Abstract
The activity and kinetic stability of a keratinolytic subtilisin-like protease from Bacillus sp. P45 was investigated in 100 mM Tris-HCl buffer (pH 8.0; control) and in buffer with addition of Ca2+ or Mg2+ (1–10 mM), at different temperatures. Addition of 3 mM Ca2+ or 4 mM Mg2+ resulted in a 26% increment on enzyme activity towards azocasein when compared to the control (100%; without added Ca2+ or Mg2+) at 55 °C. Optimal temperature for activity in the control (55 °C) was similar with Mg2+; however, temperature optimum was increased to 60 °C with 3 mM Ca2+, displaying an enhancement of 42% in comparison to the control at 55 °C. Stability of protease P45 in control buffer and with Mg2+ addition was assayed at 40–50 °C, and at 55–62 °C with Ca2+ addition. Data were fitted to six kinetic inactivation models, and a first-order equation was accepted as the best model to describe the inactivation of protease P45 with and without metal ions. The kinetic and thermodynamic parameters obtained showed the crucial role of calcium ions for enzyme stability. As biocatalyst stability is fundamental for commercial/industrial purposes, the stabilising effect of calcium could be exploited aiming the application of protease P45 in protein hydrolysis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Proteolytic enzymes are important commercial biocatalysts exploited by the food, detergent, leather, among other industries. Although animal and plant proteases are currently employed in various processes, the higher yields obtained with microorganisms amplify its significance as enzyme sources, which meet the increasing demand for proteases. The vast microbial diversity could provide proteolytic enzymes suitable for diverse applications and, among microorganisms, strains of the bacterial genus Bacillus are considered as remarkable producers of potentially valuable proteases. The major and best known proteases produced by Bacillus spp. are subtilisins and subtilisin-like proteases, which have been investigated for their structural and catalytic properties and also for evaluation of industrial applications [1–3].
Keratins are insoluble and stable proteins found as major components of the skin and its appendages such as feathers, hair and wool. These recalcitrant proteins are produced in large amounts by agroindustrial processing and are usually not degraded by common proteases such as pepsin and trypsin. Among microbial proteases, keratinases are able to hydrolyse keratins [4]. Therefore, increasing interest has been focused on the application of these enzymes for the bioconversion of keratin-rich wastes, producing protein hydrolysates for utilisation as nitrogen fertilizers and animal feed. Also, keratinases are investigated in prion degradation as a component in detergent and feed formulations and as a dehairing agent in the leather industry [5]. Production of keratinolytic enzymes is considered to be largely induced by keratinacious substrates, and maximal yields are usually achieved during the late exponential and/or stationary phase of microbial growth. Keratinases are mainly serine or metalloproteases, optimally active at 40–60 °C and at pH values of 7.0–9.0. Particularly, keratinases from Bacillus spp. are mainly characterised as subtilisin-like proteases preferring aromatic and hydrophobic amino acid residues at position P1 [6].
Enzyme activity and stability are topics directly linked with enzyme application. Various phenomena could lead to enzyme inactivation including autolysis, aggregation, coagulation, denaturation due to exposure to solvents, surfactants, salts and extremes of temperature and pH [7, 8]. Enzyme stabilisation techniques have been investigated to counteract these deleterious mechanisms, providing biocatalysts with higher efficiency that, in turn, could increase the biotechnological and economical potential of enzyme-based processes [8]. In this context, predicting enzyme inactivation is essential for enzyme characterisation from both scientific and technological perspectives. Mathematical models, consisting of equations that provide an output based on a set input data, represent a powerful and concise way to express physical behaviour in mathematical terms [9]. Therefore, adequate kinetic models and thermodynamic studies may enable a better knowledge of the enzyme behaviour during inactivation.
Bacillus sp. P45 is an effective feather-degrading microorganism isolated from the intestine of an Amazonian fish [10]. This strain produces diverse proteolytic enzymes during submerged cultivations on media containing whole feathers or feather meal as the only organic source of carbon and nitrogen [10, 11]. As fish intestinal bacteria are considered as an untouched resource for enzyme production, fish microbiota might produce enzymes well-suited for different applications. In this sense, a subtilisin-like protease was purified from culture supernatants, showing optimal azocaseinolytic activity at 50 °C and pH 8.0 [12]. This enzyme is completely inhibited by sodium dodecyl sulphate (SDS) and slightly stimulated by Triton X-100, solvents (dimethyl sulphoxide and ethanol), and some metal ions (such as Ca2+ and Mg2+). The thermostability properties of this enzyme, in addition to enzyme activity at low/moderate temperatures, might suggest the suitability of protease P45 for utilisation as a biocatalyst in energy-saving processes of protein hydrolysis/modification [12]. Therefore, aiming to further characterise the biotechnological potential of protease P45, the kinetic stability of this enzyme was investigated at different temperatures (40–62 °C), with and without the addition of metal ions (Mg2+ or Ca2+) to assay buffer, and the isothermal data was statistically treated using different enzyme inactivation models.
Materials and Methods
Enzyme Production and Purification
Production of keratinolytic protease by Bacillus sp. P45 (GenBank accession number AY962474) was performed in Erlenmeyer flasks (250 ml) containing 100 ml of mineral medium: NaCl (0.5 g l-1), K2HPO4 (0.3 g l-1), and KH2PO4 (0.4 g l-1) supplemented with feather meal (FM; 43 g l-1) and NH4Cl (1.9 g l-1) [11]. After 48 h of cultivation at 30 °C in an orbital shaker (125 rpm), the medium was centrifuged (10,000 × g for 15 min) and the supernatant was submitted to a purification protocol involving ammonium sulphate concentration (30–60% saturation) followed by gel filtration chromatography on Sephadex G-100 and anion-exchange chromatography on DEAE-Sepharose [12]. Fractions with proteolytic activity were pooled and employed in the following studies.
Enzyme Assay
Proteolytic activity was determined by a method described elsewhere using azocasein as substrate [12]. The reaction contained 100 μl of 100 mM Tris-HCl buffer (pH 8.0), 300 μl of 10 mg ml-1 azocasein (in Tris-HCl buffer) and 100 μl of conveniently diluted enzyme. After incubation (55 °C for 15 min), the reaction was stopped by adding 600 μl of 10% (w/v) trichloroacetic acid (TCA), and this mixture was centrifuged (10,000 × g for 5 min). The supernatant (800 μl) was mixed with 1.8 M NaOH (200 μl) and the absorbance at 420 nm was measured. One unit (U) of protease activity was defined as the amount of enzyme that caused an increase of 0.1 absorbance unit at the defined assay conditions.
Effects of Calcium and Magnesium on Proteolytic Activity
The effects of Ca2+ and Mg2+ were tested by adding different concentrations (0–10 mM final concentration) to the enzyme assay performed at 55 °C, which was the optimum temperature previously determined in the Tris-HCl buffer without additions [12]. Enzyme activity determined without additions was considered as 100%. Subsequently, the enzyme activity was evaluated at different temperatures (40–70 °C) in assays containing the Ca2+ or Mg2+ concentrations which resulted in the higher enzyme activity at 55 °C (above experiment). Enzyme activity determined at 55 °C without the addition of Ca2+ or Mg2+ to assay buffer was considered as 100% [12]. All assays were done in triplicate.
Enzyme Inactivation at Different Temperatures
Protease solutions in buffer (0.1 ml; 0.07 mg ml-1) without added metal ions, or containing 3 mM Ca2+ or 4 mM Mg2+, were heated in sealed tubes (1 mm of thickness, 7 mm of internal diameter and 3 cm of length) at temperatures ranging from 40 to 62 °C, in a thermostatically controlled water bath (Thermomix BM-S, B. Braun Biotech International, Melsungen, Germany). Time exposure varied between 1 and 29 min, and the tubes were immediately immersed in an ice bath, thereby stopping the heat inactivation reaction. The activity after 1 min of heating-up time (t = 0) was considered to be the initial activity, eliminating the effects of heating up [13]. Residual proteolytic activities with respect to processing time at different temperatures were fitted to several models (Table 1) through nonlinear regression using the software Statistica 7.0 (StatSoft, Tulsa, OK, USA).
Kinetic Modelling
Kinetic models commonly reported to describe enzyme behaviour during inactivation were employed in this investigation (Table 1). These models cover the one-step, parallel and series models of enzyme inactivation. In the model equations, A/A 0 represents the residual proteolytic activity at time t (min), and k (min-1) is the inactivation rate constant at a given temperature. First-order kinetics (Eq. 1) is generally employed to describe enzyme thermal inactivation [14]. The utilisation of the nth-order model (Eq. 2) is not exceptional, where n is the order of the reaction [15]. Parallel models, indicating the existence a mixture of enzymes (isoforms, isoenzymes) with different heat sensitivities (labile and resistant fractions) and/or catalytic properties [16], were checked for protease P45. Mathematical equations, which suggest that residual activity could be described by the summation of such distinct fractions, are represented by Eqs. 3–5. Residual activities for the ‘labile’ and ‘resistant’ isoenzymes are represented by A L and A R, respectively, k L and k R are the correspondent first-order reaction rate constants for each fraction, respectively, and coefficient α represents activity fraction of the thermal labile group in relation to the total activity [14, 17]. Fractional conversion, Eq. 5, refers to a first-order inactivation process and takes into account of the non-zero enzyme activity upon prolonged heating because of the presence of an extremely heat-resistant enzyme fraction [18]. The series-type model (Eq. 6) is based on a succession of two first-order steps [19]. In the first step, the protein unfolds from the native structure (E) to yield an intermediate (E 1) with lower specific activity, which is followed by the conversion of the intermediate into a final enzyme state (E 2) with possible non-zero activity. In this model, k 1 and k 2 are the first-order deactivation coefficients, and α 1 and α 2 are the ratios of specific activities of E/E 1 and E/E 2, respectively.
Comparison of Kinetic Models
For comparison of fits obtained with nonlinear regression, statistical and physical criteria were considered. Statistical criteria included coefficient of determination (r 2), chi-square (χ 2), and standard error of means (S.E.M.). These criteria have been used successfully to compare kinetics of inactivation models of several bioactive proteins [13, 20, 21].
Calculation of χ 2 is done by the equation:
S.E.M. is defined as:
where m is the number of observations, and p the number of parameters.
Estimation of negative kinetic parameter at a given temperature is a physical criterion for rejection of a model. The model with the lowest χ 2 and S.E.M. and higher r 2 for the residual proteolytic activity is the best choice from a statistical point of view [13].
Thermodynamic Analysis
The relationship between temperature and rate parameters of the inactivation models showed on Table 1 can be expressed algebraically by Arrhenius’ law (Eq. 9):
where A is the Arrhenius constant, E a is the activation energy, R is the universal gas constant (8.31 J mol-1 K-1), and T is the absolute temperature. The E a can be estimated by the slope of linear regression analysis of the natural logarithm of rate constant versus the reciprocal of the absolute temperature.
With the obtained value of E a, the activation enthalpy for inactivation (ΔH #) for each temperature was calculated by:
The free energy of inactivation (ΔG #) can be determined according to the expression:
where h (6.6262 × 10-34 J s) is the Planck’s constant, K B (1.3806 × 10-23 J K-1) is the Boltzmann’s constant, and k (s-1) is the inactivation rate constant of each temperature.
From Eqs. 10 and 11, it is possible to calculate the activation entropy of inactivation (ΔS #) by:
Data Analysis
Mean values were calculated from two independent experiments for each condition and duplicate assays of enzyme activity were performed for each experiment. Statistical analysis of the data was performed using the Statistica 7.0 software (Statsoft) and plots using Microsoft Excel 2000 (MapInfo, Troy, NY, USA). Obtained k values were compared using Tukey’s test, and a P < 0.05 was considered statistically significant.
Results and Discussion
Protease P45, a 26-kDa subtilisin-like enzyme produced by Bacillus sp. P45, was previously reported to act on various protein substrates, including casein, feather meal, azocasein, hide powder azure and keratin azure, and also to preferably hydrolyse synthetic tetrapeptides (p-nitroanilide derivatives) containing aromatic and hydrophobic residues at position P1 [12]. Such features suggest the suitability of protease P45 as a valuable biocatalyst for industrial processes. Therefore, as an essential stage for enzyme characterisation, the activity and kinetic stability of protease P45 was investigated in Tris-HCl buffer (100 mM, pH 8.0) with or without the addition of metal ions (Ca2+ or Mg2+) at different temperatures.
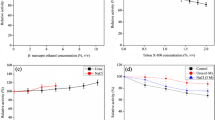
Initially, the effect of different concentrations (0–10 mM) of calcium and magnesium on P45 activity was assessed at 55 °C, which is the temperature optimum previously reported in buffer without added metal ions [12]. Enzyme activity showed to be slightly enhanced by all metal ion concentrations (Fig. 1). Maximum activity was observed with 3 mM Ca2+ (125%) or 4 mM Mg2+ (126%) in comparison to the control (100%, without metal ions) and tended to decrease with higher concentrations. Subsequently, Ca2+ (3 mM) or Mg2+ (4 mM) were added to enzyme assays performed at different temperatures (40–70 °C), and the results were compared with enzyme activities observed without the addition of metal ions to the buffer (Fig. 2). At lower temperatures (40–45 °C), the effect of metal ions was not apparent, but as the temperature increased, the presence of Ca2+ or Mg2+ tended to enhance enzyme activity. With 4 mM Mg2+, the maximum activity was reached at 55 °C, although it was 21% higher than that of the control (55 °C; 100%); and a similar activity to that of control optimum was observed at 60 °C (Fig. 2). The effect of calcium was most pronounced, with maximal activity (142% of control) observed at 60 °C; also, enzyme activity at 65 °C was higher than that of control optimum (Fig. 2). The positive effect of metal ions, particularly Ca2+, on proteolytic activity is generally reported for alkaline proteases of Bacillus spp. [22–25] and also proteolytic enzymes from other microbial sources [1, 6], such as a milk-clotting protease from Aspergillus oryzae MTCC 5341 [26], and a subtilisin-like protease produced by Streptomyces albogriseolus [27]. Enhanced activity in the presence of calcium, at temperatures in which the P45 activity without metal ions was highly diminished, suggests that Ca2+ might act improving the protein conformational stability at higher temperatures [22, 28]. Also, metal ions might contribute to stabilise the binding of the substrate and enzyme complex [29].
The influence of Ca2+ and Mg2+ ions on isothermal inactivation treatments of protease P45 in temperatures between 40 and 62 °C was then evaluated (Fig. 3), and residual activities were fitted for different inactivation models. In assays using buffer without additions and buffer with 4 mM Mg2+, enzyme inactivation was assessed at temperatures of 40 to 50 °C; however, with 3 mM Ca2+, protease P45 was stable up to 50 °C (data not shown). Due to this increased stability, temperatures employed in enzyme inactivation assays with calcium ranged from 55 to 62 °C. As expected, proteolytic activity decreased as the heating time increased as well as at higher temperatures (Fig. 3).
Residual activity of protease P45 submitted to heating, fitted to first-order model. a In 100 mM Tris-HCl buffer (pH 8.0) without added metal ions at 40 °C (○), 43 °C (□), 45 °C (Δ), 47 °C (◊), and 50 °C (×); b in buffer with 4 mM Mg2+ at 40 °C (○), 43 °C (□), 45 °C (Δ), 47 °C (◊), and 50 °C (×); and c in buffer with 3 mM Ca2+ at 55 °C (○), 56.5 °C (□), 58 °C (Δ), 60 °C (◊), and 62 °C (×). Data presented are average values of two independent experiments, and standard deviations were always lower than 4%
Although the increased thermostability of subtilisin-like proteases in the presence of calcium ions is described, few studies deal with the mathematical modelling of kinetic thermal inactivation of proteases, simply assuming a first-order kinetics from semilogarithmic plots of activity versus time. The choice of the best equation for process modelling is essential from both engineering and economical points of view, minimising errors and improving the effectiveness of the process, ultimately resulting in lower costs. Several mathematical equations have proposed to explain the behaviour of a biocompound during thermal degradation [9]. Therefore, six inactivation kinetic models were tested to fit the experimental data for heat treatments of protease P45 (Table 1). The performance of these models for the three experiments is summarised in Table 2, and the model which satisfactorily described the inactivation pattern for all experiments (with and without metal ions) was selected. For nth-order (Eq. 2), two-fraction (Eq. 4), fractional conversion (Eq. 5), and series (Eq. 6) models, negative parameter values were obtained, which is a physical criterion for rejection of the equations. In the distinct isoenzymes model (Eq. 3), equal inactivation rate parameters were calculated, excluding this model. This result is in accordance to that obtained by the two-fraction and fractional conversion models. Therefore, this indicates the absence of isoforms/isoenzymes, or different proteases with distinct stabilities and/or substrate specificities. Although microbial serine proteases are mainly monomeric enzymes, such parallel models of inactivation (Eqs. 3–5) were evaluated since the occurence of proteolytic enzymes with quaternary structures is reported [30] and, particularly, a subtilisin-like protease from Bacillus intermedius secreted by a Bacillus subtilis recombinant strain was reported to form dimers, possibly through interactions mediated by calcium ions [31].
Rejection of the series model (Eq. 6) suggests that, under the conditions presently evaluated, irreversible kinetic inactivation of protease P45 is sufficiently fast to make negligible the population of intermediate states [32]. Alternatively, the unfolded or partially unfolded enzyme might not be active, as these intermediates may not refold correctly on cooling, producing thermodynamically stable but inactive molecules [23, 33]. This seems to be particularly true for subtilisins. When the forces leading to enzyme denaturation are reversed, subtilisins are often unable to refold from denatured states. This occurs since these enzymes are produced as precursors that require its N-terminal propeptide to act as an intramolecular chaperone to yield the mature enzyme [34, 35]. Therefore, the inactivation of protease P45 at different temperatures seems to proceed through a single-step mechanism.
First-order model yielded good statistical criteria for the three conditions evaluated. The r 2 values ranged between 0.896 and 0.999; the χ2 values were low, ranging from 0.00006 to 0.0138, meanwhile S.E.M. varied between 0.007 and 0.107. These values are comparable to those calculated for model acceptance in other investigations [13, 15, 21, 36]. Hence, among the mathematical models evaluated, the first order was accepted as the best equation to describe the inactivation of protease P45 in the conditions employed. The results of experimental data fitted to first-order model are shown in Fig. 3, and an exponential behaviour could be observed. Accordingly, several investigations suggest that the thermal inactivation of proteolytic enzymes with respect to time obey first-order kinetics, as reported for Bacillus circulans [23], Chryseobacterium sp. kr6 [37], Candida buinensis [38], and Salinivibrio proteolyticus [39] proteases; however, in those studies, the first-order activity decay is assumed without model testing. An apparent first-order behaviour indicates that one of the processes leading to enzyme denaturation (probably temperature) is the controlling step, fixing the overall inactivation rate [40]. Previously, subtilisin thermal inactivation rate was reported to be characterised by a single exponential decay curve, determined by the rate of subtilisin unfolding [41].
The k values increased significantly (P < 0.05) and t 1/2 decreased with increasing temperature (Table 3), indicating a faster inactivation at higher temperatures [13]. k Values for protease P45 are significantly (P < 0.05) higher without ions than in presence of Mg+2 in the 43–50 °C range, suggesting a protective effect of this ion against protease inactivation. As the temperatures employed in the inactivation assays with Ca2+ needed to be raised, it is evident that protease P45 achieved a higher stability when compared to that observed with Mg2+ or without metal ions. Calcium is also reported to enhance the stability of various microbial proteases [1] including proteolytic enzymes from Aspergillus clavatus [42] and Thermoactinomyces sp. 27a [43]. The half-life of protease P45 without ions at 50 °C was 1.06 min, whereas that at 55 °C with calcium was 14 min (Table 3). For a Chryseobacterium sp. kr6 protease, the half-lives at 55 °C, with and without calcium, were 82 and 4 min, respectively [37]; whereas the half-life of a protease from A. clavatus at 55 °C with calcium was 6 min [42], which is related to that observed for protease P45 (Table 3). Thermal inactivation concepts (D and z values) are generally used to represent a first-order reaction. D value is the time needed for a tenfold reduction of the initial activity at a given temperature, and it is obtained by plotting the activity values on a log scale against the corresponding inactivation times. The z value is the temperature needed to reduce the D value in one log unit, and it is obtained by plotting the D values on a log scale against the corresponding temperatures [44]. The higher D values in the presence of metal ions represent the enhanced stability of protease P45 (Table 3). z Values were calculated to be 7.16 °C with no added ions, and 9.18 °C with Mg2+, indicating the slight but significant protective effect of Mg2+ against inactivation in the range 40–50 °C. The z value with added Ca2+ was 8.04 °C, in the range 55–62 °C.
Particularly, subtilases typically contain one or more calcium-binding sites, and native subtilisins are thermodynamically unstable in the absence of bound metals [35]. In subtilisins (from family A), two Ca2+-binding sites are usually present: a strong site and a weak site. The former specifically binds calcium, and the later can bind both divalent and monovalent cations [28]. In this regard, the protease P45 showed elevated homology with -subtilisin BPNʹ and related enzymes, and amino acid residues related with calcium-binding sites were conserved [12]. Calcium binding serve to tie together surface loops, reducing the flexibility of the polypeptide chain and inhibiting local unfolding, diminishing the rate of thermal inactivation of the enzyme [45]. Thermal denaturation temperature decreases with the removal of calcium from the strong binding site, and only a slight decrease is observed by removing calcium from the weak binding site; however, the portion of random coil structure increases significantly when calcium is removed from the weak as well as the strong binding sites, indicating that both sites are essential in maintaining a stable enzyme structure [46]. The effect of Mg2+ in protecting protease P45 from inactivation was less pronounced, which could be explained by the preference of calcium to bind to carboxylate and other oxygen ligands, which are the metal-binding groups most likely to be presented in external loops [45]. Mg2+ ion has an effective ionic radius considerably less than Ca2+, which might partly explain the lower degree of stabilisation by the former and, additionally, Mg2+ has a preference to bind nitrogen rather than oxygen ligands [47].
Estimation of thermodynamic parameters is an important issue to determine industrial potential of bioactive compounds and their structure–stability relationships. The data fitted well to Arrhenius equation (data not shown), enabling the calculation of thermodynamic parameters (Table 4) by the transition state theory (Eqs. 10–12). ΔG # is a measure of the spontaneity of the inactivation process, depending on ΔH # (heat change) and ΔS # (entropy change) for inactivation, which provide a measure of the number of non-covalent bonds to be broken during inactivation, and the disorder change of molecules in the system, respectively [48, 49]. Positive ΔH # values indicate the endothermic character of the inactivation process [38]. ΔG # and ΔH # decreased as the temperature increased (Table 4), suggesting the destabilisation of protease P45 at higher temperatures [44, 50]. The ΔS # values have not presented a continuous behaviour, which could result from the difficult evaluation of system disorder in such a small temperature variation.
From the analysis of the thermodynamic parameters obtained in buffer with and without metal ion additions, it is observed that values of ΔH # and ΔS # were lower, and ΔG # was higher, in the presence of metal ions (Table 4). Enzyme kinetic inactivation involves the unfolding of the protein tertiary structure to a disordered polypeptide due to the breakage of weak, non-covalent bonds [32]. As ΔG # = ΔH #-TΔS #, protein stabilisation might be improved by higher values of ΔH # (more ionic or hydrophobic interactions) or lower values of ΔS #. However, if large values of ΔH # are coupled with a large increase in the values of ΔS #, a destabilising effect could be expected. This occurs since the increase of ΔS # compensates the high inactivation barrier, causing ΔG # to be low enough, resulting in an overall less energy requirement for the inactivation process to proceed relatively fast [49, 50].
Activation energy (E a) can be seen as an energy barrier that molecules need to cross in order to be able to react, and the proportion of molecules able to do that usually increases with temperature, qualitatively explaining the effect of temperature on rates [9]. Therefore, the higher the E a values, the higher the energy barrier to be transposed for enzyme inactivation, indicating an increased stability. In subtilisins, by binding at specific sites in the enzyme structure, calcium ions contribute their binding energy to the stability of the native state, which was reported to increase the activation energy of unfolding [41]. Nevertheless, although the value of E a for inactivation of protease P45 was higher in buffer without added Ca2+ or Mg2+ than in the presence of added metal ions (Table 4), the enzyme was inactivated at slower rates in the presence of metal ions (k values; Table 3), probably due to the usual dominant role of ΔS # in the thermal inactivation of proteins in aqueous solutions [50]. Therefore, it is suggested that the decrease in the inactivation rate constant (k), or the increase in ΔG # values, are more reliable criteria to observe the enhancement of enzyme stabilisation than the increase in the E a for inactivation [51].
Investigations on thermal inactivation of proteases might become more intricate due to autolysis [7]. Although the contribution of autolysis to enzyme inactivation is minimised by employing low protease concentrations [37], the analysis of second-order plots of residual enzyme activity with respect to time might indicate if this mechanism is adding to the activity decay [47]. As the preincubation temperature decreased, both in the assays with and without metal ions, second-order plots (1/% relative activity versus time) tended to be more linear (data not shown), suggesting that autolysis might contribute to some extent to enzyme inactivation [52], even without deviation from the apparent first-order kinetics. Particularly, subtilisin BPNʹ is reported to possess two autoproteolysis sites (Ala48-Ser49 and Serl63-Thr164) in regions of high mobility localised in extended surface loops [53], which are present in protease P45 [12]. As these autolysis sites appear to be involved in calcium binding, or to be close to calcium-binding sites [28, 45], it is suggested that the decreased kinetic inactivation observed with calcium binding could also be due to a decrease in the autolysis rate, since unfolded or partially folded proteins tend to be more susceptible to proteolysis [47].
Conclusions
The kinetic inactivation of protease P45 in Tris-HCl buffer (100 mM, pH 8.0) at different temperatures, with or without the addition of Mg2+ or Ca2+, was investigated. Although the addition of Mg2+ showed a slight enhancement in the kinetic stability of this enzyme, a pronounced positive effect was observed with Ca2+, which could be a useful feature of this protease for applications involving protein hydrolysis. Despite the possible effect of autolysis, the kinetic inactivation data of protease P45 showed to be best represented by first-order kinetics when compared to the other models evaluated, corresponding to a single exponential decay in enzyme activity. The modelling of kinetic inactivation with the first-order equation, possessing only one parameter (k value), makes the prediction of activity loss easier and simpler when compared to more complex models where the intercorrelation between parameters might be difficult to analyse. As residual enzyme activities were employed in the present investigation, differentiation among distinct inactivation mechanisms is difficult. However, regardless of the specific process leading to protease inactivation, activity loss and enzyme stability are the fundamental issues from a technological standpoint, and, therefore, the modellisation of protease P45 inactivation is of remarkable importance in evaluating its biotechnological potential.
References
Gupta, R., Beg, Q. K., & Lorenz, P. (2002). Applied Microbiology and Biotechnology, 59, 15–32.
Jaouadi, B., Ellouz-Chaabouni, S., Rhimi, M., & Bejar, S. (2008). Biochimie, 90, 1291–1305.
Jaouadi, B., Ellouz-Chaabouni, S., Ben Ali, M., Ben Messaoud, E., Naili, B., Dhouib, A., et al. (2009). Biotechnology and Bioprocess Engineering, 14, 503–512.
Brandelli, A. (2008). Food and Bioprocess Technology, 1, 105–116.
Gupta, R., & Ramnani, P. (2006). Applied Microbiology and Biotechnology, 70, 21–33.
Brandelli, A., Daroit, D. J., & Riffel, A. (2010). Applied Microbiology and Biotechnology, 85, 1735–1750.
Bryan, P. N. (2000). Biochimica et Biophysica Acta, 1543, 203–222.
Iyer, P. V., & Ananthanarayan, L. (2008). Process Biochemistry, 43, 1019–1032.
van Boekel, M. A. J. S. (2008). Comprehensive Reviews in Food Science and Food Safety, 7, 144–158.
Daroit, D. J., Corrêa, A. P. F., & Brandelli, A. (2009). International Biodeterioration and Biodegradation, 63, 358–363.
Daroit, D. J., Corrêa, A. P. F., & Brandelli, A. (2011). International Biodeterioration and Biodegradation, 65, 45–51.
Daroit, D. J., Corrêa, A. P. F., Segalin, J., & Brandelli, A. (2010). Biocatalysis and Biotransformation, 28, 370–379.
Sant’Anna, V., Utpott, M., Cladera-Olivera, F., & Brandelli, A. (2010). Journal of Agricultural and Food Chemistry, 58, 3147–3152.
Ludikhuyze, L., Ooms, V., Weemaes, C., & Hendrickx, M. (1999). Journal of Agricultural and Food Chemistry, 47, 1794–1800.
Shalini, G. R., Shivhare, U. S., & Basu, S. (2008). Journal of Food Engineering, 85, 147–153.
Aymard, C., & Belarbi, A. (2000). Enzyme and Microbial Technology, 27, 612–618.
Chen, C. S., & Wu, M. C. (1998). Journal of Food Science, 63, 747–750.
Rizvi, A. F., & Tong, C. H. (1997). Journal of Food Science, 62, 1–7.
Henley, J. P., & Sadana, A. (1985). Enzyme and Microbial Technology, 7, 50–60.
Schokker, E. P., & van Boekel, M. A. J. S. (1997). Journal of Agricultural and Food Chemistry, 45, 4740–4747.
Corradini, M. G., & Peleg, M. (2004). Journal of the Science of Food and Agriculture, 84, 217–226.
Ghorbel, B., Sellami-Kamoun, A., & Nasri, M. (2003). Enzyme and Microbial Technology, 32, 513–518.
Rao, C. S., Sathish, T., Ravichandra, P., & Prakasham, R. S. (2009). Process Biochemistry, 44, 262–268.
Wan, M. Y., Wang, H. Y., Zhang, Y. Z., & Feng, H. (2009). Applied Biochemistry and Biotechnology, 159, 394–403.
Corrêa, A. P. F., Daroit, D. J., & Brandelli, A. (2010). International Biodeterioration and Biodegradation, 66, 1–6.
Vishwanatha, K. S., Rao, A. G. A., & Singh, S. A. (2010). Applied Microbiology and Biotechnology, 85, 1849–1859.
Suzuki, M., Taguchi, S., Yamada, S., Kojima, S., Miura, K. I., & Momose, H. (1997). Journal of Bacteriology, 179, 430–438.
Pantoliano, M. W., Whitlow, M., Wood, J. F., Rollence, M. L., Finzel, B. C., Gilliland, G. L., et al. (1988). Biochemistry, 27, 8311–8317.
Cao, Z. J., Zhang, Q., Wei, D. K., Chen, L., Wang, J., Zhang, X. Q., et al. (2009). Journal of Industrial Microbiology and Biotechnology, 36, 181–188.
Tiwary, E., & Gupta, R. (2010). Bioresource Technology, 101, 6103–6110.
Mikhailova, E. O., Mardanova, A. M., Balaban, N. P., Rudenskaya, G. N., Ilyinskaya, O. N., & Sharipova, M. R. (2009). Biochemistry (Moscow), 74, 308–315.
Sanchez-Ruiz, J. M. (2010). Biophysical Chemistry, 148, 1–15.
Schokker, E. P., & van Boekel, M. A. J. S. (1999). Journal of Agricultural and Food Chemistry, 47, 1681–1686.
Yabuta, Y., Subbian, E., Takagi, H., Shinde, U., & Inouye, M. (2002). Journal of Biochemistry, 131, 31–37.
Fisher, K. E., Ruan, B., Alexander, P. A., Wang, L., & Bryan, P. N. (2007). Biochemistry, 46, 640–651.
Rudra, S. G., Shivhare, U. S., Basu, S., & Sarkar, B. C. (2008). Food and Bioprocess Technology, 1, 187–195.
Silveira, S. T., Casarin, F., Gemelli, S., & Brandelli, A. (2010). Applied Biochemistry and Biotechnology, 162, 548–560.
Viana, D. A., Lima, C. A., Neves, R. P., Mota, C. S., Moreira, K. A., Lima-Filho, J. L., et al. (2010). Applied Biochemistry and Biotechnology, 162, 830–842.
Asghari, S. M., Pazhang, M., Ehtesham, S., Karbalaei-Heidari, H. R., Taghdir, M., Sadeghizadeh, M., et al. (2010). Protein Engineering, Design & Selection, 23, 599–606.
Ladero, M., Ruiz, G., Pessela, B. C. C., Vian, A., Santos, A., & García-Ochoa, F. (2006). Biochemical Engineering Journal, 31, 14–24.
Alexander, P. A., Ruan, B., & Bryan, P. N. (2001). Biochemistry, 40, 10634–10639.
Tremacoldi, C. R., Monti, R., Selistre-De-Araújo, H. S., & Carmona, E. C. (2007). World Journal of Microbiology & Biotechnology, 23, 295–299.
Zabolotskaya, M. V., Demidyuk, I. V., Akimkina, T. V., & Kostrov, S. V. (2004). The Protein Journal, 23, 483–492.
Cobos, A., & Estrada, P. (2003). Enzyme and Microbial Technology, 33, 810–818.
Smith, C. A., Toogood, H. S., Baker, H. M., Daniel, R. M., & Baker, E. N. (1999). Journal of Molecular Biology, 294, 1027–1040.
Lee, S., & Jang, D. J. (2001). Biophysical Journal, 81, 2972–2978.
Toogood, H. S., Smith, C. A., Baker, E. M., & Daniel, R. M. (2000). Biochemical Journal, 350, 321–328.
Tanaka, A., & Hoshino, E. (2002). Biochemical Journal, 364, 635–639.
Ortega, N., Diego, S., Rodríguez-Nogales, J. M., Perez-Mateos, M., & Busto, M. D. (2004). International Journal of Food Science Technology, 39, 631–639.
Bromberg, A., Marx, S., & Frishman, G. (2008). Biochimica et Biophysica Acta, 1784, 961–966.
Kazan, D., & Erarslan, A. (1997). Applied Biochemistry and Biotechnology, 62, 1–13.
Schokker, E. P., & van Boekel, M. A. J. S. (1998). The Journal of Dairy Research, 65, 261–272.
Braxton, S., & Wells, J. A. (1992). Biochemistry, 31, 7796–7801.
Acknowledgments
This study was supported by CNPq and CAPES, Brazil.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Daroit, D.J., Sant’Anna, V. & Brandelli, A. Kinetic Stability Modelling of Keratinolytic Protease P45: Influence of Temperature and Metal Ions. Appl Biochem Biotechnol 165, 1740–1753 (2011). https://doi.org/10.1007/s12010-011-9391-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-011-9391-z