Abstract
The gene encoding a thermostable β-d-xylosidase (GbtXyl43B) from Geobacillus thermoleovorans IT-08 was cloned in pET30a and expressed in Escherichia coli; additionally, characterization and kinetic analysis of GbtXyl43B were carried out. The gene product was purified to apparent homogeneity showing M r of 72 by sodium dodecyl sulfate polyacrylamide gel electrophoresis. The enzyme exhibited an optimum temperature and pH of 60 °C and 6.0, respectively. In terms of stability, GbtXyl43B was stable at 60 °C at pH 6.0 for 1 h as well as at pH 6–8 at 4 °C for 24 h. The enzyme had a catalytic efficiency (k cat/K M) of 0.0048 ± 0.0010 s−1 mM−1 on p-nitrophenyl-β-d-xylopyranoside substrate. Thin layer chromatography product analysis indicated that GbtXyl43B was exoglycosidase cleaving single xylose units from the nonreducing end of xylan. The activity of GbtXyl43B on insoluble xylan was eightfold higher than on soluble xylan. Bioinformatics analysis showed that GbtXyl43B belonging to glycoside hydrolase family 43 contained carbohydrate-binding module (CBM; residues 15 to 149 forming eight antiparallel β-strands) and catalytic module (residues 157 to 604 forming five-bladed β-propeller fold with predicted catalytic residues to be Asp287 and Glu476). CBM of GbtXyl43B dominated by the Phe residues which grip the carbohydrate is proposed as a novel CBM36 subfamily.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Xylan (main component of hemicellulose) is a linear polysaccharide consisting of a β-d-(1→4)-linked xylopyranoside backbone and various functional groups such as (1→2) and/or (1→3)-α-l-arabinofuranoside, (1→2)-α-d-glucoronic acid, and O-2- and/or O-3- acetic groups attached to the backbone. The type and nature of the attached molecules in the xylanopyranoside backbone depends on the source of biomass. Complete degradation of xylan involves complex enzymes that are responsible for hydrolyzing the various linkages in hemicellulose: endo-β-d-1,4-xylanase (EC 3.2.1.8), β-d-xylosidase (EC 3.2.1.37), α-l-arabinofuranosidase (EC 3.2.1.55), α-glucuronidase (EC 3.2.1.139), acetylxylan esterase (EC3.1.1.72), and ferulic acid esterase (EC3.1.1.73) [1–3]. Endo-β-d-1,4-xylanase degrades internal xylosidic linkages of the xylan backbone-producing xylooligosaccharides, followed by β-d-xylosidase that cleaves the nonreducing termini of xylobiose and xylooligosaccharide fragments into xylose. Both enzymes play an important role in the pulp prebleaching process to purify cellulose, the treating of animal feed to increase digestibility, the processing of food to increase clarification, and the conversion of lignocellulosic materials to feedstock and fuel. Hence, it is essential to investigate enzymes for hydrolyzing xylan, in particular enzymes possessing specific properties for industrial applications as well as environmental friendliness.
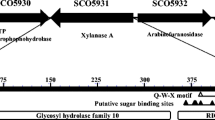
Xylanase can be widely found in bacteria, fungi, marine algae, protozoans, insects, snails, and seeds [4, 5]. The xylanase showing activity in extreme conditions such as extreme heat and cold, acidity or alkalinity, and high salinity are interesting to be investigated for many purposes. Shoham and colleagues have determined the structure of some xylanolytic enzymes from the thermostable and alkaline tolerant bacterium Geobacillus strearothermophilus T-6 which contains endo-β-d-1,4-xylanase [6], β-d-xylosidase [7], α-arabinofuranosidase [8], and α-glucuronidase [9]. We have also identified other thermostable bacterium Geobacillus thermoleovorans IT-08 producing xylanolitic enzymes isolated from the hot springs at Gunung Pancar, Bogor, West Java, Indonesia [10]. Initial investigation on the gene cluster G. thermoleovorans IT-08 by using the method of shotgun cloning shows 8,444 bp nucleotides harboring five different genes, comprising gene A (putative transposase gene), gene B (putative ABC permease), gene C (β-d-xylosidase gene, xyl43B GenBank No. DQ387047), gene D (β-d-xylosidase gene, xyl43A GenBank No. DQ345777), and gene E (α-l-arabinofuranosidase, abfa51 GenBank No. DQ387046; Fig. 1) [11]. Each of the genes B, C, D, and E has an independent ribosomal binding site. G. thermoleovorans IT-08 expresses two different β-d-xylosidases: named GbtXyl43A (encoded by gen C) and GbtXyl43B (encoded by gene D). Surprisingly, GbtXyl43B is capable of hydrolyzing oat spelt xylan, but GbtXyl43A is not. Here, we shall report further biochemical characteristics of GbtXyl43B.
Topology of GbtXyl43B in the xylanase-gene cluster. A, B, C, D, and E indicate genes: putative transposase, putative ABC permease, β-d-xylosidase (xyl43B GenBank No. DQ387047), β-d-xylosidase (xyl43A, GenBank No. DQ345777), and α-l-arabinofuranosidase (abfa51 GenBank No. DQ387046), respectively. RBS refers to a ribosomal binding site
According to the carbohydrate acting enZYme (CAZY) database (http:/www.cazy.org), β-d-xylosidases (E.C. 3.2.1.37) are classified into 10 glycoside hydrolase (GH) families: 1, 3, 30, 39, 43, 51, 52, 54, 116, or 120. They are grouped based on the structure of protein folding, reaction specificity, and reaction mechanism. A typical GH family 43 (GH43) family possesses a three-dimensional structure consisting of a five-bladed β-propeller fold [12], with shared conserved catalytic residues: Asp as a nucleophile, Glu as a donor proton, and another Asp to modulate the pK a of Glu and keep it in the correct orientation relative to the substrate and employ an inverting mechanism through a single nucleophilic displacement [7, 13]. According to the phylogenetic analysis made by Qian et al. [14], GH43 enzyme can be divided into four subfamilies: I and II consisting of proteins encoded by xylose catabolism genes related to the ABC transporter or the permease genes, as well as III and IV which consists of proteins capable of hydrolyzing carbohydrate polymers and also arabinase/arabinosidase. Unlike G. strearothermophilus T-6 expressing three different β-d-xylosidases distributed in three different GH families [15], G. thermoleovorans IT-08 produces GbtXyl43A and GbtXyl43B classified in the same GH43 family.
β-d-xylosidases have a modular structure with catalytic module connected by a linker sequence to the noncatalytic binding module (CBM). The efficiency of β-d-xylosidase to hydrolyze xylan depends on its CBM characteristics, indicated by the dominance of hydrophobic interactions between the sugar rings and aromatic residues on the carbohydrate-binding surface. Most three-dimensional structures of CBM are primarily assembled by β-strand folds, and the orientation of certain aromatic residues of the CBM is essential to ligand specificity [16]. To understand the carbohydrate binding capacity of GbtXyl43B, we expressed the xyl43B of G. thermoleovorans IT-08 in Escherichia coli, characterized the biochemical properties of recombinant GbtXyl43B, and studied the bioinformatics of GbtXyl43B. In the present study, we showed that the CBM of the GbtXyl43B, a fragment appended proximal to the C-terminus with Phe residues used to grip carbohydrate, was a novel CBM36 subfamily.
Materials and Methods
Materials
A vector cloning pTP510 was a pBluescript KS+ (Stratagene, USA) harboring gene clusters (transposase gene, permease gene, xyl43B, xyl43A, and abfa51 from G. thermoleovorans IT-08), and pET30a (Novagen, USA) was used for the expression of xyl43B. Restriction enzymes and T4 DNA polymerase were purchased from Invitrogen (USA) and used according to the manufacturer’s recommendation. E. coli TOP10 (Invitrogen) and E. coli BL21(DE3) (Promega, USA) were used as host cells for the cloning and overexpression, respectively. The substrates used were oat spelt xylan (Fluka, Switzerland), birchwood xylan (Sigma, USA), arabinan (Megazyme, Ireland), rye arabinoxylan (Megazyme), xyloglucan (Megazyme) and 1-ketose (Megazyme), p-nitrophenyl-β-d-xylopyranoside (pNPX; Sigma,), p-nitrophenyl-β-d-xylobioside (pNPXX; Sigma), and p-nitrophenyl-α-l-arabinofuranose (pNPA; Sigma). The reagents were in analytical grade.
Cloning and Sequencing of xyl43B
A full-length xyl43B was obtained from PCR of pTP510 using primers: forward primer 5′-GCGAGCTCATGACTTTACAGACGAATA-3′ (with SacI recognition side underlined) and reverse primer containing an artificial XhoI recognition sequence 5′-CGCTCGAGTTAAGTCAAAATGACATCC-3′ (with the XhoI recognition side underlined). The PCR reaction mixture in a total volume of 20 μL consisted of 10 ng pTP510, 40 pmol for each primer, 1.5 mM MgCl2, 0.3 mM dNTP (dATP, dTTP, dGTP, and dCTP), 1× PCR buffer, and 5.0 U Taq DNA polymerase. PCR was carried out in a thermal cycler (Takara DICE TP600, Japan) in one cycle at 94 °C for 5 min followed by 30 cycles of denaturation (1 min at 95 °C), annealing (45 s at 60.6 °C), and elongation (2 min at 72 °C), with a final elongation at 72 °C for 7 min. SacI and XhoI-generated DNA fragment of PCR product were cloned into pET30a (called pET-xyl43B) and sequenced by dye-end terminator in an automated DNA sequencing machine (US Biochemical Co). The sequence data were analyzed using Genetyx Mac Software ver. 11.0. The full-length xyl43B sequence was the same sequence as our previous result (GenBank No. DQ387047). The similarity of the sequence was analyzed by BLAST [17].
Overexpression and Purification of Recombinant GbtXyl43B
After introducing pET-xyl43B containing six His-Tag codons to an E. coli BL21 (DE3) cell by means of the electroporation method, for which the transformants were screened in Luria–Bertani (LB) agar plates containing the antibiotic kanamycin. To produce GbtXyl43B, the bacterial culture in LB medium containing 50 μg mL−1 kanamycin were incubated at 37 °C until optical density at 600 reach 0.6, and then induced by the addition of isopropyl-thio-β-d-thiogalactopyranoside to a final concentration of 1 mM. After 3–5 h induction, the bacterial cells were centrifuged at 7,500×g for 15 min at 4 °C, resuspended in 10 mM HEPES buffer, pH 7.5 containing 10 mM imidazole and 50 mM NaCl, and disrupted by sonication. The crude enzyme was separated from debris cells by centrifugation at 15,000×g for 30 min at 4 °C. The crude enzyme was purified on a HiTrap Chelating HP column (GE Healthcare Life Sciences) with a stepwise gradient of imidazole (10, 30, 60, 100, 250, and 500 mM) in 50 mM HEPES buffer, pH 7.4, and a flow rate of 1 mL min−1. The fractions were collected and checked by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) [18]. The relative molecular mass of pET-Xyl43B was determined by SDS-PAGE using low-weight protein markers (GE Healthcare Life Sciences) as a standard. Protein concentration was determined by the Bradford method [19] using bovine serum albumin as a standard.
Enzyme Activity Assays
Recombinant GbtXyl43B activity was determined using chromogenic substrates: pNPX or pNPA. Reaction mixtures of 1 mL containing 1 mM pNPX (or 1 mM pNPA) in 50 mM phosphate buffer, pH 6.0, and 57.1 μg mL−1 enzyme were incubated at 60 °C for 30 min, and the reaction was stopped by the addition of 0.1 mL 1 M sodium carbonate. The absorbance of the p-nitrophenol (pNP) product was measured by a UV–vis Spectrophotometer (Shimadzu UV–vis 1800) at 405 nm. The molar absorptivity of the pNP product (pH 6.0) was 4,950 M−1 cm−1. All experiments were performed in duplicates. A unit recombinant GbtXyl43B activity (International Unit; IU) was defined as the amount of enzyme required to produce 1 μmol pNP per 1 min at reaction conditions. Specific activity was given as IU per milligram of proteins.
GbtXyl43B activities on the substrates oat spelt xylan, birchwood xylan, arabinan, rye arabinoxylan, and xyloglucan were also evaluated. Total reducing sugar released was determined using the 3,5-dinitrosalicylic acid (DNS) method [20]. A reaction mixture of 1 mL containing 57.1 μg mL−1 enzyme and 75 μmol DNS was heated at 100 °C for 10 min, and then 850 μL H2O was added. The absorbance of the reaction mixture was measured at 500 nm. Xylose was used as the standard reducing sugar. Enzyme assays were performed in duplicates. One unit of enzyme activity (IU) is defined as the amount of enzyme needed to release 1 μmol reducing sugars per min under assay conditions. Specific activity of enzyme was expressed in units per milligram.
Determination of Enzyme Activity and Stability on Various Temperature and pH
The optimum working temperature was determined by measuring the activity of recombinant GbtXyl43B on substrate pNPX at a temperature range between 30 and 80 °C. Meanwhile, to determine thermal stability, recombinant GbtXyl43B in 50 mM phosphate buffer, pH 6.0 was first incubated at various temperatures (20–80 °C) for 1 h and cooled at room temperature prior to assays under standard condition. The optimum working pH on substrate pNPX was determined at a range of pH 4–10 using Britton and Robinson’s universal buffer at 60 °C for 30 min. The buffer contained 50 mM phosphoric acid and 50 mM boric acid, and adjusted with 1 M NaOH or 1 M HCl to reach the appropriate pH. To determine pH stability, recombinant GbtXyl43B in a various pH range between 4 and 9 was incubated at 4 °C for 24 h prior to enzymatic assay under standard condition.
Kinetic Study
To determine the K M and k cat of recombinant GbtXyl43B, assays were performed using pNPX with a concentration range between 9 and 27 mM. The absorbance of the pNP product was measured using a UV–vis Spectrophotometer (Biochrom BO-7000-21) at wavelength 405 nm equipped with a water-circulating device to control the temperature of the reaction at 60 °C until the reaction was completed. The molar absorpitivity of the pNP product (pH 6.0) was 5,850 M−1 cm−1. The values of K M and k cat were determined from the Lineweaver–Burk plot analysis of curve of the initial rates of reaction versus substrate concentrations.
TLC Analysis
Thin layer chromatography (TLC) on 7 cm silica gel plate (Merck, Germany) was employed to analyze the products of pNPX, pNPXX, and 1-ketose hydrolysis. Reaction mixtures consisting of 40 mM of pNPX (or 40 mM pNPXX or 10 mM 1-ketose), 0.3 mg mL−1 recombinant GbtXyl43B in 50 mM phosphate buffer, pH 6.0 were incubated at 60 °C for each interval time of 10, 20, 30, 40, and 50 min. The reactions were stopped by heating in boiled water. Ten microliter hydrolysate was spotted on a TLC plate and developed with the eluent of nitroethane/ethanol/water (1:3:1). Spots were visualized by charring with an aniline–diphenylamine reagent.
Xylan Adsorbability
The affinity of recombinant GbtXyl43B towards oat spelt xylan was studied by incubating 28 μg enzyme with 100–300 mg of oat spelt xylan at 4 °C for 1 h. After centrifugation, the amount of free enzyme in supernatant was determined by the Bradford method [19]. The bound protein was the difference between the amounts of original protein and free protein in the supernatant after binding.
Bioinformatics
Based on protein homology, a three-dimensional structure model of GbtXyl43B was build using the automatic mode modeling program SWISS MODEL [21] fold recognition server (http://www.swissmodel.com) using the β-d-xylosidase structure of Clostridium acetobutylicum (PDB No. 3KIU) as the template for the catalytic module (CM) model, and the CBM36 structure of xylanase 43A from Paenibacillus polymyxa (PDB No. 1UX7) as the template for the CBM model. Models of GbtXyl43B structure were drawn with the program PyMOL (Molecular Graphics System ver. 1.5.0.4). To analyze CBM of GbtXyl43B, a phylogenetic tree was constructed using the program MEGA ver. 5 [22] using corresponding CBM data retrieved from the CAZY database (http://www.cazy.org/CBM).
Results
Topology of GbtXyl43B
A full-length xyl43B of G. thermoleovorans IT-08 encoding GbtXyl43B consisted of 1,815 bp (GenBank No. DQ387047). The GbtXyl43B polypeptide deduced was 604 amino acid residues in length. A BLASTP search result showed that GbtXyl43B belonged to the GH43 family, containing a CBM module (134 aa) with a β-sandwich fold situated on residues 15–149 capable of binding xylan, and a CM module (447 aa) with a five-bladed β-propeller fold located at 157–604 possessing β-d-xylosidase activity (Fig. 1).
To characterize recombinant GbtXyl43B, xyl43B was PCR amplified, cloned into pET30a vector, and overexpressed in E. coli BL21 (DE3). The pET-xyl43B expressed intracellular recombinant GbtXyl43B and no inclusion body was observed. Recombinant GbtXyl43B consisted of 657 amino acid residues containing linker residues and His-Tag fusion. Table 1 showed that purified recombinant GbtXyl43B was obtained from one-step purification using HiTrap Chelating HP column with a yield of 28.2 % and purification fold of 34. Starting with 69.2 mg crude protein, the total amount of purified recombinant GbtXyl43B obtained was 0.57 mg. SDS-PAGE analysis showed that recombinant GbtXyl43B had a molecular mass of 72 kDa, which is close to the expected 74.5 kDa (Fig. 2). To confirm β-d-xylosidase activity, the SDS-PAGE zymogram of the recombinant GbtXyl43B showed a positive staining with 4-methylumbelliferyl-β-d-xyloside (data not shown).
Properties of Recombinant GbtXyl43B
To understand substrate specificity of the enzyme, the activity of recombinant GbtXyl43B was examined on various types of substrates, both synthetic (pNPX, pNPXX, and pNPA) and natural (xylan, birchwood xylan, arabinan, rye arabinoxylan, xyloglucan, and 1-ketose). Table 2 shows that recombinant GbtXyl43B displayed the highest activity on pNPX indicating β-d-xylosidase activity. To obtain further information on the hydrolysis on substrates pNPX and pNPXX, the recombinant GbtXyl43B released xylose but not xylobiose (Fig. 3). The recombinant GbtXyl43B had no activity on pNPA, which indicated that the GbtXyl43B was not arabinofuranosidase. The GbtXyl43B also had no activity on 1-ketose, indicating that the GbtXyl43B was not fructan β-(2→6) fructosidase. These are the same characteristics found in the wild type of GbtXyl43B [10]. The activities of recombinant GbtXyl43B on arabinan, rye arabinoxylan, and xyloglucan were not observed, but a little activity was observed on oat spelt xylan and birchwood xylan. This indicated that recombinant GbtXyl43B was not xylanase.
Time course of pNPX and pNPXX hydrolysis by recombinant GbtXyl43B. X 1 and X 2 are xylose and xylobiose, respectively. 1–5 pNPX hydrolysis products at 60 °C for 10, 20, 30, 40, and 50 min, respectively; 6 pNPX; 7 hydrolysis of pNPX by inactive GbtXyl43B; 8–11 pNPXX hydrolysis products at 60 °C for 20, 30, 40, and 50 min; 12 pNPXX; and 13 hydrolysis of pNPXX by inactive GbtXyl43B
The biochemical properties of recombinant GbtXyl43B such as thermal activity and stability were examined. Recombinant GbtXyl43B retained more than 50 % activity at temperature between 40 and 65 °C (Fig. 4a). The optimum activity of recombinant GbtXyl43B at 60 °C was 25.7 IU mg−1. Recombinant GbtXyl43B showed thermal stability upon heating for 1 h at up to 70 °C, and it retained more than 70 % activity. The pH characteristics of recombinant GbtXyl43B were also examined. The enzyme exhibited an optimum pH of 6.0 (24.8 IU mg−1) and remained stable under a pH environment of 6–8 (Fig. 4b). At least 50 % of recombinant GbtXyl43B activity was detectable at pH of 5–8. The optimum working temperature and pH of GbtXyl43B is similar to its counterpart GbtXyl43A.
To understand substrate affinity and catalytic efficiency, kinetic parameters of recombinant GbtXyl43B on pNPX were determined by the steady-state kinetics of Michaelis–Menten analysis. The initial rates of the reaction were determined from the kinetic curves of each reaction which contained different pNPX concentrations. The values of K M and V max of recombinant GbtXyl43B were 0.066 ± 0.022 mM and 1.9 × 10−4 ± 2.7 × 10−5 mM min−1, respectively. Since turnover number (k cat) of recombinant GbtXyl43B was 3.2 × 10−4 ± 4.76 × 10−5 s−1, the catalytic efficiency (k cat/K M) of the enzyme was 0.0048 ± 0.0010 s−1 mM−1.
To determine the role of CBM, adsorbability of recombinant GbtXyl43B at various amounts of oat spelt xylan was examined. The adsorbability of recombinant GbtXyl43B (in a range of 0–40 μg) increased linearly with the increasing of amount of substrate oat spelt xylan (in a range of 0–300 μg). The gradient of the adsorption curve indicated microgram recombinant GbtXyl43B bound per microgram oat spelt xylan. The present study showed that the adsorbability of recombinant GbtXyl43B to oat spelt xylan was 13 % (w/w).
Bioinformatics
Since no closed protein homology to overall GbtXyl43B was found at PDB (Protein Data Bank), three-dimensional models of CM and CBM of GbtXyl43B were constructed separately. To identify CM, three-dimensional model of GbtXyl43B (residues 157–604) was constructed using SWISS-MODEL with the β-xylosidase structure of C. acetobutylicum (PDB No. 3KIU) as a template. A structural model of CM of GbtXyl43B had a typical arrangement for the GH43 family containing five-bladed β-propeller fold motif (Fig. 5a). The catalytic domain was identified at the cleft on the surface close to the center of the five-bladed β-propeller fold, with the catalytic residues predicted to be Asp287 as a general base, Glu476 as a general acid, and Asp412 as a pK a modulator as well as for orientating of the general acid residue. To break the catalytic residues, site-directed mutagenesis of catalytic residues Asp287Gly, Glu476Gly, and Asp412Gly produced inactive GbtXyl43B, but the enzyme still showed substrate binding capacity (Ratnadewi, unpublished data).
To identify CBM, a three-dimensional model of GbtXyl43B (residues 15–149) was constructed using the CBM36 of xylanase 43A from P. polymyxa (PDB No. 1UX7) as a template. A structural model of CBM of GbtXyl43B had a β-sandwich of eight antiparallel β-strands with residues of Phe55, Tyr75, Phe84, and Phe108 which were predicted to be important for carbohydrate binding (Fig. 5b).
To understand the family of proteins, amino acid sequence CBM of GbtXyl43B was aligned with 103 CBMs retrieved from CAZY database and a phlyogenetic tree was constructed. Figure 6 shows a phylogenetic tree of the CBM family which was classified into six groups (group I: CBM3, 4, 20, 21, 27, 30, 33, 34, and 62; group II: CBM1, 2, 35, 40, 59, and 60; group III: CBM9, 16, 29, 31, 42, 43, 63, and 66; group IV: CBM10, 11, 12, 13, 18, 37, 49, and 51; group V: CBM5, 6, 26, 36, 39, 46, 47, 48, and 65; and group VI: CBM14, 15, 17, 19, 22, 44, 57, and 61) and an unclassified group (CBM25 and 32). The CBM of GbtXyl43B belongs to family 36 of group V, with the hydrophobic residues of Phe55, Tyr75, Phe84, and Phe108 identified as playing an important role in binding carbohydrate. This was different from CBM36 of xylanase 43A from P. polymyxa (PDB No. 1UX7) which used residues of Tyr23 and Tyr40 [23].
Discussion
The main problem of the enzymatic breakdown of plant cell walls is the opening of an intricate mixture of polysaccharides (the major components of which are cellulose, hemicelluloses, and lignin), because they are highly ordered and insoluble. β-d-xylosidase is part of an array of hemicellullases responsible for xylan hydrolysis. Screening for the powerful xylanase is essential for the biorefinery industry; particularly strains producing thermo-tolerant enzymes and acting on insoluble xylan. G. thermoleovorans IT-08 possessed a gene cluster of thermostable xylanolytic enzymes produces β-d-xylosidases acting on either soluble or insoluble xylan. To elucidate the xylanolytic activity acting on insoluble xylan, GbtXyl43B properties were explored.
Previously, GbtXyl43B is reported as a putative exoxylanase [24], and the only nucleotide sequence of the gene to have been deposited at GenBank No. DQ387047. A BLASTN search result of the xyl43B, so far, indicated no counterpart nucleotide sequence reported. However, a BLASTX search result showed that GbtXyl43B displayed the highest protein homologies to (identity percentages and protein access number in parentheses) uncharacterized glycosyl hydrolase (84 %, YP 002314901), α-N-arabinofuranosidase (71 %, YP 007457868), α-N-arabinofuranosidase (70 %, ZP 09204042), α-N-arabinofuranosidase (64 %, ZP 08191348), and α-N-arabinofuranosidase (63 %, AFC 38437). Further nucleotide sequence analysis of the xyl43B upstream showed that no signal peptide nucleotide sequence was found in the gene. In fact, the xyl43B of G. thermoleovorans IT-08 expressed wild-type GbtXyl43B extracellularly. The plausible reason for this is the role of putative transposase and putative ABC permease in the expression system of G. thermoleovorans IT-08 (Fig. 1). Some reports elucidate the ABC transporter was essential protein to transport every type molecules across cellular membrane [25–27], and Qian et al. [14] used the existence of ABC transporter or the permease genes to classify GH43 subfamilies.
The GH43 family of hemicellulolytic enzymes in general is promising candidates for the critical β-d-xylosidase activity, as they do not exhibit transglycosylation at high substrate concentration [28]. The GbtXyl43B protein sequence deduced consisted of 604 amino acid residues (but the recombinant GbtXyl43B had additional 53 amino acid residues at the N terminus owing to the expression system) containing a CBM (residues 15–149) connected through eight amino acid residues as a linker to a CM (residues 157–604). GbtXyl43B conserved catalytic residues of the GH43 family; there were predicted to beAsp287 as a general base, Glu476 as a general acid, and Asp412 as a pK a modulator and for orientation of the general acid residue (Fig. 5a). GbtXyl43B sequence shared 19 % maximum identity with XynB3 sequence from G. strearothermophilus T-6 (Genbank No. AAT98625) [15] with a significant different in the CBM but not in the CM. The optimum working pH and temperature of GbtXyl43B were 6.0 and 60 °C, respectively. GbtXyl43B was thermally stable up to 60 °C and pH stable at 6–8. These biochemical characteristics of GbtXyl43B were not much different from those of its counterpart of thermophilic XynB3 [15] as well as GbtXyl43A expressed from the synthetic gene derived from GenBank No. DQ345777 [29].
Davies et al. [30] report that most glycosidases are highly specific regarding the identity of the substrate glycon occupying the −1 subsite at the active site. Substrate specificity analysis of GbtXyl43B confirmed that the enzyme was not active toward pNPA and 1-ketose (Table 2). Some β-d-xylosidases, however, show bifunctional activities on both pNPX and pNPA, e.g., XylB from Butyrivibrio fibrisolvens [31] and XylA from Clostridium stercorarium [32]. TLC analysis showed that GbtXyl43B was active towards pNPX but not towards pNPXX (Fig. 3) indicating GbtXyl43B was truly β-d-xylosidases acting at the nonreducing sugar releasing only a single xylose molecule. This is different from XynB3 acting on both substrates pNPX and pNPXX [15]. Catalytic efficiency of GbtXyl43B (0.0048 s−1 mM−1) and XynB3 (3.3 s−1 mM−1) [15] on pNPX are relatively low. This is consistent with the commonly known fact that the catalytic efficiency of xylanolytic enzymes is rather low among hydrolases.
Enzymatic degradation of insoluble xylan is one of the most important and challenging reactions, because the inefficient enzymatic process which is attributed to either the active site of the enzymes not accessing their substrate or the enzymes not gripping the carbohydrate. So, many xylanases have evolved to append noncatalytic carbohydrate-binding module (CBMs) to facilitate grasping the enzymes to the substrates. CBM is generally localized at the C or N terminus of the protein, and its binding ability is dominated by a few aromatic amino acid residues to provide a hydrophobic surface [33]. The activity of GbtXyl43B on insoluble oat spelt xylan was eightfold higher than on soluble birchwood xylan (Table 2). This is confirmed by the adsorbability of GbtXyl43B on insoluble oat spelt xylan to reach 13 % (w/w), suggesting GbtXyl43B had a CBM.
The CBM (residues 15–149) of GbtXyl43B shared 19 % maximum identity with the CBM36 of xylanase 43A from Paenibacillus polymyxa (PDB No. 1UX7) [23]. Phylogenetic analysis of 50 CBM families has shown that CBM of GbtXyl43B belong to CBM36 family (Fig. 6). Typical folds in CBM36 family is β-sandwich platform to facilitate binding of xylan in a cleft with the aromatic residues interact with the free single polysaccharide chain [16, 23, 33]. The structure model of CBM of GbtXyl43B (Fig. 5b) contained a β-sandwich of eight antiparallel β-strands (resembling a type B CBM) that was important to grip the substrate xylan. Based on the alignment of the CBM of GbtXyl43B with the related CBM36 family, the residues forming a carbohydrate binding pocket was located between Gly41 and Gly126, with the conserved amino acid residues to be Leu54, Ala56, Asn83, Val89, Gly95, and Gly103, as well as the conservative substitutions to be Phe55 and Phe108. Compare with the CBM of xylanase 43A from P. polymyxa (PDB No. 1UX7) [23], the CBM of GbtXyl43B had more aromatic residues for gripping carbohydrate. There were predicted to be Phe55, Tyr75, Phe84, and Phe108 buried inside the β-sandwich structure. Loops of residues 76–83 and 109–105 may open or close for gripping xylan. To fasten the flat surface of the xylan chain, two pairs of residues Phe55 and Tyr75 may clamp a sugar chain of one side, along with residues Phe84 and Phe108 clamping sugars of the other side (Fig. 5). This structural reason may explain why the GbtXyl43B shows higher activity on insoluble xylan than soluble xylan. The uniqueness of CBM of GbtXyl43B was characterized by the dominance of Phe residues in carbohydrate binding (rather than Tyr residues commonly found in CBM36). Thus, the CBM of GbtXyl43B is proposed as a novel CBM36 subfamily.
Conclusion
We have cloned, expressed, and characterized a thermostable β-d-xylosidase from G. thermoleovorans IT-08. The results suggest that GbtXyl43B may form a new cluster in the CBM36 subfamily, characterized by clusters of Phe residues responsible for gripping, rather than binding, carbohydrate. Thus, further studies on designing chimeric xylanase (or other chimeric carbohydrase) with the use of this CBM to the insoluble carbohydrate target are worthy to be carried out.
References
Biely, P. (2003). Xylanolitic enzymes. In J. R. Whitacker, A. G. J. Voragen, & D. W. S. Wong (Eds.), Handbooks of food enzymology (pp. 879–915). New York: Dekker.
Saha, B. C. (2003). Hemicellulose bioconversion. Journal of Industrial Microbiology and Biotechnology, 30, 279–291.
Shallom, D., & Shoham, Y. (2003). Microbial hemicellulases. Current Opinion in Microbiology, 6, 219–228.
Polizeli, M. L. T. M., Rizzatti, A. C. S., Monti, R., Terenzi, H. F., Jorge, J. A., & Amorim, D. S. (2005). Xylanase from fungi properties and industrial applications. Journal of Applied Microbiology and Biotechnology, 67, 577–591.
Sunna, A., & Antranikian, G. (1997). Xylanolytic enzymes from fungi and bacteria. Critical Reviews in Biotechnology, 17, 39–67.
Teplitsky, A., Mechaly, A., Stojanoff, V., Sainz, G., Golan, G., Feinberg, H., et al. (2004). Structure determination of the extracellular xylanase from Geobacillus stearothermophilus by selenomethionyl MAD phasing. Acta Crystallographica, D60, 836–848.
Brüx, C., Ben-David, A., Shallom-Shezifi, D., Leon, M., Niefind, K., Shoham, G., et al. (2006). The structure of an inverting GH43 β-xylosidase from Geobacillus stearothermophilus with its substrate reveals the role of the three catalytic residues. Journal of Molecular Biology, 359, 97–109.
Hövel, K., Shallom, D., Niefind, K., Belakhov, V., Shoham, G., Baasov, T., et al. (2003). Crystal structure and snapshots along the reaction pathway of a family 51 α-l-arabinofuranosidase. EMBO Journal, 22, 4922–4932.
Golan, G., Shallom, D., Teplitsky, A., Zaide, G., Shulami, S., Baasov, T., et al. (2004). Crystal structures of Geobacillus stearothermophilus α-glucuronidase complexed with its substrate and products: mechanistic implications. Journal of Biological Chemistry, 279, 3014–3024.
Tan, I. (1999). Characterization of the thermophilic bacterium producing xylanolytic enzyme from hot spring Gunung Pancar Bogor (in Bahasa Indonesia), Master thesis, Institut Pertanian Bogor, Indonesia
Puspaningsih, N. N. T., Suwanto, A., Suhartono, M. T., Achmadi, S., Yogiara, S., & Kimura, T. (2008). Cloning, sequencing and characterization of the xylan degrading enzymes from Geobacillus thermoleovorans IT-08. Journal of Basic Science, 9, 177–187.
Nurizzo, D., Turkenburg, J. P., Charnock, S. J., Roberts, S. M., Dodson, E. J., McKie, V. A., et al. (2002). Cellvibriojaponicus α-l-arabinanase 43A has a novel five-blade β-propeller fold. Nature Structural Biology, 9, 665–668.
Sinnott, M. L. (1990). Catalytic mechanism of enzymic glycosyl transfer. Chemistry Review, 90, 1171–1202.
Qian, Y., Yamano, L. P., Preston, J. F., Aldrich, H. C., & Ingram, L. O. (2003). Cloning, characterization, and functional expression of the Klebsiella oxytoca xylodextrin utilization operon (xynTB) in E. coli. Applied and Environmental Microbiology, 69, 5957–5967.
Shallom, D., Leon, M., Bravman, T., Ben-David, A., Zaide, G., Belakhov, V., et al. (2005). Biochemical characterization and identification of the catalytic residues of a family 43 β-d-xylosidase from Geobacillus stearothermophilus T-6. Biochemistry, 44, 387–390.
Boraston, A., Bolam, D. N., Gilbert, H. J., & Davies, G. J. (2004). Carbohydrate-binding module: fine-tuning polysaccharide recognition. Biochemical Journal, 382, 769–781.
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., & Lipman, D. J. (1990). Basic Local Alignment Search Tool. Journal of Molecular Biology, 215, 403–410.
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685.
Bradford, M. M. (1976). A rapid and sensitive method for the quantisation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry, 72, 248–254.
Miller, G. L. (1959). Use of dinitrosalicylic acid reagent for determination of reducing sugar. Analytical Chemistry, 31, 426–428.
Arnold, K., Bordoli, L., Kopp, J., & Schwede, T. (2006). The SWISS-MODEL workspace: a web-based environment for protein structure homology modeling. Bioinformatics, 22, 195–201.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Neiand, M., & Kumar, S. (2011). MEGA 5: molecular evolution genetics analysis using maximum likelihood evolutionary distance and maximum parsimony method. Molecular Biology and Evolution, 28, 2731–2739.
Jamal-Talabani, S., Boraston, A. B., Turkenburg, J. P., Tarbouriech, N., Ducros, V. M.-A., & Davies, G. J. (2004). Ab initio structure determination of CBM36: a new family of calcium-dependent carbohydrate binding modules. Structure, 12, 1177–1187.
Puspaningsih, N. N. T. (2004). Characterizing of xylanolytic enzymes, and cloning of genes encoding xylosidases from Geobacillus thermoleovorans IT-08 (in Bahasa Indonesia), Ph.D Thesis, Institut Pertanian Bogor. Indonesia
Higgins, F. C. (2001). ABC transporters: physiology, structure and mechanism—an overview. Research in Microbiology, 152, 205–210.
Shulami, S., Zaide, G., Zolotnitsky, G., Langut, Y., Feld, G., Sonenshein, A. L., et al. (2007). A two-component system regulates the expression of an ABC transporter for xylo-oligoccharides in Geobacillus stearothermophilus. Applied and Environmental Microbiology, 73, 874–884.
Low, K. O., Mahadi, N. M., Rahim, R. A., Rabu, A., Bakar, F. D. A., Murad, A. M. A., et al. (2011). An effective extracellular protein secretion by an ABC transporter system in Escherichia coli: statistical modeling and optimization of cyclodextrin glucanotransferase secretory production. Journal of Industrial Microbiology and Biotechnology, 38, 1587–1597.
Jordan, D. B., & Li, X. L. (2007). Variation in relative substrate specificity of bifunctional β-d-xylosidase/α-l-arabinofuranosidase by single mutations: role substrate distortion and recognition. Biochimica et Biophysica Acta, 1774, 1192–1198.
Wagshcal, K., Heng, C., Lee, C. C., Robertson, G. H., Orts, W. J., & Wong, D. W. S. (2009). Purification and characterization of a glycoside hydrolase family 43 β-xylosidase from Geobacillus thermoleovorans IT-08. Applied Biochemistry and Biotechnology, 155, 1–10.
Davis, G. J., Wilson, K. S., & Hendrissat, B. (1997). Nomenclature for sugar binding subsite in glycosyl hydrolase. Biochemical Journal, 321, 557–559.
Utt, E. A., Eddy, C. K., Keshav, K. F., & Ingram, L. O. (1991). Sequencing and expression of the Butyrivibrio fibrisolvens xylB gene encoding a novel bifunctional protein with β-d-xylosidase and α-l-arabinofuranosidase activities. Applied and Environmental Microbiology, 57, 1227–1234.
Sakka, K., Yoshikawa, K., Kojima, Y., Karita, S., Ohmiya, K., & Shimada, K. (1993). Nucleotide sequence of Clostridium stercorarium xylA gene encoding a bifunctional protein with β-d-xylosidase and α-l-arabinofuranosidase activities and properties of the translated product. Bioscience, Biotechnology, and Biochemistry, 57, 268–272.
Guillén, D., Sánchez, S., & Rodríguez-Sanoja, R. (2010). Carbohydrate-binding domains: multiplicityof biological roles. Applied Microbiology and Biotechnology, 85, 1241–1249.
Acknowledgments
This research was partially supported from the Directorate General of Higher Education, Ministry of Education and Culture, the Republic of Indonesia through Hibah Tim Pascasarjana Program to NNTP (2011–2012), Sandwich Course Program 2011 to AAID, as well as Hibah Doktor Program to AAID. We thank Mr. Kimiya Mizutani for his assistance in the preparation of enzyme purification, Mr. Didik Huswo Utomo for molecular art work as well as Mr. Tubagus Andhika Nugraha for critical reading of this manuscript.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Ratnadewi, A.A.I., Fanani, M., Kurniasih, S.D. et al. β-d-Xylosidase from Geobacillus thermoleovorans IT-08: Biochemical Characterization and Bioinformatics of the Enzyme. Appl Biochem Biotechnol 170, 1950–1964 (2013). https://doi.org/10.1007/s12010-013-0329-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-013-0329-5