3), maltotetraose (G4), and maltopentaose (G5). Lanes 1, 2, 3, and 4 represent the hydrolytic products of maltose, maltotriose, maltotetraose, and maltopentaose after 2 h
Application of α-Amylase in Starch Saccharification and Bread Making
Wheat and sago starches were saccharified to the extent of 22.6 and 25.2 %, respectively, in 2 h. The saccharification of both raw starches further increased to 35.09 and 31.1 % when treated with amylase for 4 h. Nisha and Satyanarayana [47] reported that 39.8 ± 0.5 and 44.2 ± 1.1 % saccharification of raw wheat and corn starches were attained when treated with amylopullulanase followed by glucoamylase.
The supplementation of wheat flour with amylase of S. badius has ameliorated the texture, softness, and sweetness of bread (Fig. 6a). The bread made by supplementing the dough with amylase of S. badius DB-1 (test) was soft with higher moisture content, soluble protein, and reducing sugars (Fig. 6b) than that made using the commercial fungal α-amylase (control) (Table 8). The shelf life of test bread increased to 5 days as compared with 4 days of the control bread at room temperature due to antistaling effect. The formation of high quantity of maltooligosaccharides eliminates the increased gumminess of α-amylase-treated bread. Maltooligosaccharides exhibit high water-holding capacity, antistaling effect on bread, and prevention of sucrose crystallization. Staling of baked foods is generally defined as undesirable qualities including chewy crust and a loss of flavor. Starch retrogradation is a major factor for staling of baked goods. Amylase-producing residual dextrins (maltotriose-maltononaose) function as antistaling agents in reducing bread firming and in increasing the shelf life of bread [48].
Test and control bread made with and without α-amylase of the strain DB-1. a Test bread supplemented with the α-amylase of S. badius DB-1 has improved texture of bread as compared with that of the control and b TLC plate analysis of sugars formed in test and control bread. Lane M represents a standard mixture of glucose (G1), maltose (G2) and maltooligosaccharides maltotriose (G3), maltotetraose (G4), and maltopentaose (G5). L1 and L2 represent sugars formed in test and control bread, respectively
Conclusions
The use of OVAT and statistical approaches enabled us to formulate the amylase-production medium that supported 5.1-fold higher amylase production than that attained under unoptimized cultural conditions. The amylase production was 48.2 U mL−1, which is comparable with those reported in the literature. The pure enzyme showed affinity for raw starch and hydrolyzed it into glucose, maltose, maltotriose, and maltotetraose, suggesting its applicability in bread making. The enzyme also exhibits transglycosylation activity, thus, forms maltopentaose and maltohexaose from maltotriose, maltotetraose and maltopentaose, confirming its utility in synthesizing maltooligosaccharides that are useful in functional foods. Our efforts are underway to clone and over express the enzyme in heterologous hosts such as Escherichia coli for enhancing enzyme production.
References
Barreteau, H., Delattre, C., & Michaud, P. (2006). Production of oligosaccharides as promising new food additive generation. Food Technology and Biotechnology, 44, 323–333.
Okada, M., Nakakuki, T. (1992). In F. W. Schenck (Ed.), Oligosaccharides: production, properties, and applications. New York: VHC Publishers.
Nakakuki, T. (2003). Development of functional oligosaccharides in Japan. Trends in Glycoscience and Glycotechnology, 15, 57–64.
Subramanian, G., Ayyadurai, S., Sharma, T., Singh, S. A., Rele, M., & Kumar, L. S. (2012). Studies on a maltohexaose (G6) producing alkaline amylase from a novel alkalophilic Streptomyces species. IIOABJ, 3, 15–30.
Agger, T., Spohr, A. B., & Nielsen, J. (2001). Alpha amylase production in high cell density submerged cultivation of Aspergillus oryzae and Aspergillus nidulans. Applied Microbiology and Biotechnology, 55, 81–84.
Chakraborty, S., Raut, G., Khopade, A., Mahadik, K., & Kokare, C. (2012). Study on calcium ion independent α-amylase from haloalkaliphilic marine Streptomyces strain A3. Indian Journal of Biotechnology, 11, 427–437.
Sharma, A., & Satyanarayana, T. (2010). High maltose-forming, Ca2+-independent and acid stable α-amylase from a novel acidophilic bacterium, Bacillus acidicola. Biotechnology Letters, 32, 1503–1507.
Lonsane, B. K., & Ramesh, M. V. (1990). Production of bacterial thermostable α-amylase by solid-state fermentation: a potential tool for achieving economy in enzyme production and starch hydrolysis. Advances in Applied Microbiology, 35, 1–56.
Sharma, T. K., Bhadane, V. A., Kumar, L. S., Rele, M. V., Bhawar, G., & Rahman, I. (2013). Optimization of the production of a maltooligosaccharides producing amylase from the alkalophilic Streptomyces lonarensis strain NCL 716 using SVR modeling. Starch, 65, 179–185.
Singh, R., Kapoor, V., & Kumar, V. (2011). Influence of carbon and nitrogen sources on the α-amylase production by a newly isolated thermophilic Streptomyces sp. MSC702 (MTCC 10772). Asian Journal of Biotechnology, 3, 540–553.
Syed, D. G., Agasar, D., & Pandey, A. (2009). Production and partial purification of α-amylase from a novel isolate Streptomyces gulbargensis. Journal of Industrial Microbiology and Biotechnology, 36, 189–194.
Kar, S., Ray, R. C., & Mohapatra, U. B. (2008). α-amylase production by Streptomyces erumpens MTCC 7317 in solid state fermentation using response surface methodology (RSM). Polish Journal of Microbiology, 57, 289–296.
Mouna imen, O., & Mahmoud, K. (2015). Statistical optimization of cultural conditions of an halophilic alpha-amylase production by halophilic Streptomyces sp. grown on orange waste powder. Biocatalysis and Agricultural Biotechnology, 4, 685–693.
Shabbiri, K., Adnan, A., Noor, B., & Jamil, S. (2012). Optimized production, purification and characterization of alpha amylase by Brevibacterium linens DSM 20158, using bio-statistical approach. Annals of Microbiology, 62, 523–532.
Wenster-Botz, D. (2000). Experimental design for fermentation media development: statistical design or global random search? Journal of Bioscience and Bioengineering, 90, 473–483.
Uma Maheswar Rao J. L., & Satyanarayana, T. (2007). Improving production of hyperthermostable and high maltose-forming a-amylase by an extreme thermophile Geobacillus thermoleovorans using response surface methodology and its applications. Bioresource Technology, 98, 345–352.
Primarini, D., & Ohta, Y. (2000). Some enzyme properties of raw starch digesting amylases from Streptomyces sp. no. 4. Starch, 52, 28–32.
Kaneko, T., Ohno, T., & Ohisa, N. (2005). Purification and characterization of a thermostable raw starch digesting amylase from a Streptomyces sp. isolated in a milling factory. Bioscience, Biotechnology, and Biochemistry, 69, 1073–1081.
Yang, C. H., & Liu, W. H. (2004). Purification and properties of a maltotriose-producing α-amylase from Thermobifida fusca. Enzyme and Microbial Technology, 35, 254–260.
Kamon, M., Sumitani, J., Tani, S., Kawaguchi, T., Kamon, M., Sumitani, J., et al. (2015). Characterization and gene cloning of a maltotriose-forming exo-amylase from Kitasatospora sp. MK-1785. Applied Microbiology and Biotechnology, 99, 4743–4753.
Park, K. H. (2012). Biotechnological modification of bioactive natural compounds for food industry. Journal of Food and Drug Analysis, 20, 189–193.
Kumar, P., & Satyanarayana, T. (2008). Potential applications of microbial enzymes in improving quality and shelf-life of bakery products. In A. Koutinas, A. Pandey, & C. Larroche (Eds.), Bioprocesses in food industry (pp. 132–146). New Delhi: Asiatech Publishers Inc.
Miller, G. L. (1959). Use of dinitrosalicyclic acid reagent for determination of reducing sugar. Analytical Chemistry, 31, 426–429.
Bradford, M. M. (1976). A rapid and sensitive method for quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry, 72, 248–254.
Tripathi, G., & Rawal, S. K. (1998). Simple and efficient protocol for isolation of high molecular weight DNA from Streptomyces aureofaciens. Biotechnology Techniques, 12, 629–631.
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685.
Nisha, M., & Satyanarayana, T. (2013). Characterization of recombinant amylopullulanase (gt-apu) and truncated amylopullulanase (gt-apuT) of the extreme thermophile Geobacillus thermoleovorans NP33 and their action in starch saccharification. Applied Microbiology and Biotechnology, 97, 6279–6292.
Hansen, S. A. (1975). Thin layer chromatographic method for identification of oligosaccharides in starch hydrolysates. Journal of Chromatography, 105, 388–390.
Mishra, R., & Maheshwari, R. (1996). Amylases of the thermophilic fungus Thermomyces lanuginosus; their purification, properties, action on starch and response to heat. Journal of Bioscience, 21, 653–672.
Phillips, L. (1992). The distribution of phenotypic and genotypic characters within Streptomycetes and their relationship to antibiotic production. PhD thesis, University of Warwick.
Saddler, G. S., O’donnell, A. G., Goodfellow, M., & Minnikin, D. E. (1987). Pattern recognition in the analysis of Streptomycete fatty acids. Journal of General Microbiology, 133, 1137–1147.
Sujarit, K., Kudo, T., Ohkuma, M., Pathom-Aree, W., & Lumyong, S. (2016). Streptomyces Palmae sp. nov., isolated from oil palm (Elaeis guineensis) rhizosphere soil. International Journal of Systematic and Evolutionary Microbiology. doi:10.1099/ijsem.0.001298.
Krishnakumar, S., Bai, V. D. M., & Premkumar, J. (2015). Production of alpha amylase by salt-tolerant actinomycete Streptomyces sp.-SBU3 isolated from marine sponge. Indian Journal of Geo-Marine Sciences, 44, 1–6.
Naragani, K., Muvva, V., Munaganti, R. K., & Heema Bindu, B. S. S. N. (2015). Studies on optimization of amylase production by Streptomyces cheonanensis VUK-A isolated from mangrove habitats. Journal of Advanced Biology and Biotechnology, 3, 165–172.
Mazotto, A. M., Cedrola, S. M. L., Lins, U., Rosado, A. S., Silva, K. T., Chaves, J. Q., et al. (2010). Keratinolytic activity of Bacillus subtilis AMR using human hair. Letters in Applied Microbiology, 50, 89–96.
Narayana, K. J. P., & Vijayalakshmi, M. (2008). Production of extracelluar α-amylase by Streptomyces albidoflavus. Asian Journal of Biochemistry, 3, 194–197.
Poornima, R., Sahu, M. K., Sivakumar, K., & Pushpavalli, V. (2008). Optimization of α-amylase production by actinomycetes strain AE-19 isolated from shrimp pond. Trends in Applied Science Research, 3, 45–52.
Zhu, W., Dongmei, C., Gugue, C., Peng, Q., & Shen, P. (2007). Purification and characterization of a thermostable protease from a newly isolated Geobacillus sp. YMTC 1049. Enzyme and Microbial Technology, 40, 1592–1597.
Kammoun, R., Naili, B., & Bejar, S. (2008). Application of a statistical design to the optimization of parameters and culture medium for α-amylase production by Aspergillus oryzae CBS 819.72 grown on gruel (wheat grinding by-product). Bioresource Technology, 99, 5602–5609.
Gangadharan, D., Sivaramakrishnan, S., Nampoothiri, K. M., Sukumaran, R. K., & Pandey, A. (2008). Response surface methodology for the optimization of α-amylase production by Bacillus amyloliquefaciens. Bioresource Technology, 99, 4597–4602.
Hashemi, M., Razavi, S. H., Shojaosadati, S. A., Mousavi, S. M., Khajeh, K., & Safari, M. (2010). Development of a solid-state fermentation process for production of an alpha amylase with potentially interesting properties. Journal of Bioscience and Bioengineering, 110, 333–337.
Kumar, V., & Satyanarayana, T. (2014). Production of thermo-alkali-stable xylanase by a novel polyextremophilic Bacillus halodurans TSEV1 in cane molasses medium and its applicability in making whole wheat bread. Bioprocess and Biosystems Engineering, 37, 1043–1053.
McMahon, H. E. M., Kelly, C. T., & Fogarty, W. M. (1998). High maltose-producing amylolytic system of a Streptomyces sp. Biotechnology Letters, 21, 23–26.
Goldberg, J. D., & Edwards, C. (1990). Purification and characterization of an extracellular amylase from a thermophilic streptomycete. Journal of Applied Bacteriology, 69, 712–717.
Takasaki, Y., Kitajima, M., Tsuruta, T., Nonoghchi, M., Hayashi, S., & Imada, K. (1991). Maltotriose-producing amylase from Microbacterium imperiale. Agricultural and Biological Chemistry, 55, 687–692.
Mehta, D., & Satyanarayana, T. (2013). Dimerization mediates thermo-adaptation, substrate affinity and transglycosylation in a highly thermostable maltogenic amylase of Geobacillus thermoleovorans. PloS One, 8, e73612.
Nisha, M., & Satyanarayana, T. (2014). Characterization and multiple applications of a highly thermostable and Ca2+-independent amylopullulanase of the extreme thermophile Geobacillus thermoleovorans. Applied Biochemistry and Biotechnology, 174, 2594–2615.
Martin, M. L., & Hoseney, R. C. (1991). A mechanism of bread firming. II. Role of starch hydrolyzing enzymes. Cereal Chemistry, 68, 503–507.
Acknowledgments
The authors gratefully acknowledge financial assistance from the Department of Biotechnology and University Grants Commission, Government of India, New Delhi while carrying out the work presented in the manuscript. The authors thank Dr. Debananda Ningthoujam, Department of Biochemistry, Manipur University, Imphal (India) for providing actinobacterial cultures. The authors wish to thank Mr. Vijay Kumar Gupta (Tushar Nutritive Food Industry, New Delhi) for extending help in assessing the applicability of amylase in bread making.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
ESM 1
(DOC 1885 kb)
Rights and permissions
About this article
Cite this article
Shivlata, L., Satyanarayana, T. Characteristics of Raw Starch-Digesting α-Amylase of Streptomyces badius DB-1 with Transglycosylation Activity and Its Applications. Appl Biochem Biotechnol 181, 1283–1303 (2017). https://doi.org/10.1007/s12010-016-2284-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-016-2284-4