Abstract
Purpose
Pathology from trans-perineal template mapping biopsy (TTMB) can be used as labels to train prostate cancer classifiers. In this work, we propose a framework to register TTMB cores to advanced volumetric ultrasound data such as multi-parametric transrectal ultrasound (mpTRUS).
Methods
The framework has mainly two steps. First, needle trajectories are calculated with respect to the needle guiding template—considering deflections in their paths. In standard TTMB, a sparsely sampled ultrasound volume is taken prior to the procedure which contains the template overlaid on top of it. The position of this template is detected automatically, and the cores are mapped following the calculated needle trajectories. Second, the TTMB volume is aligned to the mpTRUS volume by a two-step registration method. Using the same transformations from the registration step, the cores are registered from the TTMB volume to the mpTRUS volume.
Results
TTMB and mpTRUS of 10 patients were available for this work. The target registration errors (TRE) of the volumes using landmarks picked by three research assistants (RA) and one radiation oncologist (RO) were on average 1.32 ± 0.7 mm and 1.03 ± 0.6 mm, respectively. Additionally, on average, our framework takes only 97 s to register the cores.
Conclusion
Our proposed framework allows a quick way to find the spatial location of the cores with respect to volumetric ultrasound. Furthermore, knowing the correct location of the pathology will facilitate focal treatment and will aid in training imaging-based cancer classifiers.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Prostate cancer (PCa) is the most prevalent form of cancer for men in both the USA and Canada [1, 2]. In the USA, in 2021, it was estimated that 148,530 new cases of PCa will emerge with 34,130 new deaths [1]. Following elevated or abnormal Prostatic-Specific Antigen (PSA) level or suspicion from Digital Rectal Examination (DRE), the standard procedure to diagnose PCa is by Transrectal Ultrasound (TRUS) guided endorectal biopsy—where 10–12 tissue cores are extracted. Due to the sparse sampling, this method has a significant false-negative rate of 21–28% [3]. Trans-perineal template-guided mapping biopsy (TTMB) is a method of extracting biopsy cores through the patient’s perineum. This technique requires general anesthesia but has a lower infection rate than standard endorectal biopsy as the needles pass through sterilized skin as opposed to the rectum; it systematically covers the entire gland, resulting in better sensitivity and staging of the disease [4]. Multi-parametric magnetic resonance imaging (mpMRI) is currently the imaging gold standard for PCa detection. But MRI is expensive and cannot be used for real-time intervention guidance. TRUS, although it is relatively cheaper and provides real-time imaging, has very poor sensitivity and specificity when it comes to imaging PCa (40% and 50%) [5]. Advanced TRUS modalities such as shear wave and strain elastography, radio-frequency time-series analysis, and contrast-enhanced ultrasound imaging have shown improvement in PCa detection [6,7,8]. Such modalities, in addition to biopsy data collected through standard procedures, can be used to train mpTRUS-based PCa classifiers. Although it would be ideal to use whole-mount histology as the ground truth for such classifiers since they would allow pixel-wise prediction, very few patients undergo radical prostatectomy (where whole mounts are obtainable) compared to patients undergoing biopsy procedures. The more accessible biopsy procedure generates more data, and improving its workflow can benefit a large patient population. To utilize this data, the spatial location of the cores needs to be correctly identified within the mpTRUS volume. For standard TTMB, the cores are roughly matched to a coarse region within the prostate since, for whole gland treatment, precise matching is not necessary. Commercially available software such as MIM Symphony (MIM Software, OH, USA) has a graphical user interface that allows users to manually label the cores onto images. This process is time-consuming as the user must manually place the cores one by one and any deflection of the needle must also be pre-calculated or cognitively estimated during this process. Furthermore, such a technique cannot be used directly as the needle template (13 \(\times \) 13 grid with 5 mm spacing used for needle guidance) position is not known with respect to the mpTRUS volume. Without the position of the template, the locations where the biopsy needles were inserted are not known and therefore, the position of the cores cannot be recovered. Other works in the literature that use biopsy labels for training PCa classifiers [8, 9] use 2D images collected in the same plane where the needle is visible before the core extraction. This means the imaging plane is already registered, and only the coordinate of the needle’s tip and its angle needs to be measured to find the position of the core within the image. This is not feasible for volumetric mpTRUS because each voxel depends on its neighbors, e.g., the shear modulus at a voxel depends on the wave pattern in its vicinity. Furthermore, the imaging data will be affected if imaging is carried out after needles have been inserted into the prostate.
In this work, we propose a framework that uses measurements taken from the standard workflow of TTMB (such as the needle insertion point and depth) to register the cores to a pre-operative mpTRUS volume. Our framework first calculates the trajectories of the needle paths to find the location of the cores with respect to the template—taking needle deflections present into the account. A registration step then aligns the template and the cores to the mpTRUS volume. To our best knowledge, no framework exists for carrying out this task.
Dataset and TTMB details
Dataset
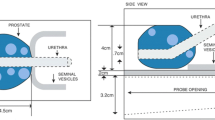
We conducted a clinical trial at BC Cancer (Vancouver Centre, Canada) to investigate the feasibility of focal low-dose-rate prostate brachytherapy (LDR-PB) for patients with early-stage PCa (REB: H12-03268-A024) [10]. With this treatment, only the region suspected of cancer is targeted with a high radiation dose. Pre-operative mpTRUS data were collected for 10 patients enrolled in this trial. After mpTRUS, TTMB was conducted on these patients. TTMB was also used for planning their treatment. Based on the cancer stage and location, eligible patients proceeded with focal LDR-PB. Both TTMB and mpTRUS were conducted with the patients in the dorsal lithotomy position and under general anesthesia. A clinical bk Pro Focus Ultrasound scanner with a bk 8848 transrectal transducer probe (bk Medical, Denmark) was used for imaging the prostate. To perform 3D volume sweeps for mpTRUS, the probe was rotated from −45\(^\circ \) to 44.5\(^\circ \) with an increment of 0.9\(^\circ \) using an in-house built robotic TRUS controller [11]. At each angle, sagittal planes of the prostate were captured using the linear array of the transducer. These 2D planes were later interpolated and scan converted into 3D volume (see ‘Target’ in Fig. 1). Additional equipment was necessary for the multi-parametric imagining (shear wave and strain elastography) and their details can be found here [6, 12]. These mpTRUS volumes are collected by streaming the raw in-phase and quadrature (I/Q) signal from the ultrasound machine. The locations of the template with respect to these volumes are not known.
Trans-perineal template mapping biopsy
In TTMB, for guiding the needles during the biopsy, a standard needle template is used. This template has 13 \(\times \) 13 grid points spaced 5 mm apart with the x-axis and y-axis coordinates spanning from A to G and 1–7, respectively (see Fig. 1). The 5 mm spacing is chosen to allow sampling of clinically significant PCa [13]. 22-mm-long tissue samples are captured using a reusable biopsy gun (Bard, Bard Inc., USA). Based on the volume of the prostate, 20–50 cores are extracted for each patient. Depending on the shape of the prostate, a single template location may have one mid-gland core or two cores (one close to the apex and the other close to the base) extracted. For reference and also for LDR-PB treatment planning (pre-planning the arrangement of radioactive sources), B-mode axial planes of the prostate starting from the base to the apex with a spacing of 5 mm are captured using the convex array of the transducer (hereinafter referred to as ‘TTMB volume’—see ‘Moving’ in Fig. 1). Transverse plane depth is controlled by rotating a knob on the stepper (where the probe is mounted) that translates the probe transversely with increments of 5 mm. For these planes, the needle template position is calibrated and overlaid on top of the ultrasound images. The distal ends of extracted cores are marked by a histology technician and deposited into separate labelled containers and sent to pathology for hematoxylin and eosin (H&E) staining and cancer identification. The extent and location of any cancer within the cores are reported by expert pathologists.
Method
To register the cores to the mpTRUS volume, our proposed framework consists of two main steps. First, the needle trajectories are calculated and the position of each biopsy core is found with respect to the template. Next, the position of the template with respect to the TTMB volume (‘moving’) is detected. This way, the cores are now mapped to the moving volume. In the second step, the moving volume is registered to the B-mode from the mpTRUS volume (‘target’). By using the same transformation from the registration step, the template and hence the cores are aligned to the target volume. The whole pipeline of this framework is shown in Fig. 2. This was implemented in MATLAB R2021a in a system with 6 core 3.2 GHz CPU and 32 GB RAM. The code is made available at https://github.com/tajwarabraraleef/TTMB-biopsy-core-registration.
Flowchart of the overall framework. The blue portions refer to the first part of the framework that calculates the needle trajectories and performs core mapping to the moving domain. The purple portion refers to the second part of the framework where the registration of cores to the target domain takes place. The green and red portions indicate the inputs and outputs of the framework
Needle trajectory calculation and mapping core to moving volume
Two parameters are taken to localize TTMB cores with respect to the template. These include the coordinate of needle insertion from the template (\(x_\mathrm{entry}\) and \(y_\mathrm{entry}\)) and the distance between the needle’s tip and base of the prostate along the z-axis (\(z_\mathrm{base2tip}\))—see Fig. 3. Although the reason for using the template is to keep the needles parallel with the point of insertion, in reality, due to tissue deformation and the bevelled tip generating a force on one side of the tip, oftentimes the needles deflect. The two components of translation/deflection experienced by the needle tip in the x and y direction can be denoted by \(\Delta x\) and \(\Delta y\), respectively (see Fig. 3). This deflection is observed from the real-time ultrasound images used by the physician during the TTMB procedure and can be easily documented along with the other two parameters. We model the trajectory as a straight line since we observed only straight needle deflections (see Fig. 3) in our data. This is likely due to the thick needle gauge (18G) and rapid throw of the biopsy gun. Furthermore, this way of modeling adds no extra time to the overall TTMB procedure as no extra measurement is required for this. More complex trajectory estimation would require imaging the needle path for every extracted core and hence adding significant clinical time and complexity to the TTMB workflow.
Figure showing how the trajectory of the needle can change based on deflection \(\Delta \)x and \(\Delta \)y. The green box shows the imaging region. \(z_0\) indicates the distance along the z-axis from the tip of the needle to the insertion point (cyan dot) on the perineum before the biopsy gun is fired. After the biopsy gun is fired, a core with a length of 22 mm is extracted as shown with the dotted part of the needle. The sagittal B-mode planes on the top right of the figure show the needle following a straight deflection (a) before and (b) after the biopsy gun is fired for a single core extraction
The coordinates of \(x_\mathrm{entry}\) and \(y_\mathrm{entry}\) which are categorized as \(x_\mathrm{entry} \subset \{A, a, B,\ldots , F\}\) and \(y_\mathrm{entry} \subset \{1, 1.5, 2,\ldots , 7\}\) are converted to mm by considering A1 as the zero point in the template and adding 5 mm with every incremental grid. To find the deflection in terms of angle in the x and y-axis, the distance from the skin (insertion point) to the tip of the needle needs to be known along the z-axis. This is given by \(z_{0}\) which is calculated with Eq. (1).
where \(z_\mathrm{start2base}\) is the distance from the start of the volume to the base of the prostate. As the image volume does not start from the skin (insertion point at the perineum), a fixed buffer of 10 mm is chosen for all patients. This 10 mm is approximated based on our dataset. However, the exact value for this can also be measured during TTMB. For our data, this was not available as this measurement is not part of the standard procedure.
With \(z_{0}\), the two angular components of deflection can be calculated in terms of \(\theta _{x}\) and \(\theta _{y}\).
Next, when the biopsy gun is fired, the needle follows the deflection angle and in an instant, captures a 22 mm core. The \(x_i\) and \(y_i\) coordinate of the core present at every plane \(z_i\) (\(z_{i} \subset [z_{0}, z_{0} + 22\text { mm}]\)) is calculated using Eq. (3).
This way, the trajectories of the needles are calculated and the locations of the cores relative to the template are found. Refer to Fig. 3 for details of the different parameters. To label these cores, information from the pathology report which contains the extent and location of cancer are used. Oftentimes, the core does not remain precisely 22 mm long but rather shrinks due to fixation and dehydration once outside the body [14]. The report also contains the observed length of the cores during pathology. To account for any shrinkage, the labels of the cores are linearly extrapolated back to 22 mm.
To know the location of the cores on the moving volume, the needle template with respect to the moving domain must be found. As the template is overlaid onto the moving slices, the grid location of the template was found automatically by performing normalized cross-correlation (NCC) between one of the moving slices and the binary mask of the template. With the location of the template found, previously calculated core locations relative to the template are mapped to the moving volume.
Detailed distribution of TRE based on landmarks picked by the evaluators for each case. For RA1-3, landmarks from both the first and second round were also plotted together with the same color coding. Here, the plots show the median (in white dot), mean (horizontal line), interquartile range (thin black box within each plot), and the outliers (points outside the thin black box). The variability between average TRE between two rounds of landmark selection for RA1-3 can be easily seen from the black line connecting the two means—higher the slope, the higher the variation
Registering cores to the target volume
As the location of the template is not known with respect to the target volume, registering the moving volume to the target domain will also align the template with it. The registration starts by identifying all planes from the base to apex with 5 mm intervals in the target volume. These planes correspond to the same planes present in the moving volume. Our framework plots the moving and target volumes side by side and allows easy scrolling through the target volume slices using the mouse wheel. This way the user can quickly select one of the mid-corresponding planes. With one plane identified, the rest of the corresponding planes can be sampled following the 5 mm intervals. The mid-corresponding plane can also be found automatically using exhaustive searching; however, matching a single plane manually is simple and it took less than 5 s per patient to do this compared to exhaustive search method that increases the registration time by 5–8 times and sometimes even fails to converge. Next, each of the corresponding planes taken from moving and target volume is aligned together following an intensity-based image registration. This is done in two steps: first, a euclidean transformation (translation and rotation) is performed that centers the two volumes followed by a similarity transformation (translation, rotation, and scaling) that further fine-tunes the alignment. For both steps, convergence was achieved with a One plus one evolutionary optimizer with Mattes mutual information as the convergence metric. Even though our data are mono-modal, a convergence metric commonly used for multi-modal data is picked since our mpTRUS volumes are converted from raw I/Q signals while the TTMB volumes are screen-captured from the Ultrasound scanner. These screen-captured B-modes are reconstructed using highly optimized techniques that are only known to the manufacturer and therefore cannot be replicated. This makes volumes between the two domains appear multi-modal. The growth factor, epsilon, initial radius, and the max iterations of the optimizer were tuned to 1.05, 1.5\(e^{-6}\), 6.25\(e^{-3}\), and 100, respectively. Finally, the average transformation of these planes is taken and applied to the template and the core locations from the moving domain—effectively registering the cores to the mpTRUS volume.
Results and discussion
To evaluate the quality of the registration, landmarks (matching observed hyperechoic or hypoechoic tissue structures) were chosen between the registered and target volumes. Three research assistants (RA1-3) with several years of expertise with ultrasound and prostate imaging and one radiation oncologist (RO) with over 35 years of experience were asked to pick landmarks for all the cases. Target registration error (TRE) was calculated for each case by finding the Euclidean distance between the landmarks. For each case, Table 1 shows the number of landmarks picked and the corresponding mean and standard deviation of TRE after just euclidean transform and then after both euclidean and similarity transform. The table also lists the time it took for running the registration framework. The number of landmarks picked for each case depended on the difficulty of locating matching landmarks in the aligned volumes and ranged between 9 and 17. It is important to note that when performing the axial imaging for TTMB volume, the probe was inserted and then retracted from the patient at 5 mm intervals which caused deformation in the prostate. This deformation can vary based on patient-specific parameters such as the patient’s weight and prostate size. Indeed, for 5 of the patients (cases 1, 2, 3, 4, and 8) the TRE was greatly improved with the addition of the similarity transform that incorporates scaling—allowing correction of deformation from the change in pressure on the prostate during probe retraction. On average, the final TRE calculated were similar between the three RA at 1.32 ± 0.8 mm, 1.41 ± 0.8 mm, and 1.23 ± 0.6 mm, respectively. For RO, the picked landmarks had a much lower average TRE of 1.03 ± 0.6 mm. Using our framework, the average duration for performing all steps including template detection, needle trajectory estimation, and registration of cores to mpTRUS volume was 97 s. In contrast, manual mapping of cores to the TTMB volume alone takes 20 ± 10 min and for mapping to mpTRUS volume, a template detection and registration algorithm is still required. Since selecting landmarks from Ultrasound is notably difficult, to evaluate the possibility of variability, RA1–3 were asked to pick landmarks again after two months of gap. Figure 4 shows a more detailed distribution of the TRE from both the first and second rounds of landmark picking for each case and each evaluator. The average TRE difference observed in the second round of landmark selection by the three RA were \(+0.16\) mm, \(-0.09\) mm, and \(+0.21\) mm—indicating not as much variability.
In Fig. 5, for Case 10, the registered biopsy cores are shown for each of the three views: axial, sagittal, and coronal. The cores are color-coded to represent their pathology. Green indicates benign cores, while blue indicates positive cores with red portions indicating the cancerous parts. In Fig. 5d, the core locations with the deflections are drawn onto a blank template based on the parameters documented during TTMB. The deflection matches with the mapped cores from our framework as seen on the axial plane in Fig. 5a.
Conclusion
We proposed a framework that registers extracted TTMB cores to mpTRUS volume. This framework is accurate, takes less time, and requires little input from the user. Such a method will enable using biopsy cores as a reference for training machine learning models for localizing PCa using advanced TRUS modalities. Finally, correctly localizing cancer from the biopsy core would also enable the possibility of successful focal-based treatments which will reduce complications seen with whole gland treatments. As the core lengths are not measured during the biopsy, there is no way to know if there was any tissue tear during core extraction. Taking measurement of the core’s length right after its extraction would allow a better estimation of whether the shrinkage is due to fixation or due to tissue tear. As this method is based on the registration of data collected before the biopsy, potential errors might arise if there is prostate motion during the needle insertions. To correct for any such motion, we suggest capturing one sagittal TRUS image with the needle in view for each core extracted. From these images, any such motion, if present, can be corrected during the needle trajectory calculation stage. Additionally, this method can also be improved by documenting the distance from the start of the imaging plane and the perineum. This information was not available to us, and therefore, an approximate value was used instead. Furthermore, we had a limited amount of data and therefore, a manual initialization of the mid-corresponding plane was quick and simple. For larger datasets, this framework can be made fully automatic by adding a segmentation network for segmenting the prostate and then finding the middle plane. Alternatively, a classification network trained to detect mid-planes can also be used to automate this. This will further improve the robustness of the framework as the registration quality is sensitive to this manual initialization.
Code availability
The code for our framework is available in the following link: https://github.com/tajwarabraraleef/TTMB-biopsy-core-registration.
References
Siegel RL, Miller KD, Fuchs HE, Jemal A (2021) Cancer statistics, 2021. CA: A Cancer J Clin 71(1):7–33
Canadian Cancer Statistics Advisory Committee in collaboration with the Canadian Cancer Society, S.C., the Public Health Agency of Canada: Canadian cancer statistics 2021. Canadian Cancer Society (2021)
Bjurlin MA, Carter HB, Schellhammer P, Cookson MS, Gomella LG, Troyer D, Wheeler TM, Schlossberg S, Penson DF, Taneja SS (2013) Optimization of initial prostate biopsy in clinical practice: sampling, labeling and specimen processing. J Urol 189(6):2039–2046
Onik G, Barzell W (2008) Transperineal 3d mapping biopsy of the prostate: an essential tool in selecting patients for focal prostate cancer therapy. In: Urologic oncology: seminars and original investigations, vol 26, pp 506–510. Elsevier
Steyerberg E, Roobol M, Kattan M, Van der Kwast T, De Koning H, Schröder F (2007) Prediction of indolent prostate cancer: validation and updating of a prognostic nomogram. J Urol 177(1):107–112
Lobo J, Baghani A, Eskandari H, Mahdavi S, Rohling R, Goldernberg L, Morris WJ, Salcudean S (2015) Prostate vibro-elastography: multi-frequency 1d over 3d steady-state shear wave imaging for quantitative elastic modulus measurement. In: 2015 IEEE international ultrasonics symposium (IUS), pp 1–4 . IEEE
Smeenge M, Mischi M, Pes MPL, de la Rosette JJ, Wijkstra H (2011) Novel contrast-enhanced ultrasound imaging in prostate cancer. World J Urol 29(5):581–587
Azizi S, Imani F, Ghavidel S, Tahmasebi A, Kwak JT, Xu S, Turkbey B, Choyke P, Pinto P, Wood B, Mousavi P, Abolmaesumi P (2016) Detection of prostate cancer using temporal sequences of ultrasound data: a large clinical feasibility study. Int J Comput Assist Radiol Surg 11(6):947-956
Javadi G, Samadi S, Bayat S, Sojoudi S, Hurtado A, Chang S, Black P, Mousavi P, Abolmaesumi P (2021) Training deep networks for prostate cancer diagnosis using coarse histopathological labels. In: International conference on medical image computing and computer-assisted intervention, pp 680–689. Springer
Mahdavi SS, Spadinger IT, Salcudean SE, Kozlowski P, Chang SD, Ng T, Lobo J, Nir G, Moradi H, Peacock M, Morris WJ (2017) Focal application of low-dose-rate brachytherapy for prostate cancer: a pilot study. J Contemp Brachyther 9(3):197
Adebar T, Salcudean S, Mahdavi S, Moradi M, Nguan C, Goldenberg L (2011) A robotic system for intra-operative trans-rectal ultrasound and ultrasound elastography in radical prostatectomy. In: International conference on information processing in computer-assisted interventions, pp 79–89. Springer
Mahdavi SS, Moradi M, Wen X, Morris WJ, Salcudean SE (2011) Evaluation of visualization of the prostate gland in vibro-elastography images. Med Image Anal 15(4):589–600
Sivaraman A, Sanchez-Salas R (2015) Transperineal template-guided mapping biopsy of the prostate. Technical Aspects of Focal Therapy in Localized Prostate Cancer, pp 101–114
Schned AR, Wheeler KJ, Hodorowski CA, Heaney JA, Ernstoff MS, Amdur RJ, Harris RD (1996) Tissue-shrinkage correction factor in the calculation of prostate cancer volume. Am J Surg Pathol 20(12):1501–1506
Funding
This work was supported by the Canadian Institutes of Health Research (CIHR MOP-1422439 and PJT-152965).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
Institutional ethics approval (REB: H12-03268-A024) was obtained for the use of clinical data in this study.
Informed consent
Informed consent was obtained from all individual participants whose data are used in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Aleef, T.A., Zeng, Q., Morris, W.J. et al. Registration of trans-perineal template mapping biopsy cores to volumetric ultrasound. Int J CARS 17, 929–936 (2022). https://doi.org/10.1007/s11548-022-02604-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-022-02604-4