Abstract
Streptomyces coelicolor A3(2) sco5931 gene was predicted to encode a putative xylanase A, a 477 amino acid protein belonging to glycoside hydrolase family 10. The entire sco5931 coding region was cloned and overexpressed in Streptomyces lividans TK24. Mature SCO5931 protein comprising 436 amino acids (47 kDa) was purified by single-step gel filtration chromatography from culture broth after ammonium sulfate precipitation, with 25.8-fold purification and yield of 30.6 %. The purified protein displayed a pronounced activity toward beechwood xylan as a substrate, but no activity was detected toward carboxymethylcellulose, Avicel, galactan, barley β-glucan, and xyloglucan, demonstrating that SCO5931 is a substrate-specific xylanase. Optimal xylanase activity was observed at 60 °C and pH 6.0. The addition of metal ions or EDTA did not affect the xylanase activity, while 4 mM MnCl2 severely inhibited the enzyme, reducing its activity by 87 %. Kinetic parameters of SCO5931 toward beechwood xylan were determined (K m = 0.24 mg/mL, V max = 6.86 μM/min). Thin layer chromatography and mass spectrometry analyses of the beechwood xylan SCO5931 hydrolysis products were conducted. Product masses corresponded to sodium adducts of xylobiose (m/z 305.24) and xylopentaose (m/z 701.59), indicating that SCO5931 specifically cleaves the β-1,4 linkage of xylan to yield xylobiose and xylopentaose.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Xylan is a component of highly complex hemicellulose and comprised by xylose units linked by β-1,4-glycosidic bond. It is the second most abundant natural biopolymer, after cellulose, accounting for 20–40 % of total plant biomass. Therefore, efficient xylan degradation has become important for the utilization of lignocellulosic materials as a sustainable biomass [1, 2]. Xylan hydrolysis has been adopted in many industrial fields, for paper, pulp, and textile production. Recently, xylooligosaccharides and d-xylose obtained by enzymatic xylan hydrolysis have come to the forefront of investigations due to their enormous biotechnical potential as functional food additives and bioenergy sources [3].
Xylanase degrades β-1,4-glycosidic bonds of the xylan backbone to yield xylooligosaccharides and d-xylose. Xylanases from various eukaryotes and prokaryotes have been isolated and characterized, but bacterial hosts have many advantages in commercial applications of xylanase owing to their ability to produce secreted extracellular enzymes, fast growth, and availability of various recently developed industrial technologies [4–6].
The genus Streptomyces covers Gram-positive soil bacteria that produce various antibiotics and grow as a spore-forming mycelium. It has been highlighted for producing valuable secondary metabolites as well as many industrially important hydrolytic enzymes including xylanases [7–9]. Streptomyces coelicolor A3(2), a best-studied model species in the genus Streptomyces, also produces many types of extracellular enzymes that hydrolyze various macromolecules, such as agar [10], xyloglucan [11], and cellulose [12]. Five ORFs were annotated as putative xylanases in the S. coelicolor A3(2) genomic sequence [13], based on comparison with annotated enzymes from other bacteria. However, their enzymatic properties have not been investigated. This study comprises a first report on the expression, purification, and characterization of a glycoside hydrolase (GH) family 10 xylanase A (SCO5931, XlnA) from S. coelicolor A3(2).
Materials and Methods
Bacterial Strains and Plasmids
Escherichia coli DH5α served as a host, and T&A cloning vector system (RBC, USA) was used in subcloning experiments. S. coelicolor A3(2) and S. lividans TK24 were acquired from the John Innes Institute, UK [14]. S. lividans TK24 and Streptomyces–E. coli shuttle vector pUWL201PW [15] were used as the host–vector system for overexpressing sco5931.
Media and Culture Conditions
E. coli was maintained on LB agar and routinely cultured with agitation in LB broth at 37 °C [16]. Streptomyces strains were grown in R2YE liquid broth at 28 °C for the preparation of protoplasts and isolation of genomic or plasmid DNA [14]. The media were supplemented with either ampicillin (50 μg/mL) or thiostrepton (25 μg/mL), as required.
Substrates and Chemicals
Carboxymethylcellulose (CMC, medium viscosity), Avicel (microcrystalline powder, 20 μm), Azurine-cross-linked (AZCL) xylan, and other fine chemicals were purchased from Sigma Chemical Co. (St. Louis, MO, USA). Barley β-glucan, potato galactan, beechwood xylan, and xylans, including xylobiose (X2) and xylotetraose (X4), were purchased from Megazyme International Ltd (Ireland). Restriction endonucleases, T4 DNA ligase, and Taq DNA polymerase were purchased from Takara Shuzo Inc., Japan. PCR primers were obtained from DyneBio Inc., Korea.
DNA Manipulations
DNA preparation and cloning were performed in E. coli using methods described by Green and Sambrook [16]. DNA samples were digested with restriction endonucleases and ligated using T4 DNA ligase, according to the supplier’s recommendations. DNA digests were analyzed by horizontal agarose gel electrophoresis in 0.04 M Tris acetate (pH 8.4)–0.001 M EDTA (TAE) buffer.
Construction of Expression Vector
The 1442-bp NdeI/HindIII fragment containing the entire sco5931 coding region (Fig. 1) was amplified by PCR using the following primers: forward, 5′-cat ATGGGCTCCTACGCCCTTCCCAGAT CAG-3′ (NdeI site is underlined and the non-coding nucleotides are written in lowercase); reverse 5′-aagctTCAGGTGCGGGTCCAGCGTTGGTTGCTGC-3′ (HindIII site is underlined and the non-coding nucleotides are written in lowercase). These primers were designed based on the nucleotide sequence deposited in the Streptomyces Genome Project webpage (http://www.sanger.ac.uk/resources/downloads/bacteria/streptomyces-coelicolor.html). PCR conditions were as previously described [10], and the digested products were cloned into T&A cloning vector: sco5931 DNA fragment double-digested with NdeI and HindIII was ligated with pUWL201PW digested with the same restriction enzymes, yielding pUWL201-5931. Recombinant plasmids were purified from E. coli and used for protoplast transformation of Streptomyces cells.
Genomic neighborhood of sco5931 and conserved domains of SCO5931 protein. Gene organization, including the ORF of the putative xylanase SCO5931, is shown with functions annotated based on the genomic sequence of S. coelicolor. SCO5931 protein (477 amino acids) contains an N-terminal glycosyl hydrolase family 10 domain (amino acids 45–341) and C-terminal Ricin-type beta-trefoil (carbohydrate-binding domain, amino acids 354–475). Conserved amino acids in putative sugar binding sites (black triangle) and Q-X-W motif (white triangle) are depicted
Bacterial Transformation
Transformations of E. coli cells were performed by the CaCl2 method [16]. Streptomyces protoplasts were prepared as described earlier [14]. The resultant protoplasts were transformed using the PEG-mediated transformation method. The transformants were selected by overlaying the transformant-containing plates with 1 mL distilled water containing thiostrepton (0.625 μg/mL).
Protein Analysis
Protein concentrations were measured with Bradford method using bovine serum albumin as standard [17]. Proteins were resolved on sodium dodecyl sulfate (0.1 %)–polyacrylamide (10 %) gel electrophoresis (SDS-PAGE), as described by Laemmli [18].
Enzyme Purification
All extracellular proteins released into the culture broth (250 mL) by S. lividans TK24/pWUL201-5931 were precipitated by the addition of solid ammonium sulfate to 75 % saturation. The precipitated proteins were recovered by centrifugation at 10,000g at 4 °C for 20 min, suspended in and dialyzed against 20 mM Tris–Cl buffer (pH 7.5). The dialyzed product (7 mL) was concentrated, 20 times, on Amicon Ultra-50 K centrifugal filter units (50 kDa cutoff). Following this, 100-μL concentrates were applied onto Superdex™ 200 HR 10/30 gel filtration column previously equilibrated with 50 mM sodium phosphate buffer (pH 7.0) containing 0.15 M NaCl. Proteins were eluted with the same buffer, at a flow rate of 0.5 mL/min. Thirty fractions (1 mL/fraction) were collected and assessed by SDS-PAGE.
SCO5931 Catalyzed Reaction and Substrate Specificity Determination
Enzyme activities with various polysaccharide substrates were determined with bicinchoninic acid (BCA) assay [19] by measuring the quantity of reducing sugars released from the substrates (5 mg/mL) during 15 min incubation. All enzymatic reactions were performed at 50 °C and pH 6.0, unless mentioned otherwise. Briefly, substrate stock solutions (40 μL, 10 mg/mL) in buffer A (50 mM sodium phosphate buffer, pH 6.0) were mixed with 30 μL buffer A, and the reactions were initiated by the addition of 10 μL (0.2 μg) enzyme solution and further incubated for 15 min. Then, 50-μL reaction mixtures were mixed with 500 μL of BCA reagent and 450 μL of distilled water and incubated at 80 °C for 40 min. The reaction mixtures were cooled and absorbance at 562 nm (OD562) was measured. d-xylose solutions (X1), 10–50 μM, were run as standards. All activities are expressed in international units, i.e., one unit of activity corresponds to the amount of enzyme releasing 1 μM reducing sugars (in xylose equivalents) per minute.
Biochemical Properties of the Xylanase SCO5931
Enzyme temperature profile was studied over 20–90 °C. Optimal pH determination was carried out in 50-mM buffer solutions with pH ranging from pH 4.0 to 11.0. Sodium citrate buffer was used for pH 4.0–5.0, sodium phosphate buffer for pH 6.0–8.0, and glycine–NaOH buffer for pH 9.0–11.0. Relative activities were defined as percentages of the maximum xylanase activity. Temperature stability was studied by incubating the xylanase at 50 °C or 60 °C in buffer A. Aliquots were taken at different time points, and activity toward beechwood xylan was determined.
The effects of metal ions and chelator on xylanase activity were investigated in the presence of the following (4 mM): CaCl2, CoCl2, CuCl2, NaCl, NiCl2, MgCl2, ZnCl2, and EDTA. Beechwood xylan (5 mg/mL) solution was incubated in the presence of each effector in buffer A and the enzyme, and the xylanase activity measured.
Kinetic parameters K m and V max for the hydrolysis of beechwood xylan by SCO5931 xylanase were determined based on the dependence of the initial rates of hydrolysis on substrate concentration (0.5–5 mg/mL) using Lineweaver–Burk coordinates [20]. A Michaelis–Menten Enzyme Kinetics Software, MM version 1.2, was used for accurate calculation (http://people.uncw.edu/hermanr/TechFiles/mm/mm.htm).
Zymogram Assay for Xylanase Activity
Cells were inoculated onto solid minimal medium [14], supplemented with 0.2 % (w/v) AZCL xylan and cultured at 28 °C for 1, 2, or 3 days. For the purified protein, 20-μL (0.4 μg) protein samples were absorbed onto paper disks, laid out on solid minimal medium containing 0.2 % AZCL xylan, and incubated at 40 °C for 1 h.
Analysis of Hydrolysis Products by Thin-Layer Chromatography and Mass Spectrometry
The SCO5931-catalyzed hydrolysis of beechwood xylan (500 μg) with the purified SCO5931 (100 units) was carried out for 96 h in 100 μL of buffer A (pH 6.0). During the hydrolysis reaction, 5-μL aliquots of the reaction mixture were withdrawn at regular intervals and spotted on a silica gel 60 plate (Merck Co., Ltd., USA). Analytical thin-layer chromatography (TLC) was performed by double-ascending method with n-butanol/acetic acid/water (2:1:1) solvent system. The resolved sugars were detected by heating the plate at 120 °C. The spots were visualized after spraying with 10 % sulfuric acid in ethanol. The 96-h hydrolysate was dried in vacuo and extracted with 100 % methanol, for mass spectrometry. Molecular masses of the products were determined using micrOTOF-Q II (Bruker Daltonics, Germany), and mass spectra were obtained in a 120–3000-m/z range.
Results
Cloning and Overexpression of sco5931 Gene
SCO5931 (GenBank accession number, WP_011030540.1) was predicted from genomic sequencing data of S. coelicolor A3(2) to encode a putative xylanase A comprising 477 amino acids, with molecular weight (Mw) of 51 kDa [13]. The amino acid sequence suggested that SCO5931 was produced as a precursor with an amino-terminal signal sequence [21] cleaved between Ala-41 and Ala-42, resulting in mature SCO5931 comprising 436 amino acids, with Mw 47 kDa (Fig. 1).
For functional validation, S. lividans TK24 was transformed with pUWL201-5931 containing the entire coding region of SCO5931, inoculated onto solid minimal medium supplemented with 0.2 % AZCL xylan and cultured at 28 °C for 3 days. A blue-colored halo formed by the hydrolysis of AZCL xylan was apparent from day 1 and becoming wider until day 3, while an empty vector-bearing control did not produce blue color, indicating that sco5931 was successfully overexpressed and SCO5931 was synthesized in active form (Fig. 2a). The xylanase activity produced by S. lividans TK24/pUWL201-5931 sharply increased and reached a maximum level (140 units/mL) at 3 days of cultivation, while that of the control at this point was negligible (less than 0.1 unit/mL) in R2YE broth.
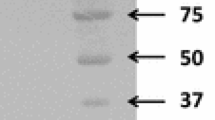
Heterologous expression and purification of SCO5931 in S. lividans TK24. a Results of zymogram plate assay with S. lividans TK24 transformants, as a function of cultivation time. Cells were grown on minimal agar medium plates containing 0.2 % (w/v) AZCL xylan for the indicated times, at 28 °C. Control: S. lividans TK24/pUWL201PW, SCO5931: S. lividans TK24/pUWL201PW-5931. b SDS-PAGE of purified proteins. SCO5931 protein overproduced in S. lividans TK24 was purified by gel filtration chromatography using Superdex™ 200 HR 10/30 column and fractionated on 0.1 % SDS-10 % PAGE. M molecular weight standards, lane 1: extracellular proteins of S. lividans TK24/pUWL201PW-5931, lane 2: extracellular proteins of S. lividans TK24/pUWL201PW control, lanes 3–4: different fractions of purified SCO5931 protein after gel filtration chromatography. The 47-kDa SCO5931 protein is depicted by a thick arrow
Purification of SCO5931
Gel permeation chromatography on Superdex™ 200 HR column allowed a rapid and efficient single-step SCO5931 purification from culture broth after 75 % ammonium sulfate precipitation. SCO5931 was purified by 25.8-fold with yield of 30.6 % from the culture broth. After SDS-PAGE, the purified protein appeared as a single band of approximately 45 kDa, slightly smaller than the expected Mw (47 kDa) of the mature form (Fig. 2a). Therefore, the purified SCO5931 was digested with trypsin and analyzed by matrix-assisted laser desorption/ionization-time-of-flight (MALDI-TOF) mass spectrometry. Nine peptides matched predicted masses and provided 21 % coverage of the entire SCO5931 (data not shown). An amino-terminal peptide (m/z 1289) corresponding to Ala-42–Arg55 was also detected. We concluded that SCO5931 was indeed correctly expressed, processed, and secreted.
Substrate Specificity of SCO5931
Specific activities of purified SCO5931 toward various cellulosic and hemicellulosic substrates were examined. No detectable hydrolyzing activity was observed with CMC, Avicel, galactan, barley β-glucan, and xyloglucan. Instead, SCO5931 displayed a pronounced activity (28,944 units/mg) toward beechwood xylan (Fig. 3a). Xylanase activity of SCO5931 was also confirmed by a zymogram assay. Purified SCO5931 created a blue region (halo) by hydrolyzing AZCL xylan in the plate zymogram assay, in contrast with no detectable activity in a protein sample prepared from culture broth of the control (data not shown). These results clearly indicated that SCO5931 is a substrate-specific xylanase.
Biochemical properties of SCO5931. a Substrate specificity of the purified SCO5931 protein. The hydrolyzing activity was investigated using carboxymethylcellulose (CMC), Avicel, galactan, β-glucan, xylan, and xyloglucan, as substrates. b Effect of temperature on beechwood xylan-hydrolyzing activity of SCO5931 xylanase. c Effect of heat treatment at 50 and 60 °C on SCO5931 stability, as determined by beechwood xylan-hydrolyzing activity. d Effect of pH on beechwood xylan-hydrolyzing activity of SCO5931 xylanase. The reaction was performed in 50-mM buffers, pH 4.0–11.0, as follows: citrate buffer, pH 4.0–5.0; potassium phosphate buffer, pH 6.0–8.0; glycine–NaOH buffer, pH 9.0–11.0. When calculating relative enzyme activities, the highest detected xylanase activity was considered as 100 %. e Effect of metal ions and chelating agent on xylanase activity of SCO5931. Beechwood xylan-hydrolyzing activity was investigated in the presence of 4 mM of each tested compound. When calculating relative enzyme activities, enzymatic activity in the absence of metal ions was considered as 100 %. f Enzyme kinetics of SCO5931 xylanase. Lineweaver–Burk plots were used to determine kinetic parameters of SCO5931 acting on beechwood xylan. All the data were obtained from mean values of three repeat experiments
Biochemical Properties of the Xylanase SCO5931
The enzyme had a maximum activity at 60 °C and retained 98 % of activity at 50 °C (Fig. 3b). The protein was relatively thermostable, maintaining 90 % of its maximum activity after 60 min incubation at 50 °C; however, 84 % activity was lost upon heat treatment at 60 °C (Fig. 3c). Optimum pH for SCO5931 was 6.0 (Fig. 3d). The enzyme was active over a wide pH range, 6.0–9.0, and its stability declined below pH 4.0 (less than 40 % maximum activity) or above pH 11.0. All subsequent reactions were carried out at pH 6.0 and 50 °C, with consideration of the enzyme’s thermostability.
Most metal ions tested, such as Ca2+, Na+, Ni2+, and Mg2+, and the chelator EDTA did not significantly affect xylanase activity when provided at 4-mM concentrations, indicating that SCO5931 did not require a metal cofactor. However, MnCl2 severely inhibited the enzyme activity, by 87 % (Fig. 3e).
Using beechwood xylan as substrate, SCO5931 K m and V max values were determined as 0.24 mg/mL and 6.86 μM/min, respectively (Fig. 3f).
Analysis of Beechwood Xylan SCO5931 Hydrolysis Products
Products released from beechwood xylan by SCO5931 were analyzed by TLC, and two distinct spots were detected, even after a long, 96-h incubation. When the spots were compared with standards, they were tentatively identified as di- and larger-than-tetra-saccharides, with decreasing Rf values. Additionally, one spot, corresponding to xylose, was detected after 72 h, but its amount was minimal (Fig. 4a). Mass spectrometry analysis of each spot revealed perfect matches with masses of sodium adducts of xylobiose (m/z 305.24) and xylopentaose (m/z 701.59) (Fig. 4b). When the enzyme reaction was performed for less than 2 h, xylobiose and a mixture of different length oligosaccharides were detected, indicating that SCO5931 acts as an endo-type glycohydrolase. Taken together, the results reported here indicated that SCO5931 is a β-1,4-endoxylanase, hydrolyzing the β-1,4 linkage of xylan to yield xylobiose and xylopentaose, a unique feature among the reported xylanases.
Thin layer chromatography (TLC) chromatogram and MALDI-TOF mass spectrogram of products of beechwood xylan hydrolysis by SCO5931. a TLC chromatogram. Products of beechwood xylan digestion by SCO5931 were analyzed on a silica gel 60 TLC plate. Lane X1: xylose, X2: xylobiose, X4: xylotetraose. Lanes 24, 48, 72, 96: SCO5931-generated hydrolysis products identified at the indicated periods of hydrolysis. b MALDI-TOF mass spectrogram. Oligosaccharides produced in a after a 96-h beechwood digestion by SCO5931 were dried in vacuo and extracted with methanol. Molecular masses were then determined using a matrix-assisted laser desorption/ionization-time-of-flight (MALDI-TOF) mass spectrometer. Molecular ions at m/z 305 (M + Na)+ and 701 (M + Na)+, corresponding to xylobiose and xylopentaose, respectively, are indicated by arrows
Discussion
SCO5931 was predicted to be a putatively secreted xylanase A (XlnAcoe) of S. coelicolor A3(2). It contains an N-terminal GH 10 superfamily domain (Conserved Domain Database [CDD] 249776, amino acids 45−341, e-value 2.77 × e−152) with putative sugar binding sites (CDD 238092; amino acids 366, 379, 381, 388, 389, 408, 419, 421, 428, 429, 449, 462, 464, and 472). It also contains a C-terminal RICIN superfamily domain (cd00161, amino acids 354–475, e-value 9.89 × e−32) (Fig. 1). RICIN superfamily domain is a carbohydrate-binding domain probably formed after a presumed gene triplication and found in a variety of molecules with diverse functions, such as enzymatic activity, inhibitory toxicity, and signal transduction [22, 23]. Three Q-X-W motifs (CDD 238092) were also well conserved in the Ricin-type beta-trefoil subdomains of XlnAcoe (amino acids 389−391, 429−431, and 472−474).
S. coelicolor A3(2) XlnAcoe (β-1,4-endoxylanase) has an apparent Mw of 45 kDa and optimum activity at pH 6.0 and 60 °C, predominantly degrading xylan to xylobiose and xylopentaose (Fig. 4). The name “xylanase A (XlnAcoe)” of S. coelicolor was based on xylanase A (XlnAliv) of S. lividans [24], because of the 99 % identity between their amino acid sequences. Enzymatic properties of XlnAliv, such as optimum pH and temperature, are similar to XlnAcoe, but the enzymes differ in apparent Mw (43 kDa on SDS-PAGE for XlnAliv) and xylan hydrolysis products (mainly xylobiose, a mixture of xylooligosaccharides, and a small amount of xylose after complete digestion by XlnAliv) [13]. Although Morosoli et al. [13] reported that XlnAliv produces a mixture of xylooligosaccharides from xylan, a more thorough investigation may be required of whether, similarly to SCO5931, this protein hydrolyzes xylan completely to xylobiose and xylopentaose. This study is the first report on enzymatic properties of xylanase A from S. coelicolor, and we expect that the enzymatic properties of many GenBank-annotated XlnA orthologs (probably including XlnAliv) will be similar to XlnAcoe.
GH family 10 xylanases perform catalysis with net retention of configuration and hydrolyze xylan into mainly xylose and xylobiose, with some extra oligosaccharides depending on enzyme [7]. For example, SoXyn10A xylanase from S. olivaceoviridis E-86 has been industrially used for producing xylose and xylobiose [25]. Contrasting to this, SCO5931 hydrolyzed Birchwood xylan into mainly xylobiose and xylopentaose, but a trace amount of xylose. A few GH 10 xylanases have been described in the genus Streptomyces including XlnAliv from S. lividans [9] and XylU from S. mexicanus [8]. Similar to XlnAliv, the XylU xylanase hydrolyzed Birchwood xylan and xylooligosaccharides (xylotriose to xylohexaose) to xylobiose as the primary degradation product and a small amount (4 %<) of xylose. In summary, all three streptomycetes GH 10 xylanases listed above have very weak activity for hydrolyzing xylan into monomeric xylose. According to Kaneko et al. [7], differences in the structure of subsite +2 of enzyme seriouly affect bond cleavage frequencies and the catalytic efficiency of xylooligosaccharide hydrolysis, influencing on production of xylose. Therefore, the relationship between xylose production and a variation in the amino acid residues comprising subsite +2 of GH 10 xylanases should be elucidated in the near future.
Five genes, namely, sco5931 (XlnA), sco2292 (XlnB), sco1883 (xylanase), sco0674 (XysA), and sco0105 (XlnC), were annotated as putative xylanase-encoding [13], but xylanase production in the S. coelicolor A(3)2 was not reported. Recently, GH family 11 xylanase C from S. coelicolor Ac-738, whose gene had one silent point mutation compared with xlnC of S. coelicolor A(3)2, was heterologously expressed and characterized [26]. It hydrolyzed xylan into, mainly, xylobiose, xylotriose, xylotetraose (minor), xylopentaose, and xylohexose, which is clearly distinct from XlnAcoe.
Xylan has been used for diverse purposes, in the food, textiles, and adhesive industries. The scope of its application is broadening to include anti-obesity agents, printing compositions, hair dying, ophthalmic solutions, and cosmetic and pharmaceutical applications [27]. Moreover, xylooligosaccharides have a high potential to lower blood sugar levels [27], exert hypolipidemic effects [27], stimulate probiotic Bifidobacterium growth [28], and stimulate immune reactions [29]. Therefore, the thermo-tolerable property of XlnAcoe and the unique production of xylobiose and xylopentaose from xylan may be useful for the development of xylan-based materials, with a huge potential in the biomedical and bioenergy fields.
References
Bajpai, P. (1997). Microbial xylanolytic enzyme system: properties and applications. Advances in Applied Microbiology, 43, 141–194.
Dashtban, M., Schraft, H., & Qin, W. (2009). Fungal bioconversion of lignocellulosic residues; opportunities & perspectives. International Journal of Biological Sciences, 5, 578–595.
Collins, T., Gerday, C., & Feller, G. (2005). Xylanases, xylanase families and extremophilic xylanases. FEMS Microbiology Reviews, 29, 3–23.
Beg, Q. K., Kapoor, M., Mahajan, L., & Hoondal, G. S. (2001). Microbial xylanases and their industrial applications: a review. Applied Microbiology and Biotechnology, 56, 326–338.
Chi, W. J., Youn, Y. S., Park, J. S., & Hong, S. K. (2015). Bacillus coreaensis sp. nov.: a xylan-hydrolyzing bacterium isolated from the soil of Jeju Island, Republic of Korea. The Journal of Microbiology, 53, 448–453.
Zhang, J., Matti, S. A., Terhi, P., Ming, T., Maija, T., & Liisa, V. (2011). Thermostable recombinant xylanases from Nonomuraea flexuosa and Thermoascus aurantiacus show distinct properties in the hydrolysis of xylans and pretreated wheat straw. Biotechnology for Biofuels, 4, 12–25.
Kaneko, S., Ichinose, H., Fujimoto, Z., Kuno, A., Yura, K., Go, M., Mizuno, H., Kusakabe, I., & Kobayashi, H. (2004). Structure and function of a family 10 beta-xylanase chimera of Streptomyces olivaceoviridis E-86 FXYN and Cellulomonas fimi Cex. Jornal of Biological Chemistry, 279, 26619–26626.
Kimdo, Y., Shin, D. H., Jung, S., Lee, J. S., Cho, H. Y., Bae, K. S., Sung, C. K., Rhee, Y. H., Son, K. H., & Park, H. Y. (2014). Biocatalytic properties and substrate-binding ability of a modular GH10 β-1,4-xylanase from an insect-symbiotic bacterium, Streptomyces mexicanus HY-14. Journal of Microbiology, 52, 863–870.
Morosoli, R., Bertrand, J. L., Mondou, F., Shareck, F., & Kluepfel, D. (1986). Purification and properties of a xylanase from Streptomyces lividans. Biochemical Journal, 239, 587–592.
Temuujin, U., Chi, W. J., Chang, Y. K., & Hong, S. K. (2012). Identification and biochemical characterization of Sco3487 from Streptomyces coelicolor A3(2), an exo- and endo-type β-agarase-producing neoagarobiose. Journal of Bacteriology, 194, 142–149.
Enkhbaatar, B., Temuujin, U., Lim, J. H., Chi, W. J., Chang, Y. K., & Hong, S. K. (2012). Identification and characterization of a xyloglucan-specific family 74 glycosyl hydrolase from Streptomyces coelicolor A3(2). Appllied and Environmental Microbiology, 78, 607–611.
Lim, J. H., Lee, C. R., Dhakshnamoorthy, V., Park, J. S., & Hong, S. K. (2015). Molecular characterization of Streptomyces coelicolor A(3) SCO6548 as a cellulose 1,4-β-cellobiosidase. FEMS Microbiology Letters, pii: fnv245.
Bentley, S. D., Chater, K. F., Cerdeño-Tárraga, A. M., Challis, G. L., Thomson, N. R., James, K. D., Harris, D. E., Quail, M. A., Kieser, H., Harper, D., Bateman, A., Brown, S., Chandra, G., Chen, C. W., Collins, M., Cronin, A., Fraser, A., Goble, A., Hidalgo, J., Hornsby, T., Howarth, S., Huang, C. H., Kieser, T., Larke, L., Murphy, L., Oliver, K., O’Neil, S., Rabbinowitsch, E., Rajandream, M. A., Rutherford, K., Rutter, S., Seeger, K., Saunders, D., Sharp, S., Squares, R., Squares, S., Taylor, K., Warren, T., Wietzorrek, A., Woodward, J., Barrell, B. G., Parkhill, J., & Hopwood, D. A. (2002). Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature, 417, 141–147.
Kieser, T., Bibb, M. J., Buttner, M. J., Chater, K. F., & Hopwood, D. A. (2000). Practical Streptomyces Genetics. John Innes Foundation, Norwich Research Park, Colney, Norwich NR4 7UH, England
Doumith, M., Weingarten, P., Wehmeier, U. F., Salah-Bey, K., Benhamou, B., Capdevila, C., Michel, J. M., Piepersberg, W., & Raynal, M. C. (2000). Analysis of genes involved in 6-deoxyhexose biosynthesis and transfer in Saccharopolyspora erythraea. Molecular Genetics and Genomics, 264, 477–485.
Green, M. R., & Sambrook, J. (2012). Molecular cloning. A laboratory manual (4th ed.). Cold Spring Harbor: Cold Spring Harbor Laboratory Press.
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry, 72, 248–254.
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685.
Percival Zhang, Y.-H., & Lynd, L. R. (2005). Determination of the number-average degree of polymerization of cellodextrins and cellulose with application to enzymatic hydrolysis. Biomacromolecules, 6, 1510–1515.
Lineweaver, H., & Burk, D. (1934). The determination of enzyme dissociation constants. Journal of the American Chemical Society, 56, 658–666.
Petersen, T. N., Brunak, S., von Heijne, G., & Nielsen, H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nature Methods, 8, 785–786.
Hazes, B. (1996). The (QxW)3 domain: a flexible lectin scaffold. Protein Science, 5, 1490–1501.
Rutenber, E., Ready, M., & Robertus, J. D. (1987). Structure and evolution of ricin B chain. Nature, 326, 624–626.
Shareck, F., Roy, C., Yaguchi, M., Morosoli, R., & Kluepfel, D. (1991). Sequences of three genes specifying xylanases in Streptomyces lividans. Gene, 107, 75–82.
Fujimoto, Z., Mizuno, H., Kuno, A., Yoshida, S., Kobayashi, H., & Kusakabe, I. (1997). Crystallization and preliminary X-ray crystallographic study of Streptomyces olivaceoviridis E-86 beta-xylanase. Journal of Biochemistry, 121, 826–828.
Lisov, A. V., Belova, O. V., Andreeva-Kovalevskaya, Z. I., Budarina, Z. I., Solonin, A. A., Vinokurova, N. G., & Leontievsky, A. A. (2014). Recombinant xylanase from Streptomyces coelicolor Ac-738: characterization and the effect on xylan-containing products. World Journal of Microbiology and Biotechnology, 30, 801–808.
Polizeli, M. L., Rizzatti, A. C., Monti, R., Terenzi, H. F., Jorge, J. A., & Amorim, D. S. (2005). Xylanases from fungi: properties and industrial applications. Applied Microbiology and Biotechnology, 67, 577–591.
Yang, J., Summanen, P. H., Henning, S. M., Hsu, M., Lam, H., Huang, J., Tseng, C. H., Dowd, S. E., Finegold, S. M., Heber, D., & Li, Z. (2015). Xylooligosaccharide supplementation alters gut bacteria in both healthy and prediabetic adults: a pilot study. Frontiers in Physiology, 6, 216.
Hansen, C. H., Frøkiær, H., Christensen, A. G., Bergström, A., Licht, T. R., Hansen, A. K., & Metzdorff, S. B. (2013). Dietary xylooligosaccharide downregulates IFN-γ and the low-grade inflammatory cytokine IL-1β systemically in mice. Journal of Nutrition, 143, 533–540.
Acknowledgements
This work was supported by the Advanced Biomass R&D Center (ABC) of Global Frontier Project funded by the Ministry of Science, ICT, and Future Planning (NRF-2015M3A6A2065700).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Enkhbaatar, B., Lee, CR., Hong, YS. et al. Molecular Characterization of Xylobiose- and Xylopentaose-Producing β-1,4-Endoxylanase SCO5931 from Streptomyces coelicolor A3(2). Appl Biochem Biotechnol 180, 349–360 (2016). https://doi.org/10.1007/s12010-016-2103-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-016-2103-y