Abstract
Background
Impairment of mitochondrial function plays an important role in obesity and the development of insulin resistance. The aim of this project was to investigate the mitochondrial DNA copy number in human omental adipose tissue with respect to obesity.
Methods
The mitochondrial DNA (mtDNA) content per single adipocyte derived from abdominal omental adipose tissue was determined by quantitative RT-PCR in a group of 75 patients, consisting of obese and morbidly obese subjects, as well as non-obese controls. Additionally, basal metabolic rate and fat oxidation rate were recorded and expressed as total values or per kilogram fat mass.
Results
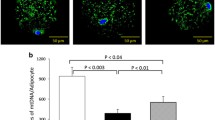
MtDNA content is associated with obesity. Higher body mass index (BMI) resulted in a significantly elevated mtDNA count (ratio = 1.56; p = 0.0331) comparing non-obese (BMI < 30) to obese volunteers (BMI ≥ 30). The mtDNA count per cell was not correlated with age or gender. Diabetic patients showed a trend toward reduced mtDNA content. A seasonal change in mtDNA copy number could not be identified. In addition, a substudy investigating the basal metabolic rate and the fasting fat oxidation did not reveal any associations to the mtDNA count.
Conclusions
The mtDNA content per cell of omental adipose tissue did not correlate with various clinical parameters but tended to be reduced in patients with diabetes, which may partly explain the impairment of mitochondrial function observed in insulin resistance. Furthermore, the mtDNA content was significantly increased in patients suffering from obesity (BMI above 30). This might reflect a compensatory response to the development of obesity, which is associated with impairment of mitochondrial function.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Human adipose tissue is now well recognized as a multifunctional organ. It is the location of lipid storage and, with its brown tissue islets, plays an important role in thermogenesis [1]. Adipose tissue, especially intra-abdominal and omental fat compartments, also has various endocrine functions [2], as it is the source of different peptide hormones and adipokines [3] such as adiponectin, IL-6, leptin, plasminogen activator inhibitor-1, resistin, or visfatin. These and other factors, in particular mitochondria, have been shown to be associated with the development of obesity, insulin resistance, and type 2 diabetes [4–8].
Mitochondria represent a major portion of the energy generating pathways, and the extent of energy expenditure or lipid storage is directly associated with the functional state of mitochondria. In the mitochondrial inner membrane, a highly regulated electron transfer chain uses a proton gradient to synthesize ATP formed from ADP and phosphate, named oxidative phosphorylation [9]. The uncoupling of this process by, e.g., uncoupling proteins leads to thermogenesis in brown adipocytes [10], as opposed to white adipocytes, which contain mitochondria of a kind different from those in brown adipose tissue [11]. It is well known that different types of cells or tissue contain different subsets of mitochondria or, rather, that mitochondria display heterogeneous morphology within individual cells [12].
The number of mitochondria per cell and the rate of mitochondrial turnover [13] also vary among different tissues [14]. While liver cells, muscle cells, or brown adipocytes contain a relatively high number of mitochondria per cell, the mitochondrial count in white adipocytes is much lower. The biogenesis of mitochondria and, thus, their numbers are controlled by different extracellular and intracellular stimuli [15], including environmental influences, such as a cold response via the peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1a), which leads to an increase in the mitochondrial mass, mitochondrial function (mt capacity), and mitochondrial DNA content [16]. Recently, temperature conditions such as the climate with its seasons have been described as a cold-response factor [17]. The mitochondrial content of cells is also controlled by fission and fusion processes [18, 19], as mitochondria are able to divide or fuse. By fusion, mitochondria share mitochondrial proteins, exchange mitochondrial DNA, and can restore function. Fusion is followed by a fission process, which leads to newly “rebuilt” or “refurbished” mitochondria. This fusion–fission process may be dysregulated in obesity. A high fusion-to-fission ratio leads to fewer mitochondria forming long interconnections [19]. On the other hand, a low fusion-to-fission ratio results in fragmentation of mitochondria, with the consequence of an increase in the mitochondrial content per cell. An association of disturbed fusion and fission processes with the development of obesity was recently detected by a reduced expression of mitofusions (e.g., mfn-2) [20], leading to a more fragmented population of mitochondria and, therefore, to a higher mitochondrial content per cell [21].
Apart from obesity, type 2 diabetes and insulin resistance have also been shown to be linked to mitochondrial function and mitochondrial density. For example, adipocytes from diabetic ob/ob mice show a decreased number of mitochondria per cell, an effect that can be reversed by treatment with PPAR-γ agonists such as rosiglitazone [22]. Thus, the development of insulin resistance appears to be associated with a reversible decrease of mitochondrial density.
Little is known about mitochondrial content and function in human omental fat in various states of obesity and the metabolic syndrome [8]. The aim of the present study was to evaluate the mitochondrial DNA (mtDNA) content per cell in human omental adipose tissue from a group of 75 participants, including 52 obese patients (BMI ≥ 30) and 23 non-obese controls (BMI < 30), and to correlate the mitochondrial count per adipocyte with the clinical parameters, “basal metabolic rate” and “fat oxidation rate.” Within our patient group, 18 of the 75 volunteers suffered from diabetes.
Patients and Methods
Patients
The study was approved by the local ethics committee. The patients participated on a voluntary basis after having given written informed consent. The patient group consisted of 52 morbidly obese patients undergoing bariatric surgery: laparoscopic proximal Roux-en-Y gastric bypass (n = 29), laparoscopic biliopancreatic diversion “duodenal switch” (n = 15, all but two after failed laparoscopic adjustable gastric banding) [23, 24], and laparoscopic sleeve resection (n = 8). Their average BMI was 42 kg/m2; 16 patients were diabetics (for more details, see Table 2). The non-obese control group consisted of 23 patients who planned to undergo elective surgery: colorectal (n = 12), pancreas (n = 5), esophagus (n = 3), stomach, cholecystectomy, and oophorectomy (n = 1 each). Their average BMI was 25 kg/m2; two of them were diabetics (Table 2). The group of non-obese patients did not include any post-bariatric surgery patient who reduced body weight (re-operation). Diabetic patients were analyzed under ongoing diabetic treatments (metformin and insulin). Patients on glitazone-based therapy were excluded, because glitazones stimulate mitochondrial biogenesis.
All data were collected prospectively. Obese patients and non-obese controls were carefully characterized prior to surgery for metabolic syndrome and other co-morbidities. In obese patients, body composition was determined by dual energy X-ray absorptiometry; energy consumption, including basal metabolic and fat oxidation rates, was measured by indirect calorimetry. The omental fat tissue samples were collected during the operation and immediately shock frozen in liquid nitrogen and stored at −78°C.
DNA Isolation
Taqman-based quantification of the mtDNA count per cell was performed with DNA prepared from frozen adipose tissue according to the standard manual (NucleoSpin DNA, Machery Nagel, Germany).
Determination of mtDNA Copy Number
The quantification of the mitochondrial DNA count per cell is an approximation to the measurement of the mitochondrial mass to bypass limitations from the small sizes of tissue biopsies. Quantitative RT-PCR was applied to determine the relative amounts of mtDNA and nuclear DNA [25], from which their relative ratio was calculated. One single-copy gene from the genome (lipoprotein lipase (LPL)) and one from the mitochondrial genome (NADH dehydrogenase subunit 1 (ND1)) were used for amplification. In addition, amplified DNA fragments from plasmids carrying either ND1 or LPL fragments were used as standards to determine accurate quantities of mtDNA content in the samples of interest. Per tissue biopsy the determination of the mtDNA count was performed in duplicate to judge the precision of measured data (see Statistics).
Primers and Probes
LPL-probe: ACA-TTC-ACC-AGA-GGG-TC with 3′ Tamra and 5′ FAM. ND1-probe: CCA-TCA-CCC-TCT-ACA-TCA-CCG-CCC with 3′ Tamra and 5′ FAM. Oligos used for amplification: LPL-fwd CGA-GTC-GTC-TTT-CTC-CTG-ATG-AT; LPL-rev TTC-TGG-ATT-CCA-ATG-CTT-CGA and ND1-fwd CCC-TAA-AAC-CCG-CCA-CAT-CT; ND1-rev GAG-CGA-TGG-TGA-GAG-CTA-AGG-T.
Statistics
First, univariate analysis was performed to analyze effect of age, gender, season, diabetes, or BMI on the mtDNA count. To compare seasonal-dependent mtDNA copy number at the time of tissue sampling, two groups “winter” (1 November to 31 March) and “summer” (1 April to 31 October) were formed to describe the seasons. The calculation of diabetes-associated influence on the mtDNA content was done by comparing diabetic patients and non-diabetics.
The univariate analysis applied to describe effects of age, gender, season, diabetes, or BMI on the mtDNA count was performed as follows: SPSS software was applied to determine the two-tailed Fischer’s exact test in case of contingency tests. Graphpad Prism software was used to calculate mean ± SEM and unpaired t test (95% confidence). In addition, the correlation analysis between mtDNA copy number and metabolic rates or fat oxidation rates was performed with Graphpad Prism software using Spearman-ratio-based correlation models. p values <0.05 were considered as significant.
Multivariate Analysis
Based on the results from the univariate statistics, an in-depth analysis was performed to detect the influence of BMI or diabetes on the number of mitochondria by using a linear, mixed-effect model. Mixed-effect models were used to evaluate repeated measure data where observations were correlated within the same subject [26]. “Precision of measured data,” “age,” “gender,” “BMI,” and “diabetes” were fixed factors; “subject” was treated as a random factor. Consequently, the results are adjusted for age, gender, precision of measured data, diabetes, and BMI. As the mitochondria counts were approximately log-normal distributed, these variables were log-transformed. Hence, back-transformed group differences from the mixed-effect model are expressed as geometric mean ratios with corresponding 95% confidence intervals.
A p value <0.05 was considered significant. Statistical analysis was performed using R (a language and environment for statistical computing).
Results
Univariate Analysis
According to the literature, age [27], gender [28], or season [17] can have an impact on the mitochondrial count or function. In our analysis these parameters did not correlate with the mtDNA count. Neither age (ratio = −0.1513; p = 0.1952; n = 75), nor gender (women 611.8 ± 53.12, n = 56 and men 438.3 ± 63.74, n = 19; p = 0.0828; n = 75), nor season (summer 584.4 ± 61.18, n = 41 and winter 548.0 ± 62.23, n = 34; p = 0.6798; Table 1) revealed any association to the mtDNA copy number.
The next point was to clarify a possible impact of the BMI on the mtDNA copy number. In a univariate analysis, all 75 patients were dichotomized according to their BMI (BMI < 30 and BMI ≥ 30 kg/m2). The obese group revealed a significantly increased mtDNA content compared to the non-obese (634.0 ± 55.54 vs. 418.4 ± 56.53; p = 0.0213; Table 2). Within the two groups of obese and non-obese patients, no differences in the distribution for gender or season could be detected (Table 2). In contrast, the non-obese group was significantly older (59.09 ± 3.276 vs. 48.44 ± 1.817; p = 0.0032; Table 2). In the obese group significantly more patients suffered from diabetes (30.8% vs. 8.7%; p = 0.0444; Table 2).
As the frequency of diabetes paralleled obesity, we divided the patient pool according to the status of diabetes. In addition, diabetes has previously been reported as a risk factor for mitochondrial mass and function [29]. When the patients were grouped into diabetics and non-diabetics, a trend toward a reduced mtDNA content in diabetics was observed; this, however, was not significant (448.7 ± 58.85 vs. 605.5 ± 53.43; p = 0.1246; Table 3). In this analysis according to diabetes, the distributions for age, gender, and season were equal (Table 3). In analogy to the association of diabetes with increased BMI as described above, the BMI showed a significantly unequal distribution (non-diabetics 35.36 ± 1.283, n = 57 and diabetics 41.07 ± 1.951, n = 18; p = 0.0273; Table 3) for the two groups.
Multivariate Analysis
Based on the observations from the univariate analyses, in-depth statistics were performed using a linear mixed-effect model. This calculation method incorporates influences from age, gender, precision of measurements, as well as BMI and diabetes. However, neither age (ratio = 0.98; p = 0.7598), gender (ratio = 0.81; p = 0.3052), nor precision of measured data (ratio = 1.06; p = 0.128; Table 4) had relevant impact on the mtDNA content. According to this linear mixed model, the mtDNA copy number was not significantly reduced in diabetics but showed a trend of a 29% reduced mtDNA content (ratio = 0.71; p = 0.1003; n = 75; Table 4).
However, grouping the participants into obese patients and non-obese controls revealed a significantly increased mtDNA count in the obese group, where the mtDNA count was elevated by 56% (ratio = 1.56; p = 0.0331; n = 75; Table 4).
The determination of the mtDNA copy number by polymerase chain reaction on DNA from tissue biopsies is an invasive method. Measuring energy expenditure could be an alternative clinical tool for the measurement of mitochondrial function and count. We were able to perform calorimetry and body composition measurements with 22 (fasting fat oxidation) or 23 obese patients (basal metabolic rate) but due to logistic reasons, not with non-obese control patients. Within this group of obese patients the mtDNA content per adipocyte showed no significant correlation with the basal metabolic rate (ratio = −0.3538; p = 0.0977; n = 23) or the basal metabolic rate per kg fat mass (ratio = 0.1038; p = 0.6376; n = 23; Table 5). In addition, correlating fasting fat oxidation rate and mtDNA content (ratio = −0.3123; p = 0.1571; n = 22) or fat oxidation rate per kilogram fat mass and mtDNA count (ratio = −0.2897; p = 0.1910; n = 22) did not reflect any associations (Table 5).
Discussion
The aim of this present study was to determine the mtDNA content of omental adipose tissue with respect to obesity as well as to investigate possible associations of the mtDNA count with different parameters such as age, season, gender, diabetes, basal metabolic rate, and fat oxidation rate. Adult human adipose tissue is present in two main compartments—white subcutaneous tissue and intra-abdominal adipose tissue. Brown adipose tissue, an important compartment in rodents, has only recently been described in humans in small amounts in the upper part of the body [30]. While white adipose tissue mainly serves as a storage of energy, the abdominal adipose tissue is, in addition, an important source of various hormones and mediators linking abdominal obesity with the metabolic syndrome [31]. Mitochondrial dysfunction leading to impaired oxidation of fatty acids has been proposed as the culprit in diabetes [32]. Diabetes, hypercholesterinemia, and arterial hypertension—the main features of the metabolic syndrome—lead to macrovascular disease, the most important cause of death in obesity. While mitochondria of white subcutaneous adipose tissue and skeletal muscle have already been studied in humans [33, 34], we here present the first study on the mtDNA content in human omental fat tissue.
In general, our data indicate that the mtDNA content of human fat omental tissue is relatively stable. This is in line with results from subcutaneous fat tissue, which also showed relative stable mitochondrial copy numbers under various conditions [35]. The number of mtDNA copies in adipose tissue is up to 10-fold smaller than that in muscle tissue [36]. This smaller number of mtDNA copies is the consequence of a reduced number of mitochondria in adipocytes. The reason for this may be the specialized function of this tissue. While muscle cells have to produce high amounts of energy for contraction, adipocytes have different functions. One main task is to store energy in form of triglycerides [3, 37–39]. Generally, it is very difficult to determine the exact number of mitochondria or mitochondrial mass from tissue samples of the small size used in this study. However, determination of the mtDNA copy number is a possibility to express changes in mitochondrial mass or number. It has been documented that increasing numbers of mtDNA copy number are associated with an increased “virtual” number of mitochondria per cell [40]. This virtual number of mitochondria per cell is regarded as a number of mitochondrial fragments forming a huge mitochondrial interconnected network or as a number for independent mitochondria [40].
First, we analyzed the data by univariate analyses to detect effects of age, gender, and season on the mtDNA content per adipocyte. No correlation could be detected between age and the mtDNA content (ratio = −0.1513; p = 0.1952; n = 75). Aging is seen as a major source of free radicals that are harmful for mitochondria [34, 35]. However, it seems that, in human omental tissue, aging does not lead to reduced mtDNA content. Separation of all patients by gender showed a weak trend toward reduced mtDNA content for male patients (women 611.8 ± 53.12, n = 56 and men 438.3 ± 63.74, n = 19; p = 0.0828; n = 75). The tendency of reduced mtDNA copy number in the male group of patients is not significant and remains unclear. It could be due to a smaller number of male patients (n = 19) than of female patients (n = 56). Therefore, further study of gender-specific mtDNA content in omental adipose tissue is needed. Finally, the grouping of patients by season of sample collection did not reflect any differences in mtDNA copy number (summer 584.4 ± 61.18, n = 41 and winter 548.0 ± 62.23, n = 34; p = 0.6798; Table 1). Cold-induced thermogenesis is a well-described mechanism, which leads to increased mitochondrial count and mass, mainly in brown adipose tissue of rodents and skeletal muscle [16], primarily controlled by activation of PGC-1a. Thus, in the present study the mtDNA copy number and the season could not be correlated to the season of tissue sampling. However, the seasons were equally distributed in the groups of obese patients and non-obese controls (Tables 1 and 2) and in diabetic and non-diabetic patients (Tables 2 and 3). It appears that omental adipose tissue is not affected by seasonal changes of the outside temperature in the range present in Switzerland. This is less surprising, as only recently cold-inducible brown adipose tissue in the upper part of the human body was detected using PET scanners [41, 42]. This tissue may be responsible for cold-induced thermogenesis [17].
Obesity is regarded to have an impact on mitochondrial function. Therefore, we distributed the participants into two BMI groups, an obese group and a group of non-obese controls. To identify a possible impact of obesity on mitochondria, the mtDNA copy numbers were calculated and compared between the non-obese control group and the obese patients. The mtDNA count (p = 0.0213; Table 2) was significantly increased in obesity. The higher mtDNA count in the obese group clearly links the mitochondrial DNA content in omental adipose tissue to the development of obesity. This may be a mechanism to compensate for mitochondrial dysfunction or other mitochondrial defects, which will be discussed later. Age also showed a significant difference in distribution (p = 0.0032, Table 2). The younger age in the obese group is primarily caused by the type of surgical intervention. Bariatric surgery is generally only applied to patients between 20 and 60 years of age, whereas in the control group the cause for abdominal surgery was not restricted to age. The two BMI groups differed neither by gender distribution (p = 0.0871, Table 2) nor by the season the samples were collected (p = 0.8058, Table 2). Within the two BMI groups, the distribution of diabetes was not equal (p = 0.0444; Table 2). This is not surprising as diabetes is associated with increased body weight.
It is well accepted that diabetes can have an impact on mitochondrial mass and function [29]. In order to investigate this point in human omental adipose tissue, we grouped the patients according to diabetes; the BMI was significantly higher in the group of diabetics (p = 0.0273, Table 3). This is in line with data distributed according to the BMI (see above) and links diabetes to obesity. The distribution of other parameters such as age (p = 0.2183), gender (p = 0.7647), or season (p = 0.7871; Table 3) did not show any inequality for the diabetes grouping (Table 3). Finally, the mtDNA count was reduced in the diabetes group but failed to be significant (p = 0.1246; Table 3). The limited number of diabetic patients, which will be discussed later, does not allow further conclusions. Further studies, including more diabetics, are needed.
Based on the findings above, showing some possible influences of age, diabetes, and BMI on the mtDNA count, a linear mixed model was applied to allow a multivariate analysis of the present data.
This multivariate interpretation revealed that the mtDNA copy number did not change with increasing age (ratio = 0.98; p = 0.7598; Table 4). This is in contrast to the general view that accumulation of free radical oxygen species during lifetime leads to impairment of mitochondria [34] and, as a consequence, to a reduced mitochondrial content in white subcutaneous tissue [35]. On the other hand, mtDNA content has been reported to be stable in skeletal muscle, a tissue with high mitochondrial activity, high production rate of reactive oxygen species, and, therefore, high risk for mitochondrial damage [36].
Although there are gender-specific trends in fat tissue distribution—women in general showing more subcutaneous fat—this does not apply in the case of the mtDNA content in human omental adipose tissue (ratio = 1.06; p = 0.128; Table 4).
Patients with type 2 diabetes showed a trend toward reduced mtDNA content per cell in their omental adipose tissue (ratio = 0.71; p = 0.1003; Table 4). These patients were not treated with glitazones, PPAR-γ agonists, such as rosiglitazone, which enhance mitochondrial biogenesis leading to increased mitochondrial count and mass [22, 25]. The development of type 2 diabetes is believed to be associated with impairment of mitochondria. This impairment could be either due to a reduced mitochondrial function or, as in our study, to a reduced mtDNA content [32, 43]. Whether this is caused by misbalanced fusion and fission processes as proposed for the development of obesity and/or other defects is not yet understood [19]. Hence, a possible association could be found between mtDNA content in omental adipose tissue and type 2 diabetes but remains subject to further analysis. In case diabetic patients exhibit a clearly reduced amount of mtDNA, this may be a plausible cause or a consequence of impaired mitochondrial function. This observation of a reduced mtDNA count in the diabetes group failed to be significant. However, there is a 29% decrease in the mean of the non-diabetes group compared to that of the diabetes group, a relatively large but not significant effect. It can be explained by the facts that we had a small diabetes group, that we had patients at different stages of disease, and that diabetes is concerning statistics a between subject effect and is, therefore, poorly estimated. Originally, it was planned to investigate the mtDNA content in obese vs. non-obese control volunteers. At that time, the study was not focused on diabetes. To study the impact of diabetes on the mtDNA content in human adipose tissue, a further analysis of diabetes patients is under way. We estimate that another 30 diabetic patients have to be tested to elucidate this observation of reduced mtDNA count in diabetes.
Interestingly, a significantly higher mtDNA copy number (ratio = 1.56; p = 0.0331; Table 4) was detected in the group of obese patients compared to that of the non-obese control group. A BMI of 30 was selected as it marks a critical point in the transition from overweight to obesity. Our observation is in contrast to a recent finding by Kaaman et al. [35], who reported a decrease in mtDNA content in subcutaneous adipose tissue in obesity. A possible explanation for this discrepancy may be the different types of adipose tissue analyzed in the two studies. While Kaaman et al. used subcutaneous adipose tissue pads for their analysis, we investigated omental adipose tissue pads. This discrepancy may be an indication for a different oxidative capacity in the two compartments of adipose tissue, as has been shown for other metabolic pathways, such as lipolytic capacity and the capacity to secrete proteins [44–47].
A further explanation for an altered mtDNA content in human omental adipose tissue of morbidly obese patients may be found in a reduced expression of mitofusins reported in obesity [20]. This leads to a more fragmented mitochondrial population [21], which could attribute to the mitochondrial content per cell.
Interestingly, diabetic patients show a decreased mtDNA count, whereas obese patients have an increased mtDNA copy number. This observation could account for different mechanisms involved in the two diseases. While mitochondrial mass—expressed by mtDNA count—of omental adipose tissue might be upregulated in obesity as a consequence of impaired function, this mechanism does not seem to take place in diabetes. The view of reduced activity of mitochondria in diabetes is supported by a series of reports, such as the link of the metabolic inflexibility from obese and type 2 diabetes patients with decreased lipid peroxidation [48]. Clearly, further studies investigating global mitochondrial function in type 2 diabetes have to be performed. In addition, the effect of PPAR-γ agonists leading to induction of mitochondrial biogenesis and resetting the mitochondrial mass demonstrates the important link between mitochondria and diabetes. The activation of mitochondrial biogenesis by PPAR-γ agonists may lead to compensation of the disturbed mitochondrial function and, thus, improve diabetes treatment.
Lymphocytes from obese patients show a reduced citrate synthase activity [49]. The same is demonstrated for muscle biopsies, showing that mitochondrial parameters, including citrate synthase activity, are reduced in obese patients [50]. In addition, a rat obesity model showed significantly elevated hepatic mitochondrial mass and area, markedly reduced citrate synthase activity but a stable number of organelles in liver from rats fed a high-fat diet [51]. There is no doubt that mitochondrial function is weakened in obesity. This, together with changes in the morphology, such as disturbed fusion and fission processes and altered expression of proteins controlling morphology (e.g., mitofusins) [20], as well as changes in the mtDNA count, clearly underline the tight association between mitochondria and obesity. It remains difficult for some of these observations to clarify what happens first: Are changes in mitochondrial count and function a cause or consequence of obesity?
Determination of the mtDNA content is based on sample biopsies and, therefore, on an invasive technique. We believe that the energy expenditure might be an alternative non-invasive way to determine mitochondrial function and mass. Energy expenditure can easily be measured with calorimetry and analysis of the body composition. In a pilot study we performed these measurements on 22 (fasting fat oxidation) or 23 (basal metabolic rate) obese patients and correlated the data with the mtDNA copy number. We failed to detect any significant association between the mtDNA content and the total basal metabolic rate, the basal metabolic rate per kg fat mass, the total fat oxidation rate, or the fat oxidation rate per kilogram fat mass (Table 5). Keeping in mind that mitochondria of different tissues may behave differently in number and function and that we measured the basal metabolic rate and fat oxidation rate of the whole organism, it is not surprising that we missed the differences in the compartment of the omental fat tissue. Furthermore, the range of BMI of the patients measured was small, as was the range in their mtDNA count. Due to logistics and other reasons we could not test non-obese patients, who are now missing for a comparative analysis with respect to BMI. Therefore, broadening the data acquisition to include non-obese patients might give further insights.
In summary, this investigation shows that omental adipose tissue has a relatively stable number of mtDNA copies per cell. Age, season, and gender have no impact. The body mass index shows a significant association with the mtDNA copy number, whereas basal metabolic rate and fat oxidation rate are not linked to mtDNA, supporting the notion that mitochondria in omental adipose tissue only exert a limited influence on whole-body energy homeostasis (basal metabolic rate and expenditure). We identified a trend toward reduced mtDNA content in diabetic patients. The number of diabetics was, however, quite low, so further studies may clarify this issue. MtDNA copy number clearly increases with BMI. Whether there is first obesity and then an increased mtDNA count, or vice versa, remains unclear and needs further investigation.
References
Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. 2004;84:277–359.
Diamond F. The endocrine function of adipose tissue. Growth Genet Horm. 2002;18(2):17–22.
Trayhurn P. Adipocyte biology. Obes Rev. 2007;8(Suppl. 1):41–4.
Maassen JA. Mitochondrial dysfunction in adipocytes: the culprit in type 2 diabetes? Diabetologia. 2006;49(4):619–20.
Choo HJ, Kim JH, Kwon OB, et al. Mitochondria are impaired in the adipocytes of type 2 diabetic mice. Diabetologia. 2006;49:784–91.
Guilherme A, Virbasius JV, Czech MP, et al. Adipocyte dysfunctions linking obesity to insulin resistance and type 2 diabetes. Nat Rev Mol Cell Biol. 2008;9:367–77.
Petersen KF, Befroy D, Shulman GI, et al. Mitochondrial dysfunction in the elderly: possible role in insulin resistance. Science. 2003;500(5622):1140–2.
Civitarese AE, Ravussin E. Minireview: mitochondrial energetics and insulin resistance. Endocrinology. 2007;149(3):950–4.
Ernster L, Schatz G. Mitochondria: a historical review. J Cell Biol. 1981;91(3):227s–55.
Kuroshima A. Brown adipose tissue thermogenesis as a physiological strategy for adaptation. Jpn J Physiol. 1993;43(43):117.
Justo R, Oliver J, Gianotti M. Brown adipose tissue mitochondrial subpopulations show different morphological and thermogenic characteristics. Mitochondrion. 2005;5(1):45–53.
Collins TJ, Bootman MD. Mitochondria are morphologically heterogenous within cells. J Exp Biol. 2003;206:1993–2000.
Menziens RA, Gold PH. The turnover of mitochondria in a variety of tissues of young adult and aged rats. J Biol Chem. 1970;246(8):2425–9.
Masuyama M, Iida R, Matsuki T, et al. Quantitative change in mitochondrial DNA content in various mouse tissues during aging. Biochim Biophys Acta. 2005;1723:302–8.
Kelly DP, Scarpulla RC. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004;18:357–68.
Wu Z, Puigserver P, Spiegelman BM, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–24.
van Marken Lichtenbelt WD, Vanhommering JW, Jaap Teule GJ, et al. Cold-activated brown adipose tissue in healthy men. N Engl J Med. 2009;360:1500–8.
Chan DC. Mitochondrial fusion and fission in mammals. Ann Rev Cell Dev Biol. 2006;22:79–99.
Detmer SA, Chan DC. Functions and dysfunctions of mitochondrial dynamics. Nat Rev Mol Cell Biol. 2007;8:870–9.
Mingrone G, Manco M, Zorzano A, et al. Could the low level of expression of the gene encoding skeletal muscle mitofusin-2 account for the metabolic inflexibility of obesity? Diabetologia. 2005;48:2108–14.
Bach D, Pich S, Zorzano A, et al. Mitofusin-2 determines mitochondrial network architecture and mitochondrial metabolism. J Biol Chem. 2003;278(19):17190–7.
Wilson-Fritch L, Nicoloro S, Corvera S, et al. Mitochondrial remodeling in adipose tissue associated with obesity and treatment with rosiglitazone. J Clin Invest. 2004;114(9):1281–9.
Peterli R, Wöllnerhanssen B, von Flüe M, et al. Prospective study of a two-stage operative concept in the treatment of morbid obesity: primary Lap-Band followed if needed by sleeve gastroectomy with duodenal switch. Obes Surg. 2007;17:334–40.
Woelnerhanssen B, Kern B, Peterli R, et al. Predictors of outcome in treatment of morbid obesity by laparoscopic adjustable gastric banding: results of a prospective study of 380 patients. Surgery of Obesity and Related Disease. 2008;4:500–6.
Bogacka I, Xie H, Smith SR, et al. Pioglitazone induces mitochondrial biogenesis in human subcutaneous adipose tissue in vivo. Diabetes. 2005;54:1392–9.
Pinheiro JC, Bates DM. Mixed-effects models in S and S-PLUS. Springer: New York; 2000.
Barazzoni R, Short KR, Sreekumaran Nair K. Effects of aging on mitochondrial DNA copy number and cytochrome c oxidase gene expression in rat skeletal muscle, liver, and heart. JBC. 2000;275(5):3343–7.
Colom B, Alcolea MP, Garcia-Palmer FJ, et al. Skeletal muscle of female rats exhibit higher mitochondrial mass and oxidative-phosphorylative capacities compared to males. Cell Physiol Biochem. 2007;19:205–12.
Mollica MP, Lionetti L, Iossa S, et al. Cold exposure differently influences mitochondrial energy efficiacy in rat liver and skeletal muscle. FEBS Lett. 2005;579:1978–82.
Needergard J, Bengtssen T. Unexpected evidence of brown adipose tissue in adult humans. Am J Physiol Endocrinol Metab. 2007;293:E444–52.
Wajchenberg BL. Subcutaneous and visceral adipose tissue: their relation to the metabolic syndrome. Endocr Rev. 2000;21(6):697–738.
Maassen JA. Mitochondrial diabetes: pathophysiology, clinical presentation, and genetic analysis. Am J Med Gen (Semin. Med. Genet.). 2002;115:66–70.
Marcuello A, González-Alonso J, Díez-Sánchez C, et al. Skeletal muscle mitochondrial DNA content in exercising humans. J Appl Physiol. 2005;99:1372–7.
Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–95.
Kaaman M, Sparks LM, Arner P, et al. Strong association between mitochondrial DNA copy number and lipogenesis in human white adipose tissue. Diabetologia. 2007;50:2526–33.
Miller FJ, Rosenfeldt FL, Nagley P, et al. Precise determination of mitochondrial DNA copy number in human skeletal and cardiac muscle by a PCR-based assay: lack of changes of copy number with age. Nucleic Acids Res. 2003;31(11):e61.
Rajala MW, Scherer PE. Minireview: the adipocyte—at the crossroads of energy homeostasis, inflammation, and atherosclerosis. Endocrinology. 2003;144(9):3765–73.
Rosen ED, Spiegelman BM. Adipocytes as regulators of energy balance and glucose homeostasis. Nat Med. 2006;444:847–53.
Ferranti SD, Mozaffarian D. The perfect storm: obesity, adipocyte dysfunction, and metabolic consequences. Clin Chem. 2008;54:945–55.
Robin ED, Wong R. Mitochondrial DNA molecules and virtual number of mitochondria per cell in mammalian cells. J Cell Physiol. 1988;136:507–13.
Cypess AM, Lehmann S, Kahn CR, et al. Identification and importance of brown adipose tissue in adult humans. N Engl J Med. 2009;360:1509–17.
Virtanen KA, Lidell ME, Nuutila P, et al. Functional brown adipose tissue in healthy adults. N Engl J Med. 2009;360:1518–25.
Befroy DE, Petersen KF, Shulman GI, et al. Impaired mitochondrial substrate oxidation in muscle of insulin-resistant offspring of type 2 diabetic patients. Diabetes. 2007;56:1376–81.
Hellmér J, Marcus C, Arner P, et al. Mechanisms for differences in lipolysis between human subcutaneous and omental fat cells. J Clin Endocrinol Metab. 1992;75(1):15–20.
van Harmelen V, Dicker A, Arner P, et al. Increased lipolysis and decreased leptin production by human omental as compared with subcutaneous preadipocytes. Diabetes. 2002;51:2029–36.
Fried SK, Bunkin DA, Greenberg AS. Omental and subcutaneous adipose tissues of obese subjects release interleukin-6: depot difference and regulation by glucocorticoid. J Clin Endocrinol Metab. 1998;83(3):847–50.
Arvidsson E, Blomqvist L, Ryden M. Depot-specific differences in perilipin mRNA but not protein expression in obesity. J Intern Med. 2004;255:595–601.
Kelley DE, Mandarino LJ. Fuel selection in human skeletal muscle in insulin resistance. Diabetes. 2000;49:677–83.
Capkova M, Houstek J, Zeman J, et al. Activities of cytochrome c oxidase and citrate synthase in lymphocates of obese and normal-weight subjects. IJO. 2002;26:1110–7.
Kim JY, Hickner RC, Houmard JA, et al. Lipid oxidation is reduced in obese human skeletal muscle. Am J Physiol Endocriol Metab. 2000;279:1039–44.
Crescenzo R, Bianco F, Iossa S, et al. Alterations in hepatic mitochondrial compartment in a model of obesity and insulin resistance. Obesity. 2008;16:958–64.
Author information
Authors and Affiliations
Corresponding author
Additional information
The authors hereby disclose any commercial interest in this present study. We thank the European Society of Pediatric Endocrinology for financial support by their donation of the 2007 IPSEN Collaborative Project Grant.
Rights and permissions
About this article
Cite this article
Lindinger, A., Peterli, R., Peters, T. et al. Mitochondrial DNA Content in Human Omental Adipose Tissue. OBES SURG 20, 84–92 (2010). https://doi.org/10.1007/s11695-009-9987-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11695-009-9987-3