Abstract
Tobacco can easily accumulate cadmium (Cd) in leaves and thus poses a potential threat to human health. Cd-stress-hydroponic-experiments were performed, and the proteomic and transcriptional features of two contrasting tobacco genotypes Yun-yan2 (Cd-tolerant) and Guiyan1 (Cd-sensitive) were compared. We identified 18 Cd-tolerance-associated proteins in leaves, using 2-dimensional gel electrophoresis coupled with mass spectrometry, whose expression were significantly induced in Yunyan2 leaves but down-regulated/unchanged in Guiyan1, or unchanged in Yunyan2 but down-regulated in Guiyan1 under 50 µM Cd stress. They are including epoxide hydrolase, enoyl-acyl-carrier-protein reductase, NPALDP1, chlorophyll a–b binding protein 25, heat shock protein 70 and 14-3-3 proteins. They categorized as 8 groups of their functions: metabolism, photosynthesis, stress response, signal transduction, protein synthesis, protein processing, transport and cell structure. Furthermore, the expression patterns of three Cd-responsive proteins were validated by quantitative real-time PCR. Our findings provide an insight into proteomic basis for Cd-detoxification in tobacco which offers molecular resource for Cd-tolerance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Tobacco (Nicotiana tabacum), being a model organism and an important cash crop in the world, can easily accumulate heavy metals in leaves, and heavy metals is then inhaled into the bronchial system during cigarette smoking (Golia et al. 2007). Consequently, smoking can be one of the major sources of heavy metals intake in humans. Among these heavy metals, cadmium (Cd) is an extreme hazardous and widespread contamination which induces various diseases such as kidney disorders, neurotoxicity and osteoporosis (Park et al. 2012). For the large-scale slightly or moderately contaminated farmlands, thus development of Cd tolerant cultivars and limiting plant Cd uptake could offer a cost-effective and practically acceptable strategy for better utilization of natural resource and safe food production. Therefore, understanding of the mechanisms underlying tobacco tolerance to Cd toxicity is necessary for developing the cultivars with improved Cd tolerance and minimized Cd accumulation.
Studies on various plant species have revealed that Cd is strongly phytotoxic and causes growth inhibition and even plant death (Wu et al. 2003, 2004). Its toxicity can be attributed to substituting elements in the active sites of proteins and breaking down disulfide bonding in proteins (Verbruggen et al. 2009). In addition, Cd will induce the generation of reactive oxygen species (ROS), which may attack cellular macromolecules leading to proteins degradation (Gallego et al. 2012). Thus the plants exposed to Cd stress will change their protein expression profiles, and the changed pattern and extent might vary with the genotypes, depending on their response to Cd stress. As a global approach, comparative proteomic analyses via two-dimensional gel electrophoresis (2-DE) with mass spectrometry (MS) and bioinformatics techniques provide an ideal way to identify the proteins expressed differently under abiotic stress. Some studies have been conducted on the responses of proteomic levels to Cd toxicity in plants such as rice (Lee et al. 2010; Cai et al. 2011), barley (Schneider et al. 2009), Arabidopsis (Sarry et al. 2006), Brassica juncea (Alvarez et al. 2009), spinach (Fagioni and Zolla 2009) and tomato (Rodríguez-Celma et al. 2010). Up-regulation of the MRP-type ABC transporter AtMRP3 was observed under Cd stress (Bovet et al., 2003). Cuypers et al. (2005) observed Cu stress induced PvPR1 and PvPR2 specific protein in bean. Ndimba et al. (2005) detected 266 protein spots significantly changed in abundance after NaCl and sorbitol treatments. Nevertheless, compared to the analysis of the plant proteome in response to abiotic stresses is still limited. Furthermore, so far there is no study on genotypic difference concerning the changes in protein pattern of tobacco under Cd stress. So that precise knowledge of the proteomic basis is required to dissect the mechanisms underlying Cd tolerance in tobacco.
The present study reports genotypic difference in Cd-induced changes in protein pattern of two tobacco genotypes differing in Cd tolerance using 2-DE. These results would be useful to understand the mechanisms of Cd tolerance in tobacco, and will open novel prospective for the improvement of Cd tolerance in plants.
Materials and methods
Plant material and experiment design
A hydroponic experiment was performed in greenhouse at Zijingang campus, Zhejiang University, Hangzhou, China. There were two tobacco genotypes Guiyan1 and Yunyan2 identified as Cd- tolerant and sensitive genotypes (Liu et al. 2014) respectively. Seeds were surface sterilized by soaking in 2 % H2O2 for 20 min, fully rinsed by deionized water, and then soaked in deionized water at 25 °C for 1 d and germinated in sterilized moist vermiculite in a growth room at 22–25 °C. After germination and growing into 3-leaf stage (approximatly 40 days old), uniform seedlings were chosen to transplant into 5-L black plastic pots fulfilled with basal nutrient solution. Each container was covered with polystyrol plates having 5 evenly spaced holes (1 plant per hole). On the 5th day after transplanting, the plants were treated with 0–50 µM Cd (CdCl2). Split plot experiment was applied with the treatments as the main plot and the genotypes as the sub plot with 5 replicates. After 5 days Cd exposure, fresh leaves were collected. The composition of the basic nutrient solution was (mg L−1): (NH4)2SO4, 48.2; MgSO4.7H2O, 154.88; K2SO4, 15.9; KNO3, 18.5; KH2PO4, 24.8; Ca(NO3)2.4H2O, 86.17; Fe-Citrate, 7.0; MnCl2.4H2O, 0.9; ZnSO4.7H2O, 0.11; CuSO4.5H2O, 0.04; H3BO3, 2.9 and H2MoO4, 0.01. The solution pH was adjusted to 6.0 ± 1 with HCl or NaOH as required, and the nutrient solution was continuously aerated with pumps.
Protein extraction and quantification
Total protein was extracted according to Carpentier et al. (2005) with some modification (Bah et al. 2010). Approximatly 3 g tobacco leaf samples were grounded in a mortar to a fine powder in liquid nitrogen and homogenized in an extraction buffer containing 30 mg PVPP. The powder was suspended in 7 ml ice-cold phenol extraction buffer (0.7 M sucrose; 0.1 M KCl; 50 mM EDTA; 0.5 M Tris–HCl; 0.1 % (w/v) DTT, pH 7.5; complete protease inhibitor cocktail (Roche Applied Science)) and added 7 ml ice-cold Tris buffered phenol immediately and vortex mixed for 15 s. Then samples were vortex mixed at 4 °C for 10 s every 5 min and repeated for 6 times. After centrifugation for 30 min at 5,000×g, under 4 °C, samples were collected and re-extracted with 15 ml of extraction buffer and vortex mixed at 4 °C for 10 s every 5 min and repeated for 6 times. After centrifugation (30 min, 5,000×g, 4 °C) the phenolic phase was collected and mixed with the phenolic phase gathered before. Then five volumes 100 mM ammonium acetate in methanol at −20 °C was added into phenolic phase and the mixed liquor were homogenized by Vortexer to precipitate at 20 °C. After centrifugation (30 min, 5,000×g, 4 °C), the supernatant was discarded to get deposit which then was washed with 0.2 % DTT in acetone for twice. At the interval between the two washing steps, samples were put at −20 °C for 60 min. After air dried, deposit was suspended in 200 μl lysis buffer (7 M urea; 2 M thiocarbamide; 4 % CHAPS; 20 mM Tris–HCl, pH 7.4, 1 % (w/v) DTT; Amersham Biosciences), then vortex mixed for 1 h at ambient temperature. Total protein concentration was measured by standard Bradford assay with bovine serum albumin as standard (Bio-Rad, Hercules, CA, USA).
2-dimensional gel electrophoresis analysis
Proteins were separated by 2-DE and then these proteins were visualized by silver staining (Bah et al. 2010; Carpentier et al. 2005). For each sample, the protein extractions were conducted twice independently, and for each protein sample two 2-DE analyses were applied. If not further specified in the text, the chemicals used were all P.A. or electrophoresis grade. All electrophoresis units used were purchased from Amersham Biosciences.
Protein visualization, image analysis and quantification
To analyze the expressed protein patterns, PowerLook1100 scanner (UMAX) were employed to scan and calibrate stained gels, followed by GE HealthCare Software (Amersham Biosciences) to analyze protein spots. The spots were quantified employing the % volume criterion. Only those with significant and reproducible changes (P < 0.05) were identified as differentially accumulated proteins. The target protein spots were automatically excised from the stained gels and digested with trypsin using a Spot Handling Workstation (Amersham Biosciences). Peptides gel pieces were put into the EP tube and washed with 1:1 mixture of 50 μl of 30 mM K3Fe(CN)6 and 100 mM NaS2O3 for 10–15 min until discolored completely, then washed with 200 μl bi-distilled water (two times for 5 min each). The washed solution was drained and washed with 50 % ACN (acetonitrile, Fisher A/0626/17) and 100 % ACN rotationally, and then incubated in 25 mM NH4HCO3 (Sigma A6141) for 5 min at 37 °C. After absorbed out of the incubation solvent, 50 % ACN and 100 % ACN was rotationally added and dried at 40 °C for 5 min respectively. Trypsin digestion was carried out as follows: sequencing-grade porcine trypsin (Promega, Madison, WI, USA) was suspended in 25 mM NH4HCO3 at a concentration of 12.5 ng/μl to rehydrate the dried gel pieces. The trypsin digestion was carried out for 16 h at 37 °C. Peptides extraction from the digest was conducted as follows for three times: 10 μl of 50 % ACN containing 0.1 % TFA (trifluoroacetic acid, GE HealthCare) was added to each tube and incubated for 5 min at 37 °C and transferred the supernatants to new EP tube. The extracts were pooled and then vacuum concentrated for about 2 h. A solution of peptides was filtrated with Millipore (Millipore ZTC18M096) and mixed with the same volume of a matrix solution consisting of saturated α-cyano-4-hydroxycinnamic acid (CHCA) in 50 % ACN containing 0.1 % TFA. The peptides were then co-crystallized with CHCA by evaporating organic solvents, and masses of tryptic-digested peptide were measured with a MALDI-TOF-TOF mass spectrometer (ABI4700 System, USA). All mass spectra were recorded in positive reflector mode and generated by accumulating data from 1,000 laser shots. The following threshold criteria and settings were used: detected mass range of 700–3,200 Da (optimal resolution for the quality of 1,500 Da), using a standard peptide mixture (des-Argl-Bradykinin Mr904.468, Angiotensin I Mr1296.685, Glul-Fihrinopeptide B Mr1570.677, ACTH (1–17) Mr2093.087, ACTH (18–39) Mr2465.199; ACTH (7–38) Mr3657.929) as an external standard calibration, with laser frequency of 50 Hz, repetition rate of 200 Hz, UV wavelength of 355 nm, and accelerated voltage of 20,000 V. Peptide mass fingerprint data were matched to the NCBInr database using Profound program under 50 ppm mass tolerance.
Peptide and protein identification by database search
Data was processed via the Data Explorer software and proteins were unambiguously identified by searching against a comprehensive non-redundant sequence database using the MASCOT software search engine (http://www.matrixscience.com/cgi/searchform.pl?FORMVER=2&SEARCH=MIS). The search parameters were shown as follows: (1) peptide quality of 800–4,000 Da, mass tolerance for the fragment ion of 0.25 Da; (2) a minimum of seven matching peptides; (3) one missed cleavage; (4) Taxonomy: Viridiplantae (green plants); and (5) allowed modifications, carbamidomethylation of Cys (complete) and oxidation of Met (partial). Moreover, in order to evaluate protein identification, we considered the percentage of sequence coverage, the observation of distribution of matching peptides (authentic hit is often characterized by peptides that are adjacent to one another in the sequence and that overlap), the distribution of error (distributed around zero), the gap in probability and score distribution from the first to other candidate; only matches with over 90 % sequence identity and a maximum e-value of 10−10 were considered.
Fold increase and decrease in Cd exposed leaves vs. unexposed plants were calculated as treated/control and -control/treated for up- and down-regulated proteins, respectively. For single-peptide identified proteins, positive/negative regulation was assigned when it was shown that the regulation factors were above 1.5 (P < 0.05).
qRT-PCR verification
Total RNA in leaves was extracted by TRIzol® reagent (Invitrogen, Carlsbad, CA, USA). All RNA samples were quantified and examined for protein contamination (A260 nm/A280 nm ratios) and reagent contamination (A260 nm/A230 nm ratios) by a Nanodrop ND 1000 spectrophotometer (Nanodrop technologies, Wilmington, DE, USA). Then, RT-PCR was applied with 1 µg total RNA and PrimeScriptTM RT reagent Kit with gDNA Eraser (TAKARA, Japan) to get single strand cDNA. The total volume of qRT-PCR mixture was 25 µl containing 12.5 µl SYBR green (SYBR®Premix Ex TaqTM (TliRNaseH Plus), TAKARA, Japan), 0.5 µl forward and reverse primers respective, 9.5 µl ddH2O and 2.0 µl cDNA. QRT-PCR was done in CFX96™ Real-Time PCR Detection System (Bio-Rad, USA) like this: 95 °C for initial denaturation, 30 s; 95 °C for 5 s and 60 °C for 30 s, 40 cycles. The primers of chosen protein gene were displayed in Table 1. Actin gene was used as control whose primers designed as shown: forward-5′-TGGCTGACGGTGAGGACA-3′, reverse-5′-CGAGGGCGACCAACTATG-3′.
Results
2-dimensional gel electrophoresis analysis
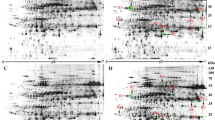
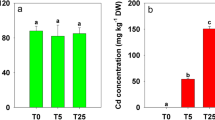
As listed in Table S1, Yuyan2 showed more tolerant to Cd stress, as recorded less Cd-induced reduction in plant height, fresh weight and SPAD value over the control compared with Guiyan1. Figure 1 shows the entire image of the silver blue-stained 2-DE gels of total extracted proteins from tobacco leaves under normal and Cd conditions. The averages of protein spots of 2-DE gels in control and Cd were 2,608 and 2,412 in Yunyan2, and 2,077 and 2,359 in Guiyan1, respectively. Comparing 2-DE gels from control and from Cd treated samples showed many differences in protein presence. A 1.5-fold quantitative change was set as the criteria (Cd vs. control). Overall, 220 and 206 protein spots were found to be altered by 50 µM Cd in Yunyan2 and Guiyan1, respectively, with 129 and 123 up-regulated by Cd, respectively, while 91 and 83 down-regulated (Fig. 2). Further comparing the data of the two genotypes, 20 protein spots appear to be related to Cd stress tolerance, whose expression were significantly up-regulated by 50 µM Cd (Cd vs. control, fold change >1.50) in Yunyan2 but unchanged/down-regulated in Guiyan2, or unchanged in Yunyan2 but down-regulated in Guiyan1. Therefore, these 20 protein spots were selected for MALDI-TOF-TOF MS analysis. All theses 20 protein spots identified as having significant changes expression levels and present in sufficient amounts to be visible on a coomassie-stained preparative gel were excised for mass spectrometric analysis. The resulting spectra of 20 protein spots were used to query databases for protein identification.
Representative 2-DE maps of Yunyan2 leaf proteins isolated from control (upper) and 50 μM Cd (below) treated plants. Total proteins were extracted and separated by 2-DE. In IEF, 100 mg proteins were loaded onto pH 4–7 IPG strips (24 cm, linear). SDS-PAGE was performed with 12.5 gels. The spots were visualized by silver staining. Arrows indicate the differentially expressed protein spots: Y1-Y18 (as shown in Table 2), whose expressions were significantly induced in Yunyan2 leaves but down-regulated (−)/unchanged in Guiyan1, or unchanged in Yunyan2 but down-regulated in Guiyan1 under 50 µM Cd stress (Cd vs. control by a factor 1.5)
Venn diagram illustrating the expression patterns of Cd stress-responsive proteins in leaves of Yunyan2 (Y) and Guiyan1 (G). The numbers of different expressed spots up- or down-regulated are shown in different segments. As to protein spots altered by 50 µM Cd stress, 1,059, 881 and 781 spots were up-regulated (up), non-changed (NC) and down-regulated (down) in Yunyan2 respectively; while 1,053, 895 and 773 spots were up-regulated, non-changed and down-regulated in Guiyan1. For the 1,059 proteins up-regulated in Yunyan2, 1,043, 14 and 2 proteins were up-regulated, non-changed and down-regulated in Guiyan1 simultaneously, and for 881 proteins non-changed in Yunyan2, the numbers were 4, 873 and 4
MALDI-TOF-TOF MS analysis and identification of proteins
From the excised protein spots, a total of 20 proteins with significantly differential expression levels were analyzed by MALDI-TOF-TOF MS. Two of them had no MS/MS data, although they could be identified by PMF data, their theoretical MW and pI did not fit well with the experimental ones. Their identities need to be further confirmed. Only 18 regulated proteins were unambiguously identified by MS and data bank analysis (Table 2; Figs. 1, 3). Parts of identified proteins had different pI value with the theoretical ones. This phenomenon is prevalent in proteomic analyses, and may be a consequence of post-translational modifications, proteolytic cleavage (Ahsan et al. 2009). The resulting spectra of the 18 protein spots were identified using MASCOT software search engine against N. tabacum and homologous proteins of other green plants in the NCBI non-redundant (nr) protein database and tobacco ESTs databases. These proteins were classified into 8 categories based on biochemical functions. The majority of the protein profile was photosynthesis (spots Y3–Y6) and metabolism (spots Y1, Y2), followed by signal transduction (spots Y9–Y11), protein processing (spots Y13–Y15) and stress response (spots Y7, Y8). Among them, two proteins involved in metabolism were epoxide hydrolases (EHs, spot Y1) and putative enoyl-acyl carrier protein (ACP) reductases (ENRs, spot Y2). Four photosynthesis related proteins were plastidic aldolase NPALDP1 (spot Y3), glyceraldehyde-3-phosphate dehydrogenase B (GAPDH subunits B, spot 5), oxygen evolving complex 33 kDa photosynthesis system II (PSII) protein (OEC33, spot Y6), and chlorophyll a–b binding protein 25 (CAB25, spot Y4). Two stress response related proteins were heat shock protein 70 (HSP70, spot Y7) and ascorbate peroxidase (APX, spot Y8). Three 14-3-3 proteins (spots Y9-11) were 14-3-3 protein 6, 14-3-3 protein isoform g and 14-3-3 protein omega1. Three protein processing/translaction proteins (Spot Y13-15) were mitochondrial processing peptidase (MPP), ATP-dependent Clp protease ATP-binding subunit clpA (ClpA) and ATP-dependent Clp protease proteolytic subunit (ClpP). The other minor groups were protein synthesis (Eukaryotic initiation factor (eIF) 4A, spot Y12), transport (Vacuolar H+-ATPase A2 subunit isoform, spot Y16); GDP dissociation inhibitor (GDI, spot Y17) and cell structure (Alpha-1,4-glucan-protein synthase (UPTG), spot Y18).
‘Spot view’ of the abundance of differentially expressed proteins in leaves of yunyan2 under 50 µM Cd for 5 days. The images of two genotypes: Yunyan2 and Guiyan1 (indicated with green circles) from control and 50 µM Cd treated plants. Protein spot ID refers to numbers in Table 2; Fig. 1. (Color figure online)
qRT-PCR verification
To verify whether the changes in protein abundance detected by 2-DE were correlated with changes at the transcript level, qRT-PCR was performed using RNA isolated from the leaves of a separate set of plants treated with 0 or 50 μM Cd (Fig. 4). The mRNA accumulation of three selected proteins, including mitochondrial processing peptidase (spot Y13), ATP-dependent Clp protease proteolytic subunit (spot Y15) and GDP dissociation inhibitor (spot Y17), were successfully found consistent with their protein levels. Following the expression trend detected by 2-DE (Table 2), spots Y15 and Y17 were up-regulated by Cd stress in Yunyan2 but down regulated in Guiyan1, while transcript level of spot Y13 was induced significantly in Yunyan2 but unchanged in Guiyan1.
The transcript levels of gene expression encoding selected proteins of two tobacco genotypes exposed to 50 µM Cd after 5 days determinated by qRT-PCR. a Mitochondrial processing peptidase (Y13); b ATP-dependent Clp protease proteolytic subunit (Y15); c GDP dissociation inhibitor (Y17). Error bars represent SD values (n = 3)
Discussion
Metabolism category
Epoxide hydrolases (EHs, spot Y1), putative enoyl-acyl carrier protein (ACP) reductases (ENRs, spot Y2) were identified associates with metabolism. EHs, involved in the metabolism of lipid, transform epoxides into diols (Newman et al. 2005). The role of EHs seems to differ from organism to organism. Overall, these enzymes have three main functions: detoxification, catabolism, and regulation of signaling molecules. EHs in plants were reported to occupy an important place in stress defense system, such as drought stress, Al stress and host/pathogen-defense (Mowbray et al. 2006; Newman et al. 2005). Therefore, Cd-induced EHs in tolerant genotype Yunyan2 is likely to be involved in Cd-tolerance. However, in order to get a better understanding of their functions in the Cd stress response, it is necessary to investigate the accumulation of related metabolite (diols) and enzymatic activities of the EHs.
ENRs are important enzymes taking part in the elongation cycle of fatty acid de novo synthesis. Fatty acids are the precursors of lipids who are the indispensable compositions of plasma membrane. Moreover, plasm membrane was reported to be a pivotal construction in preventing metals getting into the cytoplasm; Modifications in composition of membrane lipid, level of fatty unsaturated acids and fluidity of membrane lipids are significant crucial in plants to handle with toxicity of heavy metal (Meharg 1993). Up-regulated ENRs in tolerant genotypeYunyan2 may be beneficial for the synthesis of fatty acid, which was catalyzed by ENRs, to fight against Cd toxicity. But the limitation of reports about this protein in dealing with Cd calls more detailed examination.
Photosynthesis and stress response categories
Obviously, energy is essential to guarantee various biochemical and physiological activities including Cd detoxification in plants. Energy in plants originally comes from solar power stabilized by photosynthesis in chloroplast. Here, we identified 3 proteins, which was markedly down-regulated in sensitive genotype Guiyan1 after Cd stress, but remained unchanged in Yunyan2: plastidic aldolase NPALDP1 (spot Y3), GAPDH subunits B (spot 5), OEC33 (spot Y6). And 1 up-regulated protein, chlorophyll a–b binding protein 25 (CAB25, spot Y4) in Yunyan2. Plastidic aldolase NPALDP1 belongs to class I fructose-bisphosphate aldolase family. It is a crucial housekeeping protein involved in the photosynthetic carbon reduction cycle, catalyzing the synthesis of fructose-1,6-bisphosphate (FBP) into D-glyceraldehyde-3-phosphate (GAP) and dihydroxyacetone phosphate (DHAP) (Yamada et al. 2000). Yamada et al. (2000) and Rensink et al. (2005) reported that NPLDP2 but not NPLDP1 were up-regulated under salt stress in tobacco and potato. The present study is the first to examine NPALDP1 expression under Cd stress conditions. However, the function of this protein and its direct involvement in Cd tolerance are poorly understood. Therefore, the mechanisms underlying the differential expression of this protein in the two tobacco genotypes should be further explored.
Chloroplast GAPDH is an important enzyme participated in Calvin cycle to catalyze D-glyceraldehyde 3-phosphate into 3-phospho-D-glyceroyl phosphate with the presence of NADPH. This step is the junction of dark and light reaction, and it will consume assimilatory power ATP and NADPH to regenerate NADP+. With low level NADP+ in chloroplast, pseudocyclic electron transport will become the main electron transport pathway leading to the higher production of superoxide anion free radical (Michelet and Krieger-Liszkay 2012). The generation of superoxide anion free radical can cause plant cell dysfunction. Under drought, waterlogging, hypoxia and heavy metal stress, induction of GAPDH was observed (Kosová et al. 2011). The different expression in GAPDH between the two genotypes, remained unchanged in Yunayn2 while down-regulated in Guiyan1, may indicate GAPDH involved in Cd tolerance.
CAB 25, belongs to light harvest chlorophyll a–b binding proteins (LHCP), is one of the most ubiquitous proteins, taking up approximate half of chloroplast chlorophyll. LHCP functions as a light receptor, which can capture and deliver excitation energy to photosynthetic reaction centers (Xia et al. 2012). Oxygen evolving complex (OEC), member of PSII participates in water oxidation (Evlard et al. 2014; Hashimoto et al. 1997). Oxygen evolving complex 33 kDa protein (OEC33), together with oxygen evolving complex 23 kDa protein (OEC23), 17 kDa protein (OEC17) and other components, constitutes an integrated OEC (Hashimoto et al. 1997). It has been found that OEC33 can protect the reaction center-binding D1 protein from the attacking of ROS and assist in maintaining the integrity of the PSII when D1 protein is disassembled (Evlard et al. 2014). The higher expression of OEC33 in Yunyan2 over the sensitive genotype may be beneficial in fighting against ROS caused by Cd.
Another two identified protein spots are stress response related, i.e. HSP70 (spot Y7) and APX (spot Y8). Heat shock proteins are well known to be induced by stress conditions and are efficient to protecting cells against these stresses. They are molecular chaperones involving in protein folding and assembly (Newman et al. 2005). When plants poisoned by Cd, HSP70 can refold or accelerate the degradation of disrupt proteins to retard Cd damage (Neumann et al. 1994). Besides, lots of researches pointed out that Cd induced the production of ROS, includeing H2O2, to attack cellular structures such as macromolecules and plasmalemma (Gallego et al. 2012). APX functions in a process called ascorbate–glutathione (AsA-GSH) cycle, and plays an important role in the metabolism of H2O2 in higher plants. APX in combination with the effective AsA-GSH cycle functions to prevent the accumulation of toxic levels of H2O2 in photosynthetic organisms (Foyer and Shigeoka 2011). Taken together, Cd-induced high expression of these proteins in Yunyan2 leaves may be involved in Cd-detoxification mainly via efficaciously protection on photosynthesis and improvement of carbohydrate metabolism and antioxidant capacity.
Signal transduction and protein synthesis and processing categories
14-3-3 proteins (spots Y9–Y11) were identified up-regulated. As an environmental stimulus, Cd will activate comprehensive signaling transduction pathways, in which 14-3-3 proteins may own significant standings. They can not only detect the phosphorylation status of substrate, but also cause protein phosphorylation to modulate physiological processes containing stress response such as drought, cold and osmotic stresses in different plants (Denison et al. 2011). The overexpression of Hevea brasiliensis 14-3-3c protein in Escherichia coli conferred higher Co2+, Cu2+ and Zn2+ tolerance (Yang et al. 2012). Consequently, 14-3-3 proteins may be crucial in coping with heavy metals including Cd.
Cd-induced eukaryotic initiation factor 4A (eIF4A, spots Y12) protein in Yunyan2 is an ATP-dependent RNA helicase to start synthesis of protein. A pea eIF4A homologue was response to salt stress and it conferred the salinity tolerance in tobacco when this gene was introduced into tobacco (Sanan-Mishra et al. 2005). Some scholars raised a hypothesis that eIF4A is a prototypic member of DEAD box RNA helicase family. When collaborating with eIF4B, it can remove the secondary structure of mRNA caused by abiotic stresses containing Cd stress to facilitate the binding of ribosome, so that the initiation of protein synthesis can carry out normally (Vashisht and Tuteja 2006).
Besides, we identified three processing related protein spots: mitochondrial processing peptidase (MPP, spot Y13), Clp protease ATP-binding subunit ClpA (ClpA, spot Y14) and Clp protease proteolytic subunit (ClpP, spot Y15). MPP is a kind of metalloendopeptidase involved in specifically cleaving off leader sequence of the protein precursors who will be imported into mitochondrial (Sharma et al. 2001). As reported before, MPP could be induced by UV stress and salt stress in mulberry and lupine respectively (Sharma et al. 2001; Wojtyla et al. 2013). It’s the first time to find MPP induced by Cd in tobacco plants, thus it’s deserved more detailed examination of its role in Cd defense. ClpA and ClpP, having molecular chaperones and proteolytic activity severally, are the two main subunits of ATP-dependent Clp protease (Wickner et al. 1994). And ATP-dependent Clp protease aids in controlling the availability of short-lived regulatory proteins and removing abnormal or damaged proteins; the process of removing and degrade misfolding proteins are assisted by Clp protease as well (Lionaki and Tavernarakis 2013). ClpA homolog could be induced by waterlogging in Arabidopsis (Hayes and Dice 1996), and ClpP could response to Cd and Hg in Corynebacterium glutamicum (Fanous et al. 2008). Thus, we can postulate ClpA, ClpP and Clp protease occupy important places in ameliorating Cd toxicity.
Transport and cell structure categories
The tactics developed by plants to tackle with Cd include extruding it out or sequestrating it into vacuoles, which need participating of transporters and some regulatory factors (Migocka et al. 2011; Park et al. 2012). Vacuolar H+-ATPase (V-ATPase) A2 subunit isoform (spot Y16) and GDP dissociation inhibitor (GDI, spot Y17), found up-regulated in leaves of Yunyan2, are typical proteins involved in transport. V-ATPase A2 subunit isoform is a subunit of V-ATPases possessing ATP hydrolytic activity. It can also create endomembrane systems proton gradient (Morris et al. 2014). The proton motive force is an impetus for Cd deposition via the form of Cd2 +/H+ antiport activity in plant cell vacuoles (Migocka et al. 2011). And it has been previously unraveled that the abundance of V-ATPase subunit A increased with the presence of copper (Morris et al. 2014). The up-regulation of V-ATPases A2 subunit isoform may strengthen Cd2 +/H+ antiport activity to sequester Cd into vacuole.
Rab-GDP dissociation inhibitor (Rab-GDI) can modulate the distribution of Rabs between cytoplasm and membrane by the way of binding with GDP-bound state GTPases and delivering it to the cytosol (Ignatev et al. 2008). This means Rab-GDI is critically pivotal for the proper functioning of vesicular transport. It has been discovered that heterologous expressing GDI proteins in Arabidopsis increased the efflux of Al in Arabidopsis (Ezaki et al. 2000). Naturally, we can deduce that induction of GDI with the presence of Cd promotes the ability to detoxify Cd by vesicle trafficking transport.
Alpha-1,4-glucan-proteinsynthase (UPTG, spot Y18), up-regulated in Yunyan2, has the activity to synthesize cell wall polysaccharides to guarantee the completeness of cell wall (Gillet et al. 2006). Cell wall is the first line to defend Cd by binding Cd ions onto it (Zhu et al. 2013). Besides, increase of UPTG was also observed in photosynthetic algae and rice under Cd stress (Gillet et al. 2006; Lee et al. 2010). Thus we speculate that UPTG helps plants to go through Cd stress by participating in synthesizing cell wall polysaccharides.
These three Cd-induced proteins in Yunyan2 leaves cope with Cd may mainly by isolation of Cd through sequestering it into vacuole or vesicle trafficking transport as well as binding it onto cell wall.
Conclusions
Soil Cd contamination has posed a serious problems for safe food production and become a potential agricultural and environmental hazard worldwide. Tobacco can easily accumulate Cd in leaves, and Cd-contamination in tobacco has caused wide public concern. In the present study, protein profiles were used to analyze Cd stress responsive proteins in leaves of tobacco. A total of 18 differentially expressed-proteins were identified in response to Cd stress using 2-DE coupled with the MALDI-TOF-TOF MS and data bank analyses. Of these, we identified 4 photosynthesis-related proteins (Spots Y3–6), i.e.NPALDP1, CAB25, GAPDH subunits B and OEC33. Their elevated expression may be in favor of efficaciously protection on photosynthesis against Cd-toxicity. These results suggest that the effective protection of photosynthesis has a crucial role regarding Cd-tolerance in tolerant tobacco genotypeYunyan2. In addition, Cd-induced expression of metabolism related proteins EHs and ENRs (spots Y1, Y2), may generate some metabolin being great beneficial to the stability of cell in Yunyan2. Besides, other proteins identified in Yunyan2 may also occupy pivotal places in tackling with Cd toxicity. Thus, the results of this study suggest that tobacco plants cope with Cd stress in a complex manner, where protection on photosynthesis, metabolism and antioxidant capacity may play a pivotal role in this complex cellular network. Although, this study is an initial proteomic investigation into tobacco leaf response to Cd stress. This kind of study provides a good starting point in understanding the overall Cd responses of plants and the identification of proteins in response to Cd stress and provides new insights that can lead to a better understanding of the molecular basis of Cd tolerance. In any event, additional studies focused on these genes and comparative analyses may facilitate a better understanding of the mechanisms involved in Cd-tolerance in tobacco.
References
Ahsan N, Renaut J, Komatsu S (2009) Recent developments in the application of proteomics to the analysis of plant responses to heavy metals. Proteomics 9:2602–2621
Alvarez S, Berla BM, Sheffield J, Cahoon RE, Jez JM, Hicks LM (2009) Comprehensive analysis of the Brassica juncea root proteome in response to cadmium exposure by complementary proteomic approaches. Proteomics 9:2419–2431
Bah AM, Sun H, Chen F, Zhou J, Dai H, Zhang G, Wu F (2010) Comparative proteomic analysis of Typha angustifolia leaf under chromium, cadmium and lead stress. J Hazard Mater 184:191–203
Bovet L, Eggmann T, Meylan M, Polier J, Kammer P, Marin E, Feller U, Martinoia E (2003) Transcript levels of AtMRPs after cadmium treatment: induction of AtMRP3. Plant Cell Environ 26:371–381
Cai Y, Cao F, Wei K, Zhang G, Wu F (2011) Genotypic dependent effect of exogenous glutathione on Cd-induced changes in proteins, ultrastructure and antioxidant defense enzymes in rice seedlings. J Hazard Mater 192:1056–1066
Carpentier SC, Witters E, Laukens K, Deckers P, Swennen R, Panis B (2005) Preparation of protein extracts from recalcitrant plant tissues: an evaluation of different methods for two-dimensional gel electrophoresis analysis. Proteomics 5:2497–2507
Cuypers A, Koistinen KM, Kokko H, Kärenlampi S, Auriola S, Vangronsveld J (2005) Analysis of bean (Phaseolus vulgaris L.) proteins affected by copper stress. J Plant Physiol 162:383–392
Denison FC, Paul A, Zupanska AK, Ferl RJ (2011) 14-3-3 proteins in plant physiology. Semin Cell Dev Biol 22:720–727
Evlard A, Sergeant K, Ferrandis S, Printz B, Renaut J, Guignard C, Paul R, Hausman J, Campanella B (2014) Physiological and proteomic responses of different willow clones (Salix fragilis X alba) exposed to dredged sediment contaminated by heavy metals. Int J Phytoremediat 16:1148–1169
Ezaki B, Gardner RC, Ezaki Y, Matsumoto H (2000) Expression of aluminum-induced genes in transgenic Arabidopsis plants can ameliorate aluminum stress and/or oxidative stress. Plant Physiol 122:657–666
Fagioni M, Zolla L (2009) Does the different proteomic profile found in apical and basal leaves of spinach reveal a strategy of this plant toward cadmium pollution response? J Proteome Res 8:2519–2529
Fanous A, Weiss W, Görg A, Jacob F, Parlar H (2008) A proteome analysis of the cadmium and mercury response in Corynebacterium glutamicum. Proteomics 8:4976–4986
Foyer CH, Shigeoka S (2011) Understanding oxidative stress and antioxidant functions to enhance photosynthesis. Plant Physiol 155:93–100
Gallego SM, Pena LB, Barcia RA, Azpilicueta CE, Iannone MF, Rosales EP, Zawoznik MS, Groppa MD, Benavides MP (2012) Unravelling cadmium toxicity and tolerance in plants: insight into regulatory mechanisms. Environ Exp Bot 83:33–46
Gillet S, Decottignies P, Chardonnet S, Le Maréchal P (2006) Cadmium response and redoxin targets in Chlamydomonas reinhardtii: a proteomic approach. Photosynth Res 89:201–211
Golia EE, Dimirkou A, Mitsios IK (2007) Accumulation of metals on tobacco leaves (primings) grown in an agricultural area in relation to soil. B Environ Contam Toxicol 79:158–162
Hashimoto A, Ettinger WF, Yamamoto Y, Theg SM (1997) Assembly of newly imported oxygen-evolving complex subunits in isolated chloroplasts: sites of assembly and mechanism of binding. Plant Cell 9:441–452
Hayes SA, Dice JF (1996) Roles of molecular chaperones in protein degradation. J Cell Biol 132:255–258
Ignatev A, Kravchenko S, Rak A, Goody RS, Pylypenko O (2008) A structural model of the GDP dissociation inhibitor rab membrane extraction mechanism. J Biol Chem 283:18377–18384
Kosová K, Vítámvás P, Prášil IT, Renaut J (2011) Plant proteome changes under abiotic stress-contribution of proteomics studies to understanding plant stress response. J Proteomics 74:1301–1322
Lee K, Bae DW, Kim SH, Han HJ, Liu X, Park HC, Lim CO, Lee SY, Chung WS (2010) Comparative proteomic analysis of the short-term responses of rice roots and leaves to cadmium. J Plant Physiol 167:161–168
Lionaki E, Tavernarakis N (2013) Oxidative stress and mitochondrial protein quality control in aging. J Proteomics 92:181–194
Liu WX, Shang SH, FengX, Zhang GP, Wu FB (2014) Selenium reduces cadmium uptake and mitigates cadmium toxicity in the two tobacco genotypes differing in Cd tolerance. Environ Toxicol Chem, in reviewing
Meharg AA (1993) The role of the plasmalemma in metal tolerance in angiosperms. Physiol Plant 88:191–198
Michelet L, Krieger-Liszkay A (2012) Reactive oxygen intermediates produced by photosynthetic electron transport are enhanced in short-day grown plants. BBA-Bioenerg 1817:1306–1313
Migocka M, Papierniak A, Kosatka E, Kłobus G (2011) Comparative study of the active cadmium efflux systems operating at the plasma membrane and tonoplast of cucumber root cells. J Exp Bot: err180
Morris CA, Owen JR, Thomas MC, EL Hiti GA, Harwood JL, Kille P (2014) Intracellular localization and induction of a dynamic RNA-editing event of macro-algal V-ATPase subunit A (VHA-A) in response to copper. Plant Cell Environ 37:189–203
Mowbray SL, Elfström LT, Ahlgren KM, Andersson CE, Widersten M (2006) X-ray structure of potato epoxide hydrolase sheds light on substrate specificity in plant enzymes. Protein Sci 15:1628–1637
Ndimba BK, Chivasa S, Simon WJ, Slabas AR (2005) Identification of Arabidopsis salt and osmotic stress responsive proteins using two-dimensional difference gel electrophoresis and mass spectrometry. Proteomics 5:4185–4196
Neumann D, Lichtenberger O, Günther D, Tschiersch K, Nover L (1994) Heat-shock proteins induce heavy-metal tolerance in higher plants. Planta 194:360–367
Newman JW, Morisseau C, Hammock BD (2005) Epoxide hydrolases: their roles and interactions with lipid metabolism. Prog Lipid Res 44:1–51
Park J, Song WY, Ko D, Eom Y, Hansen TH, Schiller M, Lee TG, Martinoia E, Lee Y (2012) The phytochelatin transporters AtABCC1 and AtABCC2 mediate tolerance to cadmium and mercury. Plant J 69:278–288
Rensink W, Hart A, Liu J, Ouyang S, Zismann V, Buell CR (2005) Analyzing the potato abiotic stress transcriptome using expressed sequence tags. Genome 48:598–605
Rodríguez-Celma J, Rellán-Álvarez R, Abadía A, Abadía J, López-Millán A (2010) Changes induced by two levels of cadmium toxicity in the 2-DE protein profile of tomato roots. J Proteomics 73:1694–1706
Sanan-Mishra N, Pham XH, Sopory SK, Tuteja N (2005) Pea DNA helicase 45 overexpression in tobacco confers high salinity tolerance without affecting yield. Proc Natl Acad Sci USA 102:509–514
Sarry JE, Kuhn L, Ducruix C, Lafaye A, Junot C, Hugouvieux V, Jourdain A, Bastien O, Fievet JB, Vailhen D (2006) The early responses of Arabidopsis thaliana cells to cadmium exposure explored by protein and metabolite profiling analyses. Proteomics 6:2180–2198
Schneider T, Schellenberg M, Meyer S, Keller F, Gehrig P, Riedel K, Lee Y, Eberl L, Martinoia E (2009) Quantitative detection of changes in the leaf-mesophyll tonoplast proteome in dependency of a cadmium exposure of barley (Hordeum vulgare L.) plants. Proteomics 9:2668–2677
Sharma R, Sharma A, Fujimura T, Machii H (2001) Stress-responsive enhanced expression of mitochondrial processing peptidase gene after UV exposure in mulberry. Plant Sci 161:807–812
Vashisht AA, Tuteja N (2006) Stress responsive DEAD-box helicases: a new pathway to engineer plant stress tolerance. J Photochem Photobiol B 84:150–160
Verbruggen N, Hermans C, Schat H (2009) Mechanisms to cope with arsenic or cadmium excess in plants. Curr Opin Plant Biol 12:364–372
Wickner S, Gottesman S, Skowyra D, Hoskins J, McKenney K, Maurizi MR (1994) A molecular chaperone, ClpA, functions like DnaK and DnaJ. Proc Natl Acad Sci USA 91:12218–12222
Wojtyla Ł, Kosmala A, Garnczarska M (2013) Lupine embryo axes under salinity stress. II. Mitochondrial proteome response. Acta Physiol Plant 35:2383–2392
Wu FB, Zhang G, Yu J (2003) Interaction of cadmium and four microelements for uptake and translocation in different barley genotypes. Commun Soil Sci Plant Anal 34:2003–2020
Wu FB, Wu H, Zhang G, Bachir DML (2004) Difference in growth and yield in response to cadmium toxicity in cotton genotypes. J Plant Nutr Soil Sci 167:85–90
Xia Y, Ning Z, Bai G, Li R, Yan G, Siddique KH, Baum M, Guo P (2012) Allelic variations of a light harvesting chlorophyll a/b-binding protein gene (Lhcb1) associated with agronomic traits in barley. PLos One 7:e37573
Yamada S, Komori T, Hashimoto A, Kuwata S, Imaseki H, Kubo T (2000) Differential expression of plastidic aldolase genes in Nicotiana plants under salt stress. Plant Sci 154:61–69
Yang Z, Li H, Guo D, Tian W, Peng S (2012) Molecular characterization of a novel 14-3-3 protein gene (Hb14-3-3c) from Hevea brasiliensis. Mol Biol Rep 39:4491–4497
Zhu QH, Huang DY, Liu SL, Luo ZC, Rao ZX, Cao XL, Ren XF (2013) Accumulation and subcellular distribution of cadmium in ramie (Boehmeria nivea L. Gaud.) planted on elevated soil cadmium contents. Plant Soil Environ 2:57–61
Acknowledgments
We are thankful for the financial support from Guizhou Tobacco Science Institute.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xie, L., He, X., Shang, S. et al. Comparative proteomic analysis of two tobacco (Nicotiana tabacum) genotypes differing in Cd tolerance. Biometals 27, 1277–1289 (2014). https://doi.org/10.1007/s10534-014-9789-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10534-014-9789-5