Abstract
There have been considerable recent advances in the technology for rapidly detecting foodborne pathogens. However, a traditional culture method is still the “gold standard” for presumptive-positive pathogen screening although it is labor-intensive, ineffective in testing large amount of food samples, and cannot completely prevent unwanted background microflora from growing together with target microorganisms on agar media. We have developed multivariate classification models based on visible and near-infrared hyperspectral imaging for rapid presumptive-positive screening of six representative non-O157 Shiga-toxin producing Escherichia coli (STEC) serogroups (O26, O45, O103, O111, O121, and O145) on agar plates of pure and mixed cultures. The classification models were developed with spread plates of pure cultures. In this study, we evaluated the performance of the classification models with independent validation samples of mixed cultures that were not used during training and found the best classification model for differentiating non-O157 STEC colonies on spread plates of mixed cultures. A validation protocol appropriate to hyperspectral imaging of mixed cultures was developed. An additional independent validation set of 12 spread plates with pure cultures was used as positive controls to help the validation process with the mixed cultures and to affirm the model performance. One imaging experiment with colonies obtained from two serial dilutions was performed. A total of six agar plates of mixed cultures were prepared, where O45, O111 and O121 serogroups that were relatively easy to differentiate were inoculated into all six plates and then each of O26, O103 and O145 serogroups was added into the mixture of the three common bacterial cultures. The number of mixed colonies grown after 24-h incubation was 331 and the number of pixels associated with the grown colonies was 16,379. The best model found from this validation study was based on pre-processing with standard normal variate and detrending, first derivative, spectral smoothing, and k-nearest neighbor classification (kNN, k = 3) of scores in the principal component subspace spanned by 12 principal components. The results showed 95 % overall detection accuracy at pixel level and 97 % at colony level. The developed model was proven to be still valid even for the independent validation samples although the size of a validation set was small and only one experiment was performed. This study was an important first step in validating and updating multivariate classification models for rapid screening of ground beef samples contaminated by non-O157 STEC pathogens using hyperspectral imaging.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Escherichia coli (E. coli) are bacteria living in the intestine of warm-blooded animals and humans [1]. Many E. coli strains do not cause human disease; there is, however, a group of E. coli that produces Shiga toxin. Symptoms of human illnesses caused by the consumption of Shiga toxin-producing Escherichia coli (STEC) are diarrhea, stomach cramps, vomiting, and a potentially lethal kidney complication called the hemolytic uremic syndrome (HUS). The most prevalent and commonly recognized STEC serotype is E. coli O157:H7; non-O157 STEC serogroups such as O26, O45, and O103 are also increasingly recognized [2, 3]. The Center for Disease Control (CDC) in the United States (US) estimated that overall, as many as 265,000 STEC infections occur every year in the US, with about 64 % non-O157 STEC infections [4]. According to a previous study, from 1983 to 2002, about 70 % of non-O157 STEC infections were caused by six major serogroups, including O26, O45, O103, O111, O121, and O145 (called “Big Six”) [5]. In 2011, the Food Safety and Inspection Service (FSIS) of the U.S. Department of Agriculture (USDA) declared the “Big Six” non-O157 STEC serogroups as adulterants through a Federal Register Notice and announced plans to start the Hazard Analysis and Critical Control Points (HACCP) verification testing program for raw beef trim and ground beef [6]. Under the new rule, any meat that contains the six non-O157 STEC cannot be sold as raw products.
The current method of the FSIS for detecting and identifying the six non-O157 STEC takes 4 days until the non-O157 STEC bacteria are genetically and biochemically identified [7, 8]. After enrichment on the first day of analysis, multiplex real-time polymerase chain reaction (PCR) tests are performed on the second day in order to detect potential positives of these six serogroups. Samples with positive results will be further analyzed using immunomagnetic separation (IMS) beads with serogroup-specific antibodies followed by plating onto Rainbow agar O157 (in short, Rainbow agar). After 20–24 h incubation, suspicious colonies on Rainbow agar plates are visually screened for presumptive positive tests with latex agglutination on the third day before performing the PCR assays and biochemical identification test on the fourth day. Since the morphologies of the targeted STEC colonies may vary widely among strains and serogroups, the current practice of the FSIS visually identifies different colony morphologies/phenotypes and picks at least one colony from each identified colony morphology/phenotype for the presumptive positive testing, and then performs the fourth day tests only with latex agglutination positive colonies (up to five colonies per plate) [8]. Rainbow agar is a selective and differential chromogenic medium used to isolate presumptive-positive STEC. Considering the multi-day workflow of STEC detection and isolation, it is beneficial to reduce the time needed to identify presumptive-positive STEC colonies with a more objective and accurate tool. It is challenging to rapidly and accurately identify the six STEC colonies by eye due to phenotypic variability in STEC populations and/or the presence of background microflora.
The FSIS laboratories have been using Rainbow agar for presumptive positive screening of STEC O157:H7 from meat products, where it takes 5–6 days to get confirmatory test results through biochemical and genetic tests, such as latex agglutination, toxin assay, and PCR [9, 10]. It is known that STEC O157:H7 colonies appear charcoal grey–black or steel black on Rainbow agar whereas the six non-O157 STEC colonies appear purple, gray, or gray–blue on Rainbow agar [11]. Phenotypic discrimination of non-O157 serogroups on Rainbow agar has been limited with mixed results reported [12] because Rainbow agar was originally developed to isolate O157:H7 colonies and suitable selective and differential agar media are not available for non-O157 [13]. In addition, screening non-O157 STEC colonies is further complicated by the fact that ground beef harboring non-O157 STEC pathogens can potentially have high background microflora which can also grow competitively on Rainbow agar. Although time consuming and labor-intensive, plating methods still represent a field where progress is needed in order to more accurately differentiate pathogen colonies from one another or from background microflora. Rapid detection and identification of non-O157 STEC serogroups on agar media are also important for development of intervention and verification strategies for the food industry and regulatory agencies such as the FSIS and the CDC.
Hyperspectral imaging is an optical imaging technique that combines conventional imaging and vibrational spectroscopy to acquire both spatial and spectral information from every pixel in each object under test. The spectral “fingerprints” of bacteria provided by hyperspectral imaging can be used for detection and identification of pathogens grown on agar media. So far, research with hyperspectral imaging for detection and identification of pathogenic colonies has been confined to Campylobacter [14, 15] and non-O157 STEC [16–18]. In particular, a visible and near-infrared (VNIR) hyperspectral imaging technique with multivariate classification models was developed to differentiate colonies of non-O157 STEC bacteria [16–18]. The multivariate classification models were developed from spatial and spectral information obtained from non-O157 STEC colonies on Rainbow agar plates, and the models were optimized for their operating parameters. The models were based on some of popular chemometric techniques such as scatter correction, first derivative, spectral smoothing, k-nearest neighbor classification and principal component analysis (PCA), and then classification results were predicted on images. A transparent sample holder was also designed to minimize shadows cast by colonies on semi-transparent agar plates. However, the previous hyperspectral imaging studies for detecting and differentiating pathogens on agar plates were limited to pure cultures, where the identity of each colony was known based on a priori knowledge about which organism was inoculated into each Petri dish. Thus, it was necessary to study and validate the performance of the classification models with data obtained under more realistic conditions such as mixed cultures.
A mixed culture is a laboratory culture that contains two or more identified species or strains of microorganisms. Spread plates of mixed cultures may produce diverse and realistic colony populations mimicking actual microbial populations of contaminated food samples although mixed cultures are still laboratory control samples. However, in hyperspectral imaging of colonies from mixed cultures, performance of a classification model is much more difficult to validate than pure cultures because it is unknown where specific bacteria grow on an agar plate due to spreading of liquid cultures and bacterial competition for growth and survival [19, 20], and thus it is almost impractical to confirm the identity of every colony with a genetic and/or a biochemical confirmation method simply for validating classification models. This difficulty is in part because there are too many (typically about 50–300) colonies per plate. Hence, the objective of this study was (1) to develop a validation protocol appropriate for spread plates with mixed cultures of the six STEC serogroups and (2) to assess the performance of the multivariate classification models with mixed cultures.
Materials and methods
Non-O157 STEC mixed cultures
The pure cultures of the non-O157 STEC bacteria were obtained from a culture collection at the Eastern Laboratory of USDA-FSIS. A total of six non-O157 STEC strains were chosen for this study with one strain being from each representative O-serogroup (O26, O45, O103, O111, O121, and O145). The specific STEC strains were O26:H2 strain 4, O45:H2 strain 8, O103:H2 strain D, O111:H1 strain 16, O121:H19 strain A, and O145:H- strain K. The pathogenicity of all test strains was confirmed by the presence of two genetic targets: one of two stx genes (stx1 and stx2) and the intimin (eae) genes [7]. Working stocks of each culture were stored on nutrient agar slants (Becton–Dickinson, Sparks, MD, USA) at 4 °C. Cell suspensions were prepared from cultures grown overnight on Blood agar (BA, Trypticase Soy Agar with 5 % sheep blood, Remel, Lenexa, KS, USA) at 37 °C. Cells were suspended in sterile saline (0.85 %) at an initial concentration of approximately 109 CFU/mL (0.50 turbidity), with a Dade Behring MicroScan Turbidity Meter (Dade Behring, West Sacramento, CA, USA). Serial dilutions of each cell suspension were prepared in sterile saline.
Cell suspension mixtures containing equal portions (500 μL aliquots of 103 CFU/mL) of serogroups O45, O111, and O121 that were relatively easy to differentiate with the developed classification models [16–18] were prepared from the individual STEC serogroup serial dilutions. An equivalent concentration of a fourth serogroup (O26, O103 or O145) was inoculated into the three strain mixture. The reason why the mixed cultures were prepared with the mixture formula of three easy serogroups plus one difficult serogroup (not with all six serogroups) was due to its simplicity in performance validation of the developed classifiers. The aforementioned mixture formula was designed to build ground-truth maps only from the measured images. For example, when the difficult ones (O26, O103 and O145) were mixed, the identities of all colonies (typically over 100 per plate) should have been confirmed by latex agglutination and/or PCR in order to build ground-truth maps of colony identities. On the other hand, for example, when O26 was mixed with O45, O111, and O121, each colony of O45, O111, and O121 was identified with a help from both the image analysis on a computer display and the prediction using a classifier. Then, the remaining colonies on the plate belonged to O26. The resulting cell mixtures contained approximately 2.5 × 102 CFU/mL of each of four serogroups (O45, O111, O121 and O26; O45, O111, O121 and O103; or O45, O111, O121 and O145). Then for each mixture, 50 and 100 μL aliquots were spread onto individual Rainbow agar (RBA, Biolog, Inc., Hayward, CA, USA) plates (100-mm diameter).
In addition to the aforementioned mixed cultures, approximately 50 and 100 μL aliquots from serial dilutions of each pure cell suspension were inoculated onto Rainbow agar plates as positive controls by a spread plating technique. This positive control group was prepared to help to find any errors in the validation process with the mixed cultures and to affirm the model performance. All plates were incubated at 37 °C for 24 h. Following the above protocol, one experiment was carried out. Thus, a total of 6 plates (2 cell concentrations × 3 mixtures) with mixed cultures and 12 plates (2 cell concentrations × 6 serogroups) with pure cultures were used to evaluate the developed classification models.
Hyperspectral image acquisition
Hyperspectral image acquisition was performed with a push-broom line-scan visible near-infrared (VNIR) hyperspectral imager (Themis Vision Systems, Richmond, VA, USA) including a 12-bit CCD camera with 1,376 × 1,040 pixels (SensiCam QE, PCO-TECH Inc, Romulus, MI, USA), a spectrograph (ImSpector V10E with 30-μm slit, Specim-Spectral Imaging Ltd., Oulu, Finland), a C-mount objective lens (APO-Xenoplan 1.8/35-mm, Schneider Optics, Hauppauge, NY, USA) with motion control (Newark, CA, USA), a custom sample holder, and a computer. Figure 1 shows a picture of the imaging system. The imaging system acquired reflectance values for the wavelengths ranging from 368 to 1,024 nm with an average wavelength separation of 1.27 nm. Two 50-W tungsten halogen lamps (4,700 K) were used for reflectance imaging by illuminating a Petri dish at 45° from the left and right sides 43 cm apart. A transparent Acrylic box with the dimension of 33 (length) × 30 (width) × 12 (height) cm was custom-built to elevate a Petri dish to minimize colony shadows reflecting from a white Teflon plate that was placed at the bottom of the box, which was used to increase the apparent reflectance of thin-layered colonies on the semi-transparent agar. The working distance from the objective lens to the Petri dish was about 40 cm. On-camera binning was set to 2 (spatial) × 2 (spectral) with a 30-ms integration time. The integration time of the system was adjusted to maximize the apparent reflectance of a Spectralon® calibration panel (described below) without saturating. The resulting hyperspectral image data cube had the size of 688 (W) × 500 (H) × 520 (wavelengths) before removing extreme wavelength bands during image preprocessing.
Multivariate hyperspectral image analysis: pre-processing
Multivariate hyperspectral image analysis (MHIA or in short multivariate image analysis: MIA) extends the multivariate data analysis techniques widely used in chemometrics and spectroscopy to hyperspectral image analysis for segmentation, classification, detection and prediction. Pre-processing is an important first step in MHIA because hyperspectral images typically suffer from spectral and spatial abnormalities such as random noise, glints, shadows, and measurement errors. Data pre-processing methods used in this study included normalization, size and noise reduction, image mosaicing, transformation such as conversion to absorbance, feature extraction and selection such as PCA, differentiation, and correction of spectral variation. Normalization, reduction, image mosaicing and transformation operations were applied to all pixels. But, the other operations were applied to the pixels only within regions-of-interest (ROIs) confined to colonies.
Measured reflectance values were calibrated (i.e. normalized) to relative reflectance R with a 75 % reflectance Spectralon® target (13 × 13 cm, SRT-75-050, Labsphere, North Sutton, NH, USA) [14]. The spectral dimension of each image was reduced to 473 spectral bands ranging from 400 to 1,000 nm by removing extreme wavelength bands. Thus, the resulting image size became 688 (W) × 500 (H) × 473 (λ). Finally, spectral noise was reduced by a Savitzky-Golay smoothing filter (window size: 25; order of moment: 4) at each pixel position.
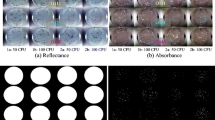
After the aforementioned operations, the calibrated hyperspectral images were stitched together into a single image mosaic. The images from the same dilution were added to each column of the mosaic from left (less cells) to right (more cells). The reflectance image mosaic was transformed to absorbance (log10(1/R)) in order to reduce non-linearity in reflectance measurements, and absorbance was used for the model development and validation. Figures 2 and 3 show each color-composite image mosaic of calibrated reflectance and absorbance images with mixed and pure cultures, respectively. In Fig. 2, the first row on each mosaic consisted of plates with mixed cultures of O26, O45, O111, and O121, the second row with O103, O45, O111, and O121, and the third row with O145, O45, O111, and O121. In Fig. 3, the hyperspectral images obtained from a positive control set of pure cultures were similarly pre-processed and arranged into a different mosaic of 12 hyperspectral data cubes with 6 rows (serogroups) and 2 columns (dilutions).
The ground-truth ROIs representing the true identity of each pixel and colony were created to build a spectral library and evaluate the predictive performance of classification models (see Figs. 2c, 3c). The ROIs were semi-automatically obtained with an interactive thresholding tool in Fiji (an open source image processing package based in ImageJ). Each image needed a different threshold value. So, the Fiji software was used to get the best segmentation result from each image by trial and error. The 428-nm image was used for colony ROI segmentation because 428 nm had good contrast between colony and background agar pixels. Each colony region was successfully segmented out by this process with some exceptions: glints and touching objects. Glint pixels with specular reflection were not included in the ROIs and a blob of touching objects was separated with the ROI tool in ENVI software (Exelis Visual Information Solutions, Boulder, CO, USA).
In the case of mixed cultures, the class of each ROI was initially predicted by multiple classification models and then adjusted manually using the ENVI software at each colony. The goal of this process was to create ground-truth ROIs from the images. The first step was to apply 80 different classification models including the ones mentioned in this paper in order to predict colony identities, the second step was to analyze the prediction results and pick the best ones, and then the final step was to manually adjust the prediction results with the ENVI software. Color, size, shape, texture and any discernible features were utilized to manually assign and re-assign a correct class to each colony (and all pixels in each colony). Positive controls of pure cultures were also referred for creating correct class labels on the images of mixed cultures. Without doubt, this heuristic process to create a ground-truth class map of the ROIs was tedious and prone to errors when a lot of new data would be presented to the classification models. As future research, one possible solution to this ROI-creation problem is to perform genetic and/or biochemical tests in order to determine true identities of only a few representative colonies. Spectra in absorbance from 400 to 1,000 nm were extracted from the ROIs on a per pixel basis with an in-house program written in MATLAB R2012a (The Mathworks, Natick, MA, USA). When extracting spectral data from pixel locations defined by the ROIs, the data were unfolded into an M × N data matrix X (predictors) whose values were associated with M observations (number of samples in pixels) in rows and N variables (wavelengths) in columns. A response vector y of class labels from 1 to 6 was also created for validation.
The data pre-treatment methods were applied as part of pre-processing to predictors X. The pre-treatment methods in this study included none (absorbance only), multiplicative scatter correction (MSC), standard normal variate and detrending (SNVD), first derivative with a gap width of 11 points, moving average smoothing with a gap width of 11 points before differentiation, and MSC-corrected first derivative, and SNVD-corrected first derivative. The application order of the pre-treatment methods was MSC (or SNVD), moving average, and differentiation when all methods were used. In addition, PCA was applied to the pre-treated data, and classification was done in the reduced feature (score) space obtained by the PCA. Thus, the number of principal components (PCs) used for classification was also considered an important operating parameter and chosen to be 12. The optimal number of PCs was studied in a previous study [18], where the minimum requirement was 6 PCs and then the prediction performance was maxed out from 12 PCs.
Multivariate hyperspectral image analysis: classification models and prediction
Multivariate classification models were applied to new independent hyperspectral images of pure cultures and mixed cultures to predict the identity of each colony from pixel-level prediction, where an image was segmented into individual colony segments with similar spatial and spectral properties. The multivariate classification models used in this study were previously developed using a training (interchangeable with calibration) set obtained from 24 spread plates of pure non-O157 STEC serogroup cultures obtained in 2011. Four classification models chosen for this study were based on (1) MSC-corrected moving average (MSC1), (2) MSC-corrected moving average and then first derivatives and (MSC2), (3) SNVD-corrected moving average (SNV1), and (4) SNVD-corrected moving average and then first derivatives (SNV2). The gap width for first derivatives and moving average was 11 points. The training set consisted of 1,421 ROIs (i.e. colonies) with 51,173 pixels (i.e. observations or samples). For both model development from a training set and validation from a test set, all spectral data were treated similarly by the aforementioned data pre-processing techniques and unfolded into X. The classification models were saved as files by only including PC scores and loadings, pre-treated mean-centered vectors of each serogroup class, pre-processing methods associated with each model and operating parameters for classifiers such as k in kNN. Multivariate hyperspectral image analysis for classification and prediction is summarized in Fig. 4. The final decision making rule was applied at colony level by the winner-take-all strategy (simple majority voting) of prediction results at pixel level.
Multivariate hyperspectral image analysis: validation
There were two independent validation (interchangeable with test) sets of the mixed and the pure cultures to measure the performance of the multivariate classification models in classification accuracy. The classification accuracy was the rate of correctly classified samples, which was assessed at each pixel and colony from a confusion matrix. Other performance metrics such as omission error, commission error, user’s accuracy and producer’s accuracy were also used to assess the performance of each serogroup. The validation set of mixed cultures was used to predict the performance of classification models in more realistic growth conditions where cells of different serogroups competed against one another. The other validation set of pure cultures was also used to affirm the predictive performance of the models.
Results and discussion
Sample size and colony morphology
The validation set of mixed cultures consisted of 331 colonies and 16,379 pixels. The sample information of the mixed-culture validation set is summarized in Table 1. The estimated size of each pixel was 0.197 mm (horizontal) × 0.211 mm (vertical). Thus, the area of each pixel was approximately 0.042 mm2. From this pixel size, the estimated average size of the colonies was 2.31 mm2 (55 pixels). The standard deviation of colony size was 0.659 mm2 (15.7 pixels). The smallest colony was 0.13 mm2 (3 pixels) and the largest was 5.75 mm2 (137 pixels). The average colony forming unit of mixed culture plates was approximately 55. The serogroup showing the largest colony size was O45 with 4.54 mm2 (108 pixels) on average per colony whose size was more than twice as large as the others. The smallest serogroup was O111 with 1.34 mm2 (32 pixels) per colony. Serogroups O26, O103, O121 and O145 were similar with about 2.1 mm2 (50 pixels) per colony.
The other validation set with the positive controls of pure cultures consisted of 854 colonies and 36,917 pixels. The sample information of the pure-culture validation set is summarized in Table 2. The average colony forming unit of pure culture plates was approximately 71. The appearances of the colonies on the plates of pure cultures were similar to mixed cultures.
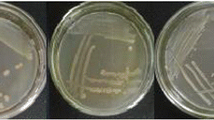
The circular forms (i.e. colony shapes) were observed from all colonies. Outer boundaries of O121 colonies were more distinctive and less fuzzy than the others. The color of O45 colonies was almost black and visually very different from the other serogroups. Thus, O45 colonies can be used as reference markers when evaluating the performance of the models against unknown colonies. Figure 5 shows the examples of colony appearance typically observed from the measured reflectance and absorbance (transformed from reflectance) images. The color of all colonies except O45 (dark green to black) and some of O111 colonies (grayish blue tone similar to the agar background) was purple varying from bright to dark. The center area of each colony was darker than the perimeter. O111 colonies were grayish color on the agar plates with less cell concentration (left column images of the mosaic) and light purple color on the agar plates with more cell concentrations (right column images of the mosaic). The detailed appearance characteristics of each colony are not discussed in this study. A further study is necessary to find the importance factors such as texture, surface causing the differences in colony appearance and to incorporate them into the multivariate classification models.
Spectral analysis
Figure 6a shows the mean spectra of the mixed cultures. As shown in the figure, all spectral responses at wavelengths longer than about 750 nm were almost identical. Serogroup O111 had almost a flat spectral response in the range from 500 to 650 nm whereas serogroups O26, O45, O103 and O145 had distinctive absorbance peaks at near 535 nm and serogroup O121 had its peak at 550 nm. Serogroup O45 had a distinctive spectral response due to its large absorbance peak at 535 nm and a broad spectral shoulder from 600 to 650 nm. Serogroups O26, O103 and O145 had similar spectral shapes but different absorbance values in the range approximately from 450 to 650 nm. The colony appearances shown in Fig. 5a confirmed the differences in absorbance of serogroups O26, O103 (lighter pink) and O145 (darker pink).
Figure 6b shows the mean spectra of each serogroup obtained from the ROIs of the training and validation sets including both pure and mixed cultures. Overall except O26, the spectral responses of the two validation sets were more similar than the training set, which confirmed the previous study finding that replication of experiments was the largest uncertainty to the predictive performance of the classification models. The 600–700 nm shoulders of the pure O26 cultures disappeared when O26 was mixed. One possible explanation for why O26 in mixed cultures showed the spectral difference between 600 and 700 nm was the bacterial competition for survival and growth [19, 20]. Although no quantitative analysis was made to measure the differences or variability between the training and validation sets, we assumed, from Fig. 6b, that the differences in mean-spectral responses between the training and validation sets were not large enough to re-train the models.
Prediction results
A total of 16 prediction models (interchangeable with classification models) from four preprocessing methods (MSC1, MSC2, SNV1 and SNV2), two classifiers (Mahalanobis distance and kNN), and two detection levels (pixel and colony) were evaluated with classification accuracy against the two validation sets of pure and mixed cultures, respectively. Table 3 shows the overall classification accuracies of each prediction model against the pure cultures. All 16 prediction models produced over 97 % prediction performance. The average classification accuracy was 98.31 %. The colony-level decision making algorithm was approximately 2 % better than the pixel-level decision making. The performance variability among the four preprocessing models was less than 1 %. The performance difference between two classifiers was trivial (0.23 %). The best model was SNV1 or SNV2 with kNN and colony-level decision making.
When the performance of the same 16 models was measured against the mixed culture data, the best performance was obtained from the model adopting SNV2 (SNVD-corrected first derivative with a gap width of 11 points, moving average with a gap width of 11 points) and kNN (k = 3). First, the performance of pixel-level classification is summarized in Table 4. The overall classification accuracy at pixel level was about 95.6 % with Kappa coefficient 0.9457. All ROI pixels of serogroups O45, O111 and O121 were classified with 100 % accuracy. The classification accuracy for serogroup O103 was the worst at 88.54 %. About 11 % among ROI pixels of serogroup O103 were incorrectly classified as serogroup O26. About 8 % among ROI pixels of serogroup O145 were also misclassified as serogroup O26. In terms of average accuracy that was an average of user’s and producer’s accuracies, the performance for serogroup O26 was the worst with 89 % average accuracy whereas both serogroups O103 and O145 showed about 92 % average accuracies, respectively (Table 5). The average accuracy of serogroups O45, O111, and O121 was 100, 98, and 100 %, respectively. Second, the decision making algorithm using colony-level classification misclassified 8 colonies out of 331 (97.6 % of overall classification accuracy), as shown in Fig. 7. Five colonies were falsely predicted as serogroup O26 on the agar plates that were not supposed to have O26 colonies. Two O103 colonies and one O145 colony were falsely predicted as well. Figure 7b showed that most misclassification errors were from the 100 μL plates. Although it is largely unknown why this happened and whether this trend will be repeated with more experiments, one possible explanation is morphological or phenotypic changes of the colonies due to the more bacterial competition in higher-density cell populations than 50 μL plates.
Conclusion
This study showed the potential of visible and near-infrared hyperspectral imaging for rapidly identifying colonies of the big six non-O157 STEC serogroups on Rainbow agar plates inoculated with mixed cultures. Spatial and spectral data analysis revealed that the differences in appearance of the six non-O157 STEC serogroup colonies were mainly due to the differences in absorption bands and color tones. Other features affecting colony morphology might have influenced the differences in colony appearance but this topic was not pursued in the study. Spectral characteristics at near-infrared wavelengths from 750 to 1,000 nm were almost identical and thus the discrimination power of the near-infrared spectral range was trivial. The color was the major feature exploited in the classification model. In this study, the multivariate classification models developed and optimized using a training set of pure spread plates were validated against two independent test sets with pure and mixed cultures. Sixteen different models were compared and produced over 97 % classification accuracies against the validation set of pure cultures whereas the average classification accuracy of the models on the training set was about 95 % at colony level. The best model, based on SNVD, first derivative, moving average, and k-nearest neighbor classification (k = 3) of scores in the principal component subspace spanned by 12 PC, was selected and applied to the validation set of mixed cultures. The classification accuracy of the model against the mixed cultures was 95 % at pixel level and 97 % at colony level. The developed model was proven to be still valid even for the independent samples although the size of a validation set with mixed cultures was small and only one experiment was performed. The validation was based on a heuristic image analysis that manually determined the ground-truth identification of each colony. Thus, further research is needed to validate the classification models in terms of positively identified colonies using confirmatory testing such as latex agglutination tests or PCR. A few more experiments need to be conducted. Also, the classification models need to be validated with bacterial cultures directly extracted from a food matrix such as ground beef.
References
CDC, E. coli general information, http://www.cdc.gov/ecoli/general/index.html. Accessed 8 March 2013
J.M. Bosilevac, M. Koohmaraie, Prevalence and characterization of non-O157 Shiga toxin producing Escherichia coli isolates from commercial ground beef in the United States. Appl. Environ. Microbiol. 77, 2103–2112 (2011)
P.M. Griffin, in Escherichia coli 0157:H7 and other Shiga toxin-producing E. coli strains (ASM Press, Washington, DC, 1998)
E. Scallan, R.M. Hoekstra, F.J. Angulo, R.V. Tauxe, M.A. Widdowson, S.L. Roy, J.L. Jones, P.M. Griffin, Foodborne illness acquired in the United States-major pathogens. Emerg. Infect. Dis. 17, 7–15 (2011)
J.T. Brooks, E.G. Sowers, J.G. Wells, K.D. Greene, P.M. Griffin, R.M. Hoekstra, N.A. Strockbine, Non-O157 Shiga toxin-producing Escherichia coli infections in the United States, 1983–2002. J. Infect. Dis. 192, 1422–1429 (2005)
USDA, FSIS, Shiga toxin-producing Escherichia coli in certain raw beef products, Fed. Regist. 77, 31975–31981 (2012)
USDA, FSIS, Microbiology Laboratory Guidebook (MLG) 5B.03, Detection and isolation of non-O157 Shiga toxin-producing Escherichia coli (STEC) from meat products (2012), http://www.fsis.usda.gov/Science/Microbiological_lab_guidebook/index.asp. Accessed 8 March 2013
USDA, FSIS, MLG 5B.03 Appendix 4.01, FSIS laboratory flow chart for detection and isolation of non-O157 Shiga toxin-producing Escherichia coli (STEC) from meat products (2012), http://www.fsis.usda.gov/Science/Microbiological_lab_guidebook/index.asp. Accessed 8 March 2013
USDA, FSIS, MLG 5.06, Detection, isolation and identification of Escherichia coli O157:H7 from meat products (2012), http://www.fsis.usda.gov/Science/Microbiological_lab_guidebook/index.asp. Accessed 8 March 2013
USDA, FSIS, MLG 5 Appendix 1.00, E. coli O157:H7 method flow chart (2012), http://www.fsis.usda.gov/Science/Microbiological_lab_guidebook/index.asp. Accessed 8 March 2013
K.A. Bettelheim, Studies of Escherichia coli cultured on Rainbow Agar O157 with particular reference to enterohaemorrhagic Escherichia coli (EHEC). Microbiol. Immunol. 42, 265–269 (1998)
M.A. Grant, C. Hedberg, R. Johnson, J. Harris, C.M. Logue, J. Meng, J.N. Sofos, J.S. Dickson, The significance of non-O157 Shiga toxin-producing Escherichia coli in food. Food Prot. Trends 31, 33–45 (2011)
M.B. Medina, W.L. Shelver, P.M. Fratamico, L. Fortis, G. Tillman, N. Narang, W.C. Cray, E. Esteban, A. Debroy, Latex agglutination assays for detection of non-O157 Shiga toxin producing Escherichia coli serogroups O26, O45, O103, O111, O121, and O145. J. Food Prot. 75, 819–826 (2012)
S.C. Yoon, K.C. Lawrence, G.R. Siragusa, J.E. Line, B. Park, P.W. Feldner, Hyperspectral reflectance imaging for detecting a foodborne pathogen: campylobacter. Trans. ASABE 52, 651–662 (2009)
S.C. Yoon, K.C. Lawrence, J.E. Line, G.R. Siragusa, P.W. Feldner, B. Park, W.R. Windham, Detection of Campylobacter colonies using hyperspectral imaging. J. Sens. Instrum. Food Qual. Saf. 4, 35–49 (2010)
W.R. Windham, S.C. Yoon, S.R. Ladely, G.W. Heitschmidt, K.C. Lawrence, B. Park, N. Narang, W.C. Cray, The effect of regions of interest and spectral pre-processing on the detection of non-0157 Shiga-toxin producing Escherichia coli serogroups on agar media by hyperspectral imaging. J. Near Infrared Spectrosc. 20, 547–558 (2012)
S.C. Yoon, W.R. Windham, S.R. Ladely, G.W. Heitschmidt, K.C. Lawrence, B. Park, N. Narang, W.C. Cray. Hyperspectral imaging for detection of non-O157 Shiga-toxin producing Escherichia coli (STEC) serogroups on spread plates of mixed cultures, SPIE Conference on Sensing for Agriculture and Food Quality and Safety, in Proceedings of SPIE 8369, 09 (2012)
S.C. Yoon, W.R. Windham, S.R., Ladely, G.W. Heitschmidt, K.C. Lawrence, B. Park, N. Narang, W.C. Cray, Hyperspectral imaging for differentiating colonies of non-O157 Shiga-toxin producing Escherichia coli (STEC) serogroups on spread plates of pure cultures, J. Near Infrared Spectrosc. (in press)
A.G. Fredrickson, Behavior of mixed cultures of microorganisms. Ann. Rev. Microbiol. 31, 63–87 (1977)
M.E. Hibbing, C. Fuqua, M.R. Parsek, S.B. Peterson, Bacterial competition: surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 8, 15–25 (2010)
Acknowledgments
The authors would like to thank Jerrie Barnett and Peggy Feldner in Quality and Safety Assessment Research Unit in Athens for their assistant for this research. This project was supported in part by a grant from National Institute for Hometown Security.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yoon, S.C., Windham, W.R., Ladely, S. et al. Differentiation of big-six non-O157 Shiga-toxin producing Escherichia coli (STEC) on spread plates of mixed cultures using hyperspectral imaging. Food Measure 7, 47–59 (2013). https://doi.org/10.1007/s11694-013-9137-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11694-013-9137-4