Abstract
Low temperature is an important abiotic stress for plant development and has serious effects on crop production. Because tobacco is sensitive to low temperature, it is suitable for analyzing the mechanisms of cold response in plants. In the current study, NC567 and Taiyan8, two cultivars with different sensitivities to low temperature, were used in Isobaric Tags for Relative and Absolute Quantitation (iTRAQ)-based proteomics to uncover their different mechanisms in response to cold stress. A total of 4317 distinct proteins were identified and the differentially expressed proteins in four comparison sets were used for further analysis. The gene ontology (GO) analysis indicated that the majority of differentially expressed proteins were in the categories involved in metabolic and cellular processes. Surprisingly, there were 55 proteins decreased in NC567, but increased in Taiyan8 in response to cold, while the levels of 42 proteins were lower in Taiyan8 than NC567 at normal temperature, but higher in Taiyan8 than NC567 under cold treatment, suggesting different responses to cold stress in these cultivars. The levels of polypeptides involved in protein synthesis and degradation, photosynthesis, and respiration, as well as ROS scavenging, were different in the comparison sets, implying that protein and energy metabolisms may be important for the establishment of cellular environment at low temperature. In conclusion, our study identified the potential pathways involved in low-temperature response of tobacco and provides hints for the further improvement of cold tolerance in crops.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cold stress is a critical abiotic stress which has effects on the productivity and geographical distribution of many important crops. Low temperature has serious effects on physiological processes and results in damages of plant cells (Beck et al. 2004). Plants have evolved precise mechanisms to adapt to low temperature and the cold response is associated with changes in the levels of transcripts, proteins, and metabolites (Chinnusamy et al. 2007). Proteins involved in different physiological events are important players in cold stress response and development after exposure to low temperature. There are several types of proteins critical in cold response, such as structural, regulatory, and protective proteins (Thomashow 1999). For instance, the cold response is mediated by a complex protein cascade that ICE1 (inducer of CBF expression 1) activates the expression of CBFs (C-repeat binding factors) for downstream gene transcription (Chinnusamy et al. 2003). The variation of signaling pathway components or downstream compositions may contribute to the alternative tolerance to low temperature. There are significant variations in cold tolerance between genotypes dependent on differential expression levels of responsive genes (Chen et al. 2015). Therefore, characterization of the variation of protein compositions under low temperature in different genotypes of plants may provide possibility to uncover the cold response mechanism.
The response to cold stress in plants includes the alternations in the composition of the transcriptome, proteome, and metabolome. The mechanisms of cold tolerance have been studied in a range of plants, such as arabidopsis, barley and wheat, via transcriptome analysis to identify genes upregulated or downregulated in response to cold (Fowler and Thomashow 2002; Winfield et al. 2010; Greenup et al. 2011). However, these quantitative RNA data cannot absolutely be used to infer the protein levels in cold stress (Bogeat-Triboulot et al. 2007), resulted from the regulation of protein translation and degradation. Instead, proteome provides a tool to uncover the complete protein composition and more direct evidence to understand the physiological processes in this situation (Kosová et al. 2013). Recently, the proteomic data from rice, wheat, barley, and Arabidopsis elucidated the physiological mechanisms underlying low-temperature tolerance (Yan et al. 2006; Hlavackova et al. 2013; Rocco et al. 2013; Gharechahi et al. 2014). Compared with the control plants, the photosynthesis components are downregulated, but the glycolysis-related proteins are upregulated after cold acclimation (Han et al. 2013). Meanwhile, proteins involved in defense such as chaperones, dehydrins and ROS scavenging enzymes are accumulated after cold treatment (Xu et al. 2013). These changes of protein levels may be critical for the creation of cellular environment under low temperature.
Because tobacco is a model plant sensitive to low temperature, it provides a good system for studying the mechanism of cold response in plants. In recent years, comparative proteomics has been applied to reveal the protein changes in Cd tolerance, drought stress, and glandular trichome development in tobacco (Amme et al. 2005; Xie et al. 2014; Gharechahi et al. 2015). Following the rapid development of proteomic technologies, the liquid chromatography coupled with tandem mass spectrometry (MS) detection with isobaric tags for relative and absolute quantitaion (ITRAQ) has been broadly used for quantitative identification of proteins (Ross et al. 2004). Here, we use this powerful approach to compare the levels of proteins during low-temperature stress in two tobacco cultivars with different cold sensitivities. This work would provide important hints for understanding the mechanism of cold response in plants.
Materials and methods
Plant growth conditions
The tobacco cultivars NC567 and Taiyan8 were germinated on Murashige and Skoog (MS) plates with 2% (w/v) sucrose for around 3 weeks and were transferred to soil for around three additional weeks until the 6–8 leaf stage at 25 °C, with 16 light/8 h dark photoperiod (600 µmol/m−2s−1) and 80% of relative humidity. For cold treatment, the plants were moved to an incubator at 4 °C for 24 h, and plants growing under 25 °C were used as control. The third leaves from three independent plants were collected together as a sample for one biological replication in proteomic analysis. Two biological replications were performed.
Protein isolation
The leaf samples were treated in liquid nitrogen and ground with a pestle and mortar, and five times volume of TCA/acetone (1:9) was mixed with the sample by vortex. Then, it was kept at − 20 °C for 4 h, and spun down at 4 °C with 6000×g for 40 min. The precipitation was washed for three times using pre-cooled acetone. After air-dried, 20–30 mg sample was resuspended in 30 times volume of SDT buffer (150 mM Tris–HCl pH8.0, 4% SDS, and 100 mM DTT) and kept at 100 °C for 5 min. The sample was sonicated and then incubated at 100 °C for additional 15 min. After spun down for 40 min at 14000×g, 0.22 µm filters were used for filtration, and BCA Protein Assay Kit (Bio-Rad, USA) was applied for protein quantification. The proteins were stored at − 80 °C for further analysis.
Protein digestion and iTRAQ labeling
Following the previous protocol (Wisniewski et al. 2009), 200 μg of proteins was prepared in 30 μl SDT buffer. The contaminants were removed via ultrafiltration with UA buffer (150 mM Tris–HCl pH 8.0, 8 M Urea). Reduced cysteine residues were blocked via adding 100 μl 0.05 M iodoacetamide and stayed for 20 min in dark. After washing using UA buffer for three times and DS buffer (50 mM triethylammoniumbicarbonate, pH 8.5) for twice, the sample was treated overnight with 2 μg trypsin (Promega) at 37 °C in 40 μl DS buffer. The concentration of peptides was measured via 280 nm absorbance with extinction coefficient of 1.1 of 0.1% (g/l) solution (Wisniewski et al. 2009). The peptides then were, respectively, labeled using iTRAQ reagents (Applied Biosystems) (Sample1)—114, (Sample2)—115, (Sample3)—116, and (Sample4)—117. After labeling, the samples were multiplexed and dried with a vacuum.
Strong cation exchange (SCX) for peptide fractionation
SCX chromatography was performed using the AKTA Purifier system (GE Healthcare) with a PolySULFOETHYL 4.6 × 100 mm column. The samples were acidified in 2 ml buffer A (10 mM KH2PO4 in 25% of ACN, pH 2.7) for further chromatography and eluted at 1 ml/min with different concentrations of buffer B (10 mM KH2PO4 in 25% of ACN, pH 2.7, 500 mM KCl) (2 min with 0–10%, 25 min with 10–20%, 5 min with 20–45%, 5 min with 50–100% buffer B). The samples were measured at wavelength 214 nm and collected every minute. Around 30 fractions were combined to ten pools for desalting on C18 Cartridges (Empore™ SPE Cartridges C18, Sigma). The samples were concentrated using vacuum centrifugation and prepared in 40 µl of 0.1% (v/v) trifluoroacetic acid for further identification.
Liquid chromatography (LC)—electrospray ionization (ESI) tandem MS (MS/MS) analysis
An Easy nanoLC-coupled Q exactive mass spectrometer (Proxeon Biosystems) was used for analysis. 10 μl of sample (5 μg) was injected to a C18-reversed phase column (3 μm resin, 75 μm inner diameter, 10 cm long, Thermo Scientific) in 0.1% Formic acid and separated with a gradient of 80% acetonitrile at 250 nl/min for more than 140 min. A data-dependent top10 method which chooses the most abundant precursor ions dynamically from survey scan (300–1800 m/z) for HCD fragmentation was used for data acquisition, and predictive Automatic Gain Control (pAGC) was used to determine the target value. The resolution for survey scan was set to 70,000 at m/z 200 and the resolution for HCD spectra was set to 17,500 at m/z 200. Dynamic exclusion durations were set to 60 s. The underfill ratio was defined at 0.1% and normalized collision energy was defined at 30 eV. The machine was run with peptide recognition mode enabled. MASCOT engine (version 2.2, Matrix Science) embedded into Proteome Discoverer 1.3 (Thermo Electron) was used to search in the NCBI_nicotianoideae database. Following options were used in protein identification: MS/MS tolerance = 0.1 Da; peptide mass tolerance = 20 ppm; enzyme = Trypsin; missed cleavage = 2; variable modification = oxidation (M); Fixed modification = carbamidomethyl (C); FDR ≤ 0.01.
Results
Two tobacco cultivars display different sensitivities to low temperature
To dissect the mechanism of cold stress response in tobacco, a screening of tobacco cultivars sensitive or insensitive to low temperature was performed in a field experiment. As a result, NC567 displayed a cold insensitive phenotype and Taiyan8 was sensitive to cold stress. The cold sensitivity of these cultivars was confirmed by 24 h treatment at 4 °C (Fig. 1a); in contrast to NC567, the leaves of Taiyan8 started to wilt. The freezing tolerance of these cultivars was also tested at − 1 °C. After treatment, the Taiyan8 tobaccos wilted and were completely dead after recovery for 4 days at normal temperature. However, the growth of NC567 was not affected by the low-temperature treatment (Fig. 1b). These results supported that Taiyan8 is more sensitive to low temperature than NC567. To understand the reason for their different responses to low temperature, a comparative proteomics approach was applied. The plant materials from NC567 and Taiyan8 at 25 °C or treated at 4 °C for 24 h were collected for iTRAQ analysis. The experiment was designed as four comparisons: N24/N0 (NC567 for 24 h treatment vs. NC567 control); T24/T0 (Taiyan8 for 24 h treatment vs. Taiyan8 control); T0/N0 (Taiyan8 control vs. NC567 control); and T24/N24 (Taiyan8 for 24 h treatment vs. NC567 for 24 h treatment) (Fig. 1c).
Cold treatment of two tobacco cultivars for proteomic analysis. a Phenotypes of NC567 and Taiyan8 under the cold condition. After 4 °C treatment for 0, 4 and 24 h, the photos were taken. b Phenotypes of NC567 and Taiyan8 after freezing treatment. The plants after − 1 °C treatment for 3 days (top panel) and after 4-day recovery (bottom panel) are shown. c Comparison sets for the levels of differentially expressed proteins in NC567 and Taiyan8 with or without 24 h treatment at 4 °C. N24/N0 (NC567 for 24 h treatment vs. NC567 control); T24/T0 (Taiyan8 for 24 h treatment vs. Taiyan8 control); T0/N0 (Taiyan8 control vs. NC567 control); T24/N24 (Taiyan8 for 24 h treatment vs. NC567 for 24 h treatment)
Differential protein levels in NC567 and Taiyan8 in response to cold stress
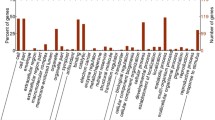
After the sample collection after treatment, proteins were analyzed by a strategy based on iTRAQ-based proteomics. A total of 4317 distinct proteins were identified and we focused on the proteins which were upregulated or downregulated more than 1.2 folds in both two biological independent experiments (Online Resource 1a). The detailed identification and quantification of these proteins are listed in Online Resource 1. After low-temperature treatment, 194 proteins were upregulated and 215 proteins were downregulated in NC567 (Online Resource 1b), while 205 proteins were upregulated and 109 proteins were downregulated in Taiyan8 (Online Resource 1c). It is also interesting whether the protein contents are different in these cultivars even under normal temperature. The analysis showed that the levels of 162 proteins were higher, but 213 proteins were lower in Taiyan8 than those in NC567 (Online Resource 1d). After 24 h treatment at 4 °C, the levels of 166 proteins were higher, but 123 proteins were lower in Taiyan8 than those in NC567 (Online Resource 1e). These data are summarized in Fig. 2a, suggesting that the protein composition is different in these genotypes in both normal and low temperature. The differentially expressed proteins were functionally categorized based on their GO annotations. In the current study, proteins were classified as biological processes, molecular functions, or cellular components. The GO patterns were similar in these four comparisons and the majority of differentially expressed proteins were in the categories involved in metabolic and cellular process, the proteins with catalytic and binding activity, the proteins reside in cytosol, and organelle (Fig. 2b, c, d, the detailed information is listed in Online Resource 2).
GO analysis of the proteins with differential levels in the proteomic data. a Number of differentially expressed proteins in comparison sets. Upregulated in white and downregulated in black. b GO analysis of all differentially expressed proteins by biological process. BP1: biological regulation, BP2: metabolic process, BP3: growth, BP4: localization, BP5: signaling, BP6: cellular component organization or biogenesis, BP7: single-organism process, BP8: multi-organism process, BP9: developmental process, BP10: response to stimulus, BP11: multicellular organismal process, BP12: reproductive process, BP13: cellular process, BP14: reproduction. c GO analysis of all differentially expressed proteins by molecular function. MF1: molecular transducer activity, MF2: structural molecule activity, MF3: transporter activity, MF4: catalytic activity, MF5: nucleic acid binding transcription factor activity, MF6: binding, MF7: receptor activity, MF8: enzyme regulator activity. d GO analysis of all differentially expressed proteins by cellular component. CC1: organelle, CC2: cell, CC3: macromolecular complex, CC4: extracellular region, CC5: membrane-enclosed lumen, CC6: membrane
The overlaps of protein comparisons in NC567 and Taiyan8 in low-temperature response
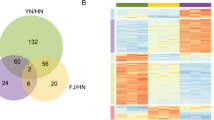
To study the protein candidates involved in low-temperature response, we analyzed the overlapping proteins in different comparison sets. The overlapping proteins with significant changes in both N24/N0 and T24/T0, as well as in both T0/N0 and T24/N24 were collected for further analysis. First, the proteins with significant changes in both N24/N0 and T24/T0 sets were compared (Fig. 3a, the detailed protein information is listed in Online Resource 3a). Surprisingly, there were 55 proteins decreased in N24/N0, but increased in T24/T0, indicating that the levels of these proteins are decreased in NC567, but increased in Taiyan8 in response to cold. This result implied that the mechanisms in response to low temperature may be dramatically different between NC567 and Taiyan8. Interestingly, the abscisic acid receptor PYL2 (Melcher et al. 2009), as well as the chromatin regulation-related factors SWI3D (Sarnowski et al. 2005) and HDA19 (Chen and Wu 2010) were found in this overlapping data set.
Overlap analysis of protein comparisons in NC567 and Taiyan8 in low-temperature response. The overlapping proteins with significant level changes in both N24/N0 and T24/T0, as well as in both T0/N0 and T24/N24 were collected. The number of proteins overlapped in the indicated comparison sets is shown. The upregulated group is indicated by an up arrow, and the downregulated group is indicated by a down arrow. a Overlaps between N24/N0 and T24/T0. b Overlaps between T0/N0 and T24/N24
The overlapping proteins in the T0/N0 and T24/N24 were also analyzed to show whether the difference in protein composition between these cultivars changes during cold stress (Fig. 3b, the detailed protein information is listed in Online Resource 3b). The levels of 19 proteins were increased and the levels of 19 proteins were decreased in both T0/N0 and T24/N24 sets, suggesting that some differentially expressed proteins in NC567 and Taiyan8 is not affected by low temperature. However, the levels of 42 proteins were lower in Taiyan8 at normal temperature, but higher in Taiyan8 than NC567 under cold treatment. Interestingly, the abscisic acid receptor PYL2 (Melcher et al. 2009) was also found in this overlapping data set.
The activities of protein and energy metabolism pathways may be different in NC567 and Taiyan8 in cold stress
To elucidate the potential mechanism for the different response to low temperature, the pathways associated with the differentially expressed proteins were analyzed. The proteins with level changes were involved in many pathways including carbon and energy metabolism, protein synthesis and processing, as well as signaling transduction. The majority of candidates were involved in protein and energy metabolism. Many proteins associated with protein synthesis, such as ribosome proteins, elongation factors, and tRNA ligases, were found in the comparison sets (Fig. 4a, Online Resource 4). 25 proteins involved in protein biogenesis were upregulated in N24/N0 (Online Resource 4a), as well as 13 proteins in this pathway were downregulated in T24/T0 (Online Resource 4b), suggesting that low temperature enhances protein synthesis in NC567, but inhibits it in Taiyan8. The data from T0/N0 and T24/N24 indicated that protein synthesis may be more active in NC567 under either normal or cold condition. The T0/N0 comparison also indicated that the levels of some proteins involved in protein transport, such as exocyst complex components, are decreased, but the levels of some chaperonins, such as heat shock proteins, are increased in Taiyan8 under normal temperature (Fig. 4a, Online Resource 4c). The data from T24/T0 showed that the polypeptides associated with protein degradation, such as ubiquitin ligases and proteasome subunits, are upregulated in Taiyan8 during low-temperature treatment (Fig. 4a, Online Resource 4b).
Protein and energy metabolism pathways may be different in NC567 and Taiyan8 in cold stress. The numbers of differentially expressed proteins associated with the indicated biological processes in different comparison sets are shown. The upregulated group is labeled in an up arrow, and the downregulated group is labeled in a down arrow. a Protein metabolism pathway: protein synthesis, degradation, transport, and chaperonin. b Energy metabolism pathway: photosynthesis, respiration, and anti-oxidation
The proteins involved in energy metabolisms such as photosynthesis and respiration were also abundant in the comparison sets (Fig. 4b, Online Resource 4). The data from N24/N0 and T24/T0 suggested that the more proteins associated with photosynthesis, such as chlorophyll binding proteins, were upregulated in NC567 (Online Resource 4a), but downregulated in Taiyan8 (Online Resource 4b) in response to cold stress. However, the numbers of upregulated proteins involved in photosynthesis are both more in T0/N0 and T24/N24 data sets (Online Resource 4c, d). The data from N24/N0 and T24/T0 indicated that more proteins associated with respiration, such as enzymes in glycolysis and electron transport chain, are increased in both NC567 and Taiyan8 in cold treatment (Online Resource 4a, b). The result from T0/N0 showed that the proteins involved in the respiration pathway are a little more in Taiyan8 than NC567 under normal condition (Online Resource 4c). The number of proteins related to anti-oxidation pathway is also higher in Taiyan8 in either normal or cold condition. Surprisingly, the proteins with the highest increase in both T0/N0 and T24/N24 (Online Resource 4c, d) were superoxide dismutase (SOD) and thioredoxin-like protein CDSP32 (Carmel-Harel and Storz 2000; Alscher et al. 2002), which are involved in ROS scavenging.
Discussion
Low-temperature stress is an important environment stresses affecting plant distribution and performance. In recent years, advances have been made to discover the cold signaling and regulatory pathways (Miura and Furumoto 2013). We tried to reveal the mechanisms how plants are in response to cold stress using tobacco, because compared with other model plants, tobacco is more sensitive and there are many cultivars with different sensitivity to low temperature, providing a possibility to reveal the fine-tuning mechanism. Two tobacco cultivars used in this study displayed difference under low temperature and are suitable for mechanism comparison. Because proteins are critical performers in various processes involved in stress responses, protein composition is important for understanding the differential strategies in these cultivars. The iTRAQ-based proteomics provides quantitative protein information for direct analysis on the potential biological pathways associated with cold stress. The GO analysis indicated that the majority of differentially expressed proteins are involved in catalytic metabolism and cellular process, implying that metabolism may be critical for creation of intracellular environment under low temperature (Bohnert and Sheveleva 1998).
The results from the overlapping analysis between four comparison sets showed that 55 proteins are downregulated in NC567 but upregulated in Taiyan8 under cold treatment, suggesting that the response mechanisms in these cultivars are different. The chromatin remodeling factor SWI3D and the histone deacetylase HDA19 were found in this overlapping data set. SWI3D is a component in the SWI/SNF chromatin remodeling complex (Sarnowski et al. 2005), which is involved in gene transcriptional regulation in various stress processes, while the activity of HDA19 in Arabidopsis is associated with development and salt tolerance (Tanaka et al. 2008; Chen and Wu 2010); therefore, their opposite response in these cultivars in response to low temperature implied that epigenetics may be important for downstream gene regulation in these situations. Both SWI3D and HDA19 mutants in Arabidopsis display defects in development, but their roles in cold response are unknown, and it would be interesting to test their functions under low temperature in further study. At the same time, the data from T0/N0 and T24/N24 suggested that the levels of 42 proteins are higher in NC567 at normal temperature, but lower in NC567 after cold treatment, compared with Taiyan8. These results suggested that the proteins with changed levels may be important for the establishment of the cellular environment under cold stress. Surprisingly, an ABA receptor PYL2 was found in this overlapping data set. Given that ABA is a critical hormone for signaling transduction in abiotic stresses (Tuteja 2007), while PYL proteins have been identified as ABA receptors in Arabidopsis (Melcher et al. 2009), the higher expression of PYL2 in NC567 at normal temperature may increase the ABA signaling perception and easily establish a cellular environment in response to low temperature.
It is very interesting that the majority proteins varying between different samples are associated with protein and energy metabolisms. Because proteins are direct performers in various biological processes, the protein synthesis and degradation may be related to the general activities of macromolecular machines for maintenance of cellular environment during stress treatment (Hahn and Walbot 1989). For instance, low temperature significantly affects proteins synthesis in wheat, possibly through increasing the levels of amino acid synthases, ribosome-related proteins, and RNA-binding proteins (Herman et al. 2006). In our proteomics data, the polypeptides involved in protein synthesis, such as ribosome subunits, elongation factors, and tRNA amidotransferases, are upregulated in NC567, but downregulated in Taiyan8 under cold treatment, suggesting that protein synthesis may be more active in NC567. A previous study showed that Arabidopsis ribosomal maturation factors are required for leaf growth in cold (Schmidt et al. 2013); therefore, downregulation of ribosome subunits in Taiyan8 may be a reason for its cold sensitivity. In addition, more polypeptides in protein degradation pathway, such as proteasome subunits, are upregulated in Taiyan8 at low temperature, possibly attenuating the efficiency of protein machines involved in cold response. Taken together, from our differential data in NC567 and Taiyan8, the balance between protein synthesis and degradation may be a critical regulatory mechanism for the plant performance in cold stress.
Given that cold adaption is an active and energy-demanding process, the regulation on the processes associated with energy production may be required in response to low temperature (Paredes and Quiles 2015). The previous study in rice showed that photosynthesis is downregulated, but glycolysis is upregulated when exposed to low temperature (Yan et al. 2006), suggesting a potential mechanism on the balance of energy in stress. Interestingly, under normal condition, more proteins involved in both photosynthesis and respiration are upregulated in Taiyan8, implying that the level of the basic energy metabolism may be higher in this cultivar. For instance, the protein levels of Rubisco large and small subunits, as well as components in photosystems I and II, are increased in Taiyan8. During cold stress, photosynthesis may be attenuated, but respiration may be even more active, resulting in imbalance of energy flow in Taiyan8. In addition, higher level of photosynthesis is associated with the accumulation of ROS (Foyer and Shigeoka 2011), resulting in oxidative damage, which contributes to stress injury. Therefore, the scavenging system may be activated in plant cells at low temperature to escape from the damage induced by ROS (Gill and Tuteja 2010). For instance, the levels of ROS scavengers are dramatically increased by cold stress in Arabidopsis and rice (Lee et al. 2007; Fanucchi et al. 2012). In our study, the levels of ROS scavenging enzymes, such as superoxide dismutase, thioredoxin-like protein, and chaperonin (Carmel-Harel and Storz 2000; Alscher et al. 2002), are much higher in Taiyan8 than those in NC567 under both normal and cold conditions, suggesting that the ROS accumulation may be higher in Taiyan8 and it may be another possible reason for the cold sensitivity in this cultivar. Taken together, our data support that protein metabolism and energy balance may be critical for the different performance of two tobacco cultivars in cold stress response. In the further study, more physiological and genetics evidence are needed to support this conclusion and it would be helpful for molecular design of cold tolerant crops in the future.
Author contribution statement
RH, ZL, CY, and JL conceived the project and designed the research; RH, SX, XZ, XZ, ZL, and LZ prepared samples for proteomics; RH, YC, CY, and JL analyzed the data; and JL wrote the manuscript.
References
Alscher RG, Erturk N, Heath LS (2002) Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J Exp Bot 53:1331–1341
Amme S, Rutten T, Melzer M, Sonsmann G, Vissers JP, Schlesier B, Mock HP (2005) A proteome approach defines protective functions of tobacco leaf trichomes. Proteomics 5:2508–2518
Beck EH, Heim R, Hansen J (2004) Plant resistance to cold stress: mechanisms and environmental signals triggering frost hardening and dehardening. J Biosci 29:449–459
Bogeat-Triboulot MB, Brosche M, Renaut J, Jouve L, Le Thiec D, Fayyaz P, Vinocur B, Witters E, Laukens K, Teichmann T, Altman A, Hausman JF, Polle A, Kangasjarvi J, Dreyer E (2007) Gradual soil water depletion results in reversible changes of gene expression, protein profiles, ecophysiology, and growth performance in Populus euphratica, a poplar growing in arid regions. Plant Physiol 143:876–892
Bohnert HJ, Sheveleva E (1998) Plant stress adaptations—making metabolism move. Curr Opin Plant Biol 1:267–274
Carmel-Harel O, Storz G (2000) Roles of the glutathione- and thioredoxin-dependent reduction systems in the Escherichia coli and saccharomyces cerevisiae responses to oxidative stress. Annu Rev Microbiol 54:439–461
Chen LT, Wu K (2010) Role of histone deacetylases HDA6 and HDA19 in ABA and abiotic stress response. Plant Signal Behav 5:1318–1320
Chen H, Chen X, Chen D, Li J, Zhang Y, Wang A (2015) A comparison of the low temperature transcriptomes of two tomato genotypes that differ in freezing tolerance: Solanum lycopersicum and Solanum habrochaites. BMC Plant Biol 15:132
Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong X, Agarwal M, Zhu JK (2003) ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev 17:1043–1054
Chinnusamy V, Zhu J, Zhu JK (2007) Cold stress regulation of gene expression in plants. Trends Plant Sci 12:444–451
Fanucchi F, Alpi E, Olivieri S, Cannistraci CV, Bachi A, Alpi A, Alessio M (2012) Acclimation increases freezing stress response of Arabidopsis thaliana at proteome level. Biochem Biophys Acta 1824:813–825
Fowler S, Thomashow MF (2002) Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14:1675–1690
Foyer CH, Shigeoka S (2011) Understanding oxidative stress and antioxidant functions to enhance photosynthesis. Plant Physiol 155:93–100
Gharechahi J, Alizadeh H, Naghavi MR, Sharifi G (2014) A proteomic analysis to identify cold acclimation associated proteins in wild wheat (Triticum urartu L.). Mol Biol Rep 41:3897–3905
Gharechahi J, Hajirezaei MR, Salekdeh GH (2015) Comparative proteomic analysis of tobacco expressing cyanobacterial flavodoxin and its wild type under drought stress. J Plant Physiol 175:48–58
Gill SS, Tuteja N (2010) Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem 48:909–930
Greenup AG, Sasani S, Oliver SN, Walford SA, Millar AA, Trevaskis B (2011) Transcriptome analysis of the vernalization response in barley (Hordeum vulgare) seedlings. PLoS One 6:e17900
Hahn M, Walbot V (1989) Effects of cold-treatment on protein synthesis and mRNA levels in rice leaves. Plant Physiol 91:930–938
Han Q, Kang G, Guo T (2013) Proteomic analysis of spring freeze-stress responsive proteins in leaves of bread wheat (Triticum aestivum L.). Plant Physiol Biochem 63:236–244
Herman EM, Rotter K, Premakumar R, Elwinger G, Bae H, Ehler-King L, Chen S, Livingston DP 3rd (2006) Additional freeze hardiness in wheat acquired by exposure to − 3 °C is associated with extensive physiological, morphological, and molecular changes. J Exp Bot 57:3601–3618
Hlavackova I, Vitamvas P, Santrucek J, Kosova K, Zelenkova S, Prasil IT, Ovesna J, Hynek R, Kodicek M (2013) Proteins involved in distinct phases of cold hardening process in frost resistant winter barley (Hordeum vulgare L.) cv Luxor. Int J Mol Sci 14:8000–8024
Kosová K, Vítámvás P, Urban MO, Klíma M, Roy A, Prášil IT (2013) Biological networks underlying abiotic stress tolerance in temperate crops—a proteomic perspective. Int J Mol Sci 16:20913–20942
Lee DG, Ahsan N, Lee SH, Kang KY, Lee JJ, Lee BH (2007) An approach to identify cold-induced low-abundant proteins in rice leaf. CR Biol 330:215–225
Melcher K, Ng LM, Zhou XE, Soon FF, Xu Y, Suino-Powell KM, Park SY, Weiner JJ, Fujii H, Chinnusamy V, Kovach A, Li J, Wang Y, Li J, Peterson FC, Jensen DR, Yong EL, Volkman BF, Cutler SR, Zhu JK, Xu HE (2009) A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature 462:602–608
Miura K, Furumoto T (2013) Cold signaling and cold response in plants. Int J Mol Sci 14:5312–5337
Paredes M, Quiles MJ (2015) The effects of cold stress on photosynthesis in hibiscus plants. PLoS One 10:e0137472
Rocco M, Arena S, Renzone G, Scippa GS, Lomaglio T, Verrillo F, Scaloni A, Marra M (2013) Proteomic analysis of temperature stress-responsive proteins in Arabidopsis thaliana rosette leaves. Mol BioSyst 9:1257–1267
Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteom 3:1154–1169
Sarnowski TJ, Rios G, Jasik J, Swiezewski S, Kaczanowski S, Li Y, Kwiatkowska A, Pawlikowska K, Kozbial M, Kozbial P, Koncz C, Jerzmanowski A (2005) SWI3 subunits of putative SWI/SNF chromatin-remodeling complexes play distinct roles during Arabidopsis development. Plant Cell 17:2454–2472
Schmidt S, Dethloff F, Beine-Golovchuk O, Kopka J (2013) The REIL1 and REIL2 proteins of Arabidopsis thaliana are required for leaf growth in the cold. Plant Physiol 163:1623–1639
Tanaka M, Kikuchi A, Kamada H (2008) The Arabidopsis histone deacetylases HDA6 and HDA19 contribute to the repression of embryonic properties after germination. Plant Physiol 146:149–161
Thomashow MF (1999) PLANT COLD ACCLIMATION: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol 50:571–599
Tuteja N (2007) Abscisic acid and abiotic stress signaling. Plant Signal Behav 2:135–138
Winfield MO, Lu C, Wilson ID, Coghill JA, Edwards KJ (2010) Plant responses to cold: transcriptome analysis of wheat. Plant Biotechnol J 8:749–771
Wisniewski JR, Zougman A, Nagaraj N, Mann M (2009) Universal sample preparation method for proteome analysis. Nat Methods 6:359–362
Xie L, He X, Shang S, Zheng W, Liu W, Zhang G, Wu F (2014) Comparative proteomic analysis of two tobacco (Nicotiana tabacum) genotypes differing in Cd tolerance. Biometals 27:1277–1289
Xu J, Li Y, Sun J, Du L, Zhang Y, Yu Q, Liu X (2013) Comparative physiological and proteomic response to abrupt low temperature stress between two winter wheat cultivars differing in low temperature tolerance. Plant Biol 15:292–303
Yan SP, Zhang QY, Tang ZC, Su WA, Sun WN (2006) Comparative proteomic analysis provides new insights into chilling stress responses in rice. Mol Cell Proteom 5:484–496
Acknowledgements
This work was supported by the grants from the Key Science and Technology Project of China National Tobacco Company (110201302002), the Key Science and Technology Project of Hunan Provincial Tobacco Company (13-17ZDAa01), the National Natural Science Foundation of China 31400314, the University Innovation Program from the Department of Education of Guangdong province (2014), and Guangzhou Scientific and Technological Program (201607010377).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Stobiecki.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hu, R., Zhu, X., Xiang, S. et al. Comparative proteomic analysis reveals differential protein and energy metabolisms from two tobacco cultivars in response to cold stress. Acta Physiol Plant 40, 19 (2018). https://doi.org/10.1007/s11738-017-2582-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11738-017-2582-7