Abstract
Purpose
This paper addresses localization of needles inserted both in-plane and out-of-plane in challenging ultrasound-guided interventions where the shaft and tip have low intensity. Our approach combines a novel digital subtraction scheme for enhancement of low-level intensity changes caused by tip movement in the ultrasound image and a state-of-the-art deep learning scheme for tip detection.
Methods
As the needle tip moves through tissue, it causes subtle spatiotemporal variations in intensity. Relying on these intensity changes, we formulate a foreground detection scheme for enhancing the tip from consecutive ultrasound frames. The tip is augmented by solving a spatial total variation regularization problem using the split Bregman method. Lastly, we filter irrelevant motion events with a deep learning-based end-to-end data-driven method that models the appearance of the needle tip in ultrasound images, resulting in needle tip detection.
Results
The detection model is trained and evaluated on an extensive ex vivo dataset collected with 17G and 22G needles inserted in-plane and out-of-plane in bovine, porcine and chicken phantoms. We use 5000 images extracted from 20 video sequences for training and 1000 images from 10 sequences for validation. The overall framework is evaluated on 700 images from 20 sequences not used in training and validation, and achieves a tip localization error of 0.72 ± 0.04 mm and an overall processing time of 0.094 s per frame (~ 10 frames per second).
Conclusion
The proposed method is faster and more accurate than state of the art and is resilient to spatiotemporal redundancies. The promising results demonstrate its potential for accurate needle localization in challenging ultrasound-guided interventions.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Real-time and accurate localization of handheld needles is vital for the success of percutaneous ultrasound (US)-guided interventions such as biopsies and regional anesthesia. However, when the needle shaft and tip have low intensity, needle localization is difficult. For in-plane insertions (where the US probe axis and the needle are colinear, and the whole needle is ideally visible as a hyperechoic line), small diameter needles usually produce an invisible shaft even if the needle is well aligned with the scan plane. Moreover, the needle tip may not exhibit a high-intensity feature. For out-of-plane insertions (where the US probe axis and the needle are orthogonal), only the tip or a cross section of the shaft is visible.
Several hardware-based approaches have been proposed to improve needle visibility. Mechanical needle guides are attached to US probes to align the needle trajectory to the ultrasound beam [1]. However, needle guides are not efficient in procedures where fine needle trajectory adjustments are required. Technical changes to the design of needles, such as embedding sensors at the tip [2, 3] and echogenic coats [4], enhance needle localization, but such needles are costly. 3D/4D US offers bi-planar visualization but suffers from poor resolution and a low frame rate [5]. Electromagnetic (EM)/optical tracking systems [6, 7] improve needle visualization but require specialized needles and probes. Moreover, EM systems are affected by metal objects in the operating environment. Robotic systems have been integrated with US imaging to facilitate autonomous or semiautonomous needle insertion [8]. However, robotic systems are expensive. Considering limitations associated with all these advancements, handheld US remains the gold standard in clinics.
On the other hand, image processing methods do not require additional hardware to the conventional US system. Some of these methods rely on full or partial brightness of the needle shaft and tip [9,10,11,12,13]. When the shaft or tip is not conspicuous, these methods are untenable. Moreover, [10,11,12,13] focus on in-plane insertion, yet in some procedures, in-plane trajectories may be impractical because of the need to go around critical anatomy.
There have been attempts to localize needles from dynamic intensity changes that arise from needle movement in the US image [9, 14, 15]. These utilize optical flow, which works best if intensity changes associated with needle motion exhibit a smooth transition. Further, optical flow assumes that neighboring points in an image always belong to the same feature and move together. These two assumptions reduce reliability of optical flow for dynamic handheld needle localization: Spatiotemporal redundancies reduce localization efficiency, and US images are sensitive to speckle and susceptible to artifacts arising from abrupt changes in probe motion, patient movement, hyperechoic anatomy or physiological events such as pulsation and breathing.
Recently, deep learning-based methods using convolutional neural networks (CNNs) have shown promise for detection of the needle in static 2D US [16] and 3D US data [17, 18]. In our recently published work [16], we demonstrated a robust approach for needle detection. However, it was difficult to detect all needle pixels, and thus, tip localization required a computationally expensive post-processing step. Hence, although the needle detection step could be performed in real time, the overall needle localization process could not.
In this paper, we propose a robust needle tip localization strategy in 2D US that combines a computationally efficient tip enhancement framework and a deep learning approach that captures the expected tip shape and variation. By learning the expected features associated with the needle tip, the model successfully localizes the tip in the presence of motion artifacts arising from low-amplitude perturbations. The main contributions of this paper are: (1) a novel digital subtraction algorithm that performs differencing of consecutive image frames within the US sequence. Thus, we are able to extract salient motion from temporal relationships in the US sequence with a dynamic background model; (2) an augmentation technique for the needle tip, in which we extend the split Bregman approach to solve a spatial total variation (TV) problem for the tip-enhanced image; and (3) a single-shot detector deep learning framework optimized for needle tip detection from end-to-end learning. The detector learns contextual patterns associated with the needle tip and outputs static bounding boxes, from which the needle tip position is estimated.
The proposed method achieves both in-plane and out-of-plane needle localizations, as well as localization of thin needles since it does not depend on full needle visibility. This is achieved at significantly faster computational accuracy than state of the art. Our method is suitable for minimally invasive procedures where there is minimum tissue/organ motion, for example in spinal/epidural and peripheral nerve blocks. Since the method is resilient to low-amplitude perturbations, it would work if there is motion from breathing during needle insertion. It is also insensitive to high-intensity artifacts from anatomical structures or other instruments. The proposed method could be utilized in a smart computer-assisted interventional system to facilitate needle localization in challenging US-guided interventions.
Methods
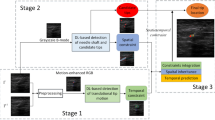
The proposed method is designed for handheld 2D US probes during in-plane and out-of-plane needle insertion. The problem of motion-based needle localization is split into two main components: (1) detecting moving objects in each frame and (2) associating the detections corresponding to the needle over time. Consequently, the proposed method consists of three main stages illustrated in Fig. 1: (1) we detect scene changes caused by needle motion in the US image scene (“Needle tip enhancement model” section). In each frame of the US sequence, the needle tip is treated as the foreground, while the rest of the image is designated as background data. Needle enhancement is performed from logical subtraction of the dynamic reference US frame from the current US frame. This step does not require a priori knowledge of needle insertion side or angle; (2) we augment the appearance of the enhanced needle tip, obtained from step 1, using a spatial regularization filter (“Needle tip augmentation” section); and (3) we localize the needle tip using a deep learning approach adapted from the YOLO architecture [19] (“Needle tip detection” section). Next, we describe how these three major processes are achieved.
Needle tip enhancement model
Consider a US frame sequence with temporal continuity, represented by the function \( p\left( {x,y,t} \right) \), where \( t \) denotes the position in the time sequence and \( \left( {x,y} \right) \) are the spatial coordinates. We propose a dynamic background subtraction model which quickly adapts to changes in the US scene based on logical differencing between adjacent frames. For the first frame, the background is denoted as: \( b\left( {x,y,t_{0} } \right) = p\left( {x,y,t_{0} } \right) \). For all subsequent frames, the background is modeled as the previous frame in the sequence, i.e., \( b\left( {x,y,t_{n} } \right) = p\left( {x,y,t_{n - 1} } \right). \) We then determine the bitwise complement of the background image. Considering only spatial variation, for \( b\left( {x,y} \right) = \left( {x,y} \right)|b\left( {x,y} \right) \ne 0 \), the complement is \( b^{c} \left( {x,y} \right) = \left( {x,y} \right) \in {\mathbb{Z}}^{2} |\left( {x,y} \right) \notin b\left( {x,y} \right) \). For an 8-bit image, the complement of each pixel (an unsigned integer) is equal to itself subtracted from 255. For any current frame \( p\left( {x,y} \right) \), the needle-enhanced image is given by:
where \( \wedge \) denotes the pointwise AND logical operation. (1) yields only the objects in the US data that moved between two successive frames and thus gives an enhanced current tip location. Although it is plausible that tissue surrounding the needle tip moves concurrently, we consider collocated motion of the tissue and tip to be more significant than any other motion. Depending on the needle visibility profile, \( q\left( {x,y} \right) \) may also contain shaft pixels.
Needle tip augmentation
The output of (1) \( q\left( {x,y} \right), \) may contain artifacts caused by brightness variations, motion artifacts and speckle. We need to further enhance \( q\left( {x,y} \right) \) to minimize the effect of this noise. This step is crucial before the employment of the deep learning framework explained in “Needle tip detection” section. Without it, our model may attempt to overfit the noise at the expense of needle features. Therefore, we first devise means of denoising \( q\left( {x,y} \right) \). First, \( q\left( {x,y} \right) \) is passed through a median filter with an 8 × 8 kernel. We denote the resulting image as \( r\left( {x,y} \right) \). While speckle noise is multiplicative, we formulate an additive noise model to aggregate the effect of speckle, motion artifacts and any other stochastic or deterministic noise sources: \( r\left( {x,y} \right) = e\left( {x,y} \right) + n\left( {x,y} \right) \), i.e., a sum of two components; the desired image \( e\left( {x,y} \right) \) and the aggregate noise, \( n\left( {x,y} \right) \). We consider \( e\left( {x,y} \right) \) to be a function of bounded variation. Going forward, we will adopt a notation where the images are represented by vectors. The image restoration model becomes:
where \( {\mathbf{e}} \in {\mathbb{R}}^{mn \times 1} \) is the desired augmented needle tip image (of size \( m \times n \)), \( {\mathbf{r}} \in {\mathbb{R}}^{mn \times 1} \) is the corrupted image obtained from the previous step, while \( {\mathbf{n}} \in {\mathbb{R}}^{mn \times 1} \) is the noise. In this notation, \( {\mathbf{r}} \), e and n are vectors containing all the pixel values in the respective image matrices in lexicographic order. Conceptually, this problem necessitates recovering low-rank matrices from under-sampled measurements, and it can be solved using total variation (TV)-based methods [20, 21]. Problems of this nature are ill-conditioned and solving them directly is difficult due to noise sensitivity. Since pixels in the segmented image have spurious detail and possibly high TV, we formulate a TV regularization problem of the form:
where \( \lambda \) is a regularization parameter and \( \left\| {\mathbf{e}} \right\|_{{TV}} = \left\| {\varvec{D}_{\varvec{x}} {\mathbf{e}}} \right\|_{1} + \| {\varvec{D}_{\varvec{y}} {\mathbf{e}}}\|_{1} \) is the anisotropic TV norm, defined by \( \varvec{D}_{\varvec{x}} \) and \( \varvec{D}_{\varvec{y}} \), the spatial first-order forward finite difference operators along the horizontal and vertical directions, respectively. (3) is a constrained formulation of a non-differentiable optimization problem. This problem can be efficiently solved with the split Bregman approach [22], in which the main problem is reduced to a sequence of unconstrained optimization problems and variable updates. We first transform (3) into a constrained equivalent problem by introducing intermediate variables \( {\mathbf{v}} \) and \( {\mathbf{w}} \), i.e.,
The formulation in (4) can be converted into an unconstrained convex optimization problem (5) by use of augmented Lagrangian and split Bregman techniques [21], where the constraints in (4) are weakly enforced by introducing quadratic penalties:
where \( \nu \) is an additional regularization parameter, and \( \varvec{b}_{1} \) and \( \varvec{b}_{2} \) are Bregman relaxation variables which are determined through Bregman iteration. Inclusion of the last two augmented Lagrangian terms in (5) improves algorithm robustness since we do not have to strictly reinforce the equality constraint. (5) can be split into three subproblems, solved by fixing one variable and minimizing over the other in turn:
(6) and (7) decouple over space and have closed-form solutions as vectorial shrinkages (soft thresholding):
(8) is a simple least square problem (Tikhonov regularization) which can be solved analytically using a gradient descent algorithm. First, we derive the pertinent normal equation:
(10) is solved using LSMR [23], an iterative least squares solver. \( \varvec{b}_{1} \) and \( \varvec{b}_{2} \) are initialized to zero and updated between every consecutive iteration of the subproblems: \( \varvec{b}_{1}^{i + 1} = \varvec{b}_{1}^{i} + \varvec{D}_{\varvec{x}} {\mathbf{e}} - {\mathbf{v}} \), \( \varvec{b}_{2}^{i + 1} = \varvec{b}_{2}^{i} + \varvec{D}_{\varvec{y}} {\mathbf{e}} - {\mathbf{w}}. \) The enhancement process is summarized in Algorithm 1. Figure 2 illustrates the result of needle tip augmentation.

Needle augmentation in three consecutive frames with in-plane insertion of a 17G needle in a bovine tissue phantom (I–III) and one frame with out-of-plane insertion of a 17G needle in a porcine shoulder phantom (IV). a Original images before tip enhancement and augmentation. Identifying the needle in these images is difficult. b Tip-augmented image \( e\left( {x,y} \right) \) (color-coded). Circle surrounds the augmented tip. The proposed method achieves accurate enhancement of the tip despite low tip intensity in the original image or the presence of high-intensity artifacts
Needle tip detection
From the preceding sections, we have achieved a needle tip-enhanced image \( e\left( {x,y} \right) \) in which the tip exhibits a high intensity. However, we still need to localize the tip. Usually, the needle tip will not move in each US frame because the speed of needle actuation by hand may not match the US frame rate or the operator may intermittently stop moving the needle. Therefore, we need to identify frames in which no significant motion has occurred. Further, despite the prior enhancement process, there could still be high-intensity interfering artifacts not associated with needle motion. Therefore, we cannot rely on the tip to always exhibit the highest intensity in \( e\left( {x,y} \right). \) For these reasons, we sought to formulate a deep learning framework for efficient needle tip detection. Next, we describe elements of the deep learning framework that are unique to our method.
CNN architecture The proposed deep learning framework is shown in Fig. 3 and is built based on YOLO [19], a state-of-the-art single-shot object detection CNN architecture. The framework outputs 2D bounding box predictions consisting of five components: \( x,y,w,h \) and \( \eta \), where \( \left( {x,y} \right) \) coordinates represent the center of the box, \( w \) and \( h \) are the width and height, respectively, and \( \eta \) is the confidence that the box contains an object and that the object is the needle tip. The new framework consists of a 256 × 256 image input layer, unlike the one in [19] which has a 416 × 416 input. To further reduce computational complexity toward real-time performance, we use only eight convolutional layers. We implement a pixel-level fusion layer in which the current US image \( p\left( {x,y} \right) \) and its tip-enhanced counterpart \( e\left( {x,y} \right) \) are concatenated before inputting to the CNN. Since the needle tip is a fine-grained feature, we configure the convolution layers to maintain spatial dimensions of the respective inputs, thus mitigating reduction in resolution. More so, CNN neurons at deeper layers always have large receptive fields that will ensure incorporation of image-level context pertinent to needle tip appearance.
Uniquely, each of the first seven convolution layers is followed by an exponential linear unit (ELU) [24] with \( \alpha = 0.5 \). The YOLO implementation in [19] utilizes leaky rectified linear unit (leakyReLU) activations. In [24], it is shown that with ELU, activations close to zero mean and unit variance always converge toward zero mean and unit variance even under the presence of noise and perturbations. This informed our choice of ELU. In Sect. 3, we will present comparative analysis of the proposed model’s performance with and without ELU. The first five convolution layers are followed by a 2 × 2 max pooling layer with a stride of 2. All the other physical attributes of the YOLO architecture in [19] are unchanged. At test time, the model is malleable to any input size. Two advantages accrue from treating our challenge as a detection problem. Inherently, needle tip features will be learned end to end, thus eliminating the need to explicitly encode them. It is expected that frames where no needle tip has moved will exhibit no detectable features, while the learned model will accurately extract the tip when it is present.
Training details The model is initialized with weights derived from training on the PASCAL VOC dataset [25]. The ground-truth bounding box labels are defined using an EM tracking system and an expert radiologist with over 30 years of experience in interventional radiology. The ground-truth tip location becomes the center of the bounding box \( \left( {x,y} \right) \), and the thickness \( w \times h \) is chosen to be at most 20 × 20 pixels in all images. We use an initial learning rate of 10−4, a batch size of 4 and train for 60 epochs. Our choice of optimizer is Adam.
Data acquisition and experimental validation
To train and evaluate our model, we collected a dataset of 2D B-mode US images using materials and settings specified in Table 1. Two imaging systems: SonixGPS (Analogic Corporation, Peabody, MA, USA) with a handheld C5-2/60 curvilinear probe and 2D handheld wireless US (Clarius C3, Clarius Mobile Health Corporation, Burnaby, BC, Canada) were used. Experiments were performed on a freshly excised bovine tissue, a porcine shoulder phantom and chicken breast, with insertion of a 17G (1.5 mm diameter, 90 mm length) Tuohy epidural needle (Arrow International, Reading, PA, USA), a 17G SonixGPS vascular access needle (Analogic Corporation, Peabody, MA, USA) and a 22G spinal Quincke-type needle (Becton, Dickinson and Company, Franklin Lakes, NJ, USA). In all our experiments, the probe was handheld. Small amplitude perturbations not associated with needle motion were simulated by manually pressing the probe against the imaging medium and rotating it slightly about its long axis. Further, the chicken breast overlaid on a lumbosacral spine model was immersed in a water bath during needle insertion to simulate fluid motion in the imaging medium. With the SonixGPS needle, we collected ground-truth needle tip localization data using an EM tracking system (Ascension Technology Corporation, Shelburne, VT, USA). In-plane insertion was performed at 40°–70°, and the needle was inserted up a depth of 70 mm. Fifty (35 in-plane, 15 out-of-plane) sequences of US images, each containing more than 400 frames, were collected.
Performance of the proposed method was evaluated by comparing the automatically detected tip location (center of the detected bounding box) to the ground truth determined from the EM tracking system for data collected with the SonixGPS needle. For data collected with needles without tracking capability, the ground truth was determined by our expert radiologist. By retrospectively inspecting the frame sequences, we obtained the ground-truth tip location from intensity changes and tissue deformation. (This is more difficult in the real-time clinical setting.) To account for large EM tracking errors (since the sensor does not reach the needle tip), the radiologist performed manual labeling of the dataset obtained with the SonixGPS needle and compared the EM data with the manual data. In scenarios where the tip intensity is low, the EM system provides annotation on the US frames which acts as a visual cue to the approximate tip location, and the expert used this information to label the tip. If the difference in tip localization was 4 pixels (~ 0.7 mm) or greater, the localizations were not included in our computation. Tip localization accuracy was determined from the Euclidean distance between the ground truth and the localization from our method.
We implemented our methods on an NVIDIA GeForce GTX 1060 6 GB GPU, 3.6 GHz Intel(R) Core™ i7 16 GB CPU Windows PC. The needle tip enhancement and augmentation methods were implemented in MATLAB 2018a. For the subproblems in (9) and (10), we empirically determined \( \nu = 2 \) and \( \lambda = 5 \) as optimum values. Throughout the validation experiments, these values were not changed. The tip detection framework was implemented in Keras 2.2.4 (on the Tensorflow 1.1.2 backend). In total, 5000 images from 20 video sequences were used for training, while 1000 images from 10 other sequences were used for validation. Lastly, 700 images from 20 sequences not used in training or validation were used for testing. The images were purposely selected from continuous sequences where there is needle motion.
Experimental results and discussion
Qualitative Results Figure 4 shows needle detection results for four consecutive frames for both in-plane and out-of-plane insertions. Note that the tip is accurately localized despite the presence of other high-intensity interfering artifacts in the B-mode US data. In case there is a point cloud arising from partial enhancement of the shaft, the detection CNN learns to automatically identify the tip at the distal end of the cloud in the enhanced image \( e\left( {x,y} \right). \) For out-of-plane insertion, the temporal window for needle tip visibility is limited, but our method can be useful for tracking small movements of the needle tip close to the target. Meanwhile, our method is agnostic to the type and size of needle used if the tip appears in the enhanced US image and needle motion is available in the B-mode data. However, increasing the training data size for each needle type would improve the performance of the proposed method.
Needle detection and localization in four consecutive frames with a in-plane insertion of the 17G SonixGPS needle into chicken breast tissue and b out-of-plane insertion of the 22G needle into the porcine shoulder phantom. (I) Original image. The white box is the annotated ground-truth label, determined with an electromagnetic tracking system for (a) and an expert sonographer for (b). (II) Detection result with bounding box (white) overlaid on enhanced image \( e\left( {x,y} \right) \). The inset number alongside the box annotation is the detection confidence. (III) Localized tip, the center of the detected bounding box (red star) overlaid on original image. Our method achieves high detection and localization accuracy
Model comparison Ablation studies, where the structural configuration of a deep learning framework is altered to assess the impact on model performance, are used to justify design choices. In line with this standard approach, we compare efficiency of our needle tip detection framework to that from alternative implementation approaches. We evaluate accuracy of detection using the mean average precision (mAP) metric on the validation dataset. mAP is calculated as the average value of the precision across a set of 11 equally spaced recall levels [25], yielding one value that depicts the shape of the precision–recall curve. Table 2 shows the mAP for different configurations of the detection CNN. First, we examine performance of the proposed CNN with the raw US image \( p\left( {x,y} \right) \) as an input. As expected, the detection efficiency is very low (20.2%). This is because without our tip enhancement algorithm, tip features are barely discernible and are overshadowed by other high-intensity artifacts in the cluttered US image. We also consider only the enhanced image \( e\left( {x,y} \right) \) as the input. A high mAP of 86.7% is achieved, showing that our enhancement algorithm is efficient. Furthermore, we show that fusion of \( e\left( {x,y} \right) \) and \( p\left( {x,y} \right) \) achieves the highest mAP of 94.6%
With the fusion input, and with other hyperparameters maintained constant, we compare performance of the proposed method against a similar model with leakyReLU activation layers as is the case in [19] instead of ELU. The proposed method outperforms this configuration. It is worth mentioning that we chose a batch size of 4 in all our experiments because of the memory constraints of the GPU. It is expected that a bigger batch size would have resulted in an even higher mAP from the proposed model.
Runtime performance On the NVIDIA GeForce GTX 1060 GPU, our framework runs at 0.094 ± 0.01 s per frame (0.014 s for enhancement, 0.06 for augmentation and 0.02 s for detection). This is ~ 10 frames per second (fps), and to the best of our knowledge, the fastest needle tip localization framework reported so far. Certainly, the processing speed can be increased with more computing resources. In frames where the needle tip is salient, the augmentation step is unnecessary, and the runtime speed increases to 29 fps.
Mitigating false detections Since YOLO is a multi-object detection framework, it is possible that several bounding boxes with different confidence scores can be detected on the same input image. We sought to minimize these false positives by selecting the bounding box with the highest confidence score and using a hard threshold of 0.35 for the score, a value which was empirically determined and kept constant throughout validation. With this threshold, we achieved an overall sensitivity and specificity of 98% and 91.8%, respectively. It is expected that the robustness of tip detection would further be improved if a bigger training dataset was used. To mitigate the effect of the false positives on tip localization, we estimate the needle trajectory using the technique illustrated in Fig. 5. We assume that the tip detection framework has already accurately localized two previous spatial positions \( A\left( {x_{1} ,y_{1} } \right) \) and \( B\left( {x_{2} ,y_{2} } \right) \) in successive frames that are at least 30 pixels (~ 5 mm) apart. From A and B, we approximate the needle trajectory \( \alpha_{1} = \tan^{ - 1} \left( {\left| {\left( {y_{2} - y_{1} } \right)/\left( {x_{2} - x_{1} } \right)} \right|} \right) \). Then for each subsequent detection with a bounding box at \( F\left( {x_{\text{f}} ,y_{\text{f}} } \right) \), we estimate the trajectory angle \( \alpha_{2} \) using points A and F, with A as a static reference. If \( \left| {\alpha_{1} - \alpha_{2} } \right| > 10^\circ \), the new detection is deemed to be skewed from the correct trajectory (and thus a false positive), and the localization result is not utilized in calculating localization error. During the localization process, false positives and true negatives lead to maintenance of the current tip position. In so doing, our method is robust to spatiotemporal redundancies.
Tip localization accuracy Overall, the tip localization error was 0.72 ± 0.4 mm. Direct and fair comparison to state-of-the-art methods is difficult since our dataset is collected to suit evaluation of a method that does not require initial needle visibility. Although the method in [14] localizes imperceptible in-plane inserted needles with good accuracy (0.82 mm), their computation time of 1.18 s per frame (~ 1 fps) is significantly inferior to ours (10 fps).
We compared the proposed method to the method in [16] by evaluating the two on the same set of 200 randomly selected US images with only in-plane needle insertion. The results are shown in Table 3. Note that the proposed method outperforms the method in [16] in both tip localization accuracy and computational efficiency. For a fair comparison, localization errors above 2 mm (56% of the data) were discarded. A one-tailed paired t test shows that the difference between the localization errors from the proposed method and the method in [16] is statistically significant (p < 0.005). The localization accuracy obtained from [16] is worse than previously reported because we used a more challenging dynamic dataset with very low shaft intensity, unlike [16] where static US images were used for validation. We also compared the proposed method to an intensity-based method that directly localizes the needle tip using the Hough transform and RANSAC [10]. This method achieved success in only 18% of the dataset (neglecting errors > 2 mm), with an overall localization error of 1.2 ± 0.32 mm.
Furthermore, we determined needle localization from the maximum intensity in \( e\left( {x,y} \right) \), i.e., the proposed method without the tip detection step. The results are shown in Table 4 and demonstrate that the localization accuracy is worse without the detection framework. This is expected because without the benefit of implicitly learning heuristic features associated with the tip via deep learning, there is a higher likelihood of localizing artifacts with similar intensity to the tip.
Conclusions
We have demonstrated a novel approach for needle tip localization in 2D US, suitable for challenging imaging scenarios in which the needle is not continuously visible. The main strength of our work is in the robust and accurate tip localization at a close to real-time processing rate of 10 fps. This is better than reported in previous methods [9,10,11,12,13,14,15,16,17,18]. The proposed method does not require the needle to appear as a high-intensity, continuous linear structure in the US image. Therefore, both in-plane and out-of-plane needle localizations are possible. We used the thinner 22G needle in our experiments to demonstrate the robustness of our method. Typically, such thin needles are prone to bending and the shaft has limited visibility, but this did not affect the accuracy of tip localization. Therefore, it is possible that our method can localize bending needles. However, we will further investigate this in our future work.
The detection component in our method mitigates motion artifacts arising from small amplitude perturbations simulated from probe pressure, probe rotation and fluid motion. Generally, any method reliant on motion detection is prone to drastic motion between consecutive frames, for example due to abrupt changes in probe alignment or rapid physiological motion, such as pulsation and breathing. In the clinical scenario, needle advancement is often paused if major probe re-orientation is to be undertaken, so this would not be a major hindrance to our method. Further, while we postulate that our method is robust to physiological activity such as breathing and pulsation, we will further investigate this in future clinical studies.
References
Elsharkawy H, Babazade R, Kolli S, Kalagara H, Soliman ML (2016) The Infiniti plus ultrasound needle guidance system improves needle visualization during the placement of spinal anesthesia. Korean J Anesthesiol 69(4):417–419
Lu H, Li J, Lu Q, Bharat S, Erkamp R, Chen B, Drysdale J, Vignon F, Jain A (2014) A new sensor technology for 2D ultrasound-guided needle tracking. MICCAI 17(Pt. 2):389–396
Xia W, West S, Finlay M, Mari J, Ourselin S, David A, Desjardins A (2017) Looking beyond the imaging plane: 3D needle tracking with a linear array ultrasound probe. Sci Rep 7(1):3674
Miura M, Takeyama K, Suzuki T (2014) Visibility of ultrasound-guided echogenic needle and its potential in clinical delivery of regional anesthesia. Tokai J Exp Clin Med 39(2):80–86
Arif M, Moelker A, van Walsum T (2018) Needle Tip Visibility in 3D Ultrasound Images. Cardiovasc Interv Radiol 41(1):145–152
Fevre MC, Vincent C, Picard J, Vighetti A, Chapuis C, Detavernier M, Allenet B, Payen JF, Bosson JL, Albaladejo P (2018) Reduced variability and execution time to reach a target with a needle GPS system: comparison between physicians, residents and nurse anaesthetists. Anaesth Crit Care Pain Med 37(1):55–60
Stolka PJ, Foroughi P, Rendina M, Weiss CR, Hager GD, Boctor EM (2014) Needle guidance using handheld stereo vision and projection for ultrasound-based interventions. MICCAI 17(Pt.2):684–691
Priester AM, Natarajan S, Culjat MO (2013) Robotic ultrasound systems in medicine. IEEE EEE Trans Ultrason Ferroelectr Freq Control 60:507–523
Ayvali E, Desai J (2014) Optical flow-based tracking of needles and needle-tip localization using circular hough transform in ultrasound images. Ann Biomed Eng 43(8):1828–1840
Zhao Y, Cachard C, Liebgott H (2013) Automatic needle detection and tracking in 3D ultrasound using an ROI-based RANSAC and Kalman method. Ultrason Imaging 35(4):283–306
Hacihaliloglu I, Beigi P, Ng G, Rohling RN, Salcudean S, Abolmaesumi P (2015) Projection-based phase features for localization of a needle Tip in 2D curvilinear ultrasound. MICCAI 9349:347–354
Hatt CR, Ng G, Parthasarathy V (2015) Enhanced needle localization in ultrasound using beam steering and learning-based segmentation. Comput Med Imaging Graph 41:46–54
Mwikirize C, Nosher JL, Hacihaliloglu I (2018) Signal attenuation maps for needle enhancement and localization in 2D ultrasound. Int J CARS 13(3):363–374
Beigi P, Rohling R, Salcudean S, Ng G (2017) CASPER: computer-aided segmentation of imperceptible motion-a learning-based tracking of an invisible needle in ultrasound. Int J CARS 12(11):1857–1866
Beigi P, Rohling R, Salcudean SE, Ng GC (2016) Spectral analysis of the tremor motion for needle detection in curvilinear ultrasound via spatiotemporal linear sampling. Int J CARS 11(6):1183–1192
Mwikirize C, Nosher JL, Hacihaliloglu I (2018) Convolution neural networks for real-time needle detection and localization in 2D ultrasound. Int J CARS 13(5):647–657
Pourtaherian A, Ghazvinian Zanjani F, Zinger S, Mihajlovic N, Ng G, Korsten H, With P (2017) Improving needle detection in 3D ultrasound using orthogonal-plane convolutional networks. MICCAI 2:610–618
Pourtaherian A, Ghazvinian Zanjani F, Zinger S, Mihajlovic N, Ng G, Korsten H, With P (2018) Robust and semantic needle detection in 3D ultrasound using orthogonal-plane convolutional neural networks. Int J CARS 13(9):1321–1333
Redmon J, Farhadi A (2016) Yolo9000: better, faster, stronger. arXiv:1612.08242
Afonso M, Bioucas-Dias J, Figueiredo M (2010) Fast image recovery using variable splitting and constrained optimization. IEEE Trans Image Process 19(9):2345–2356
Chan S, Khoshabeh R, Gibson K, Gill P, Nguyen T (2011) An augmented lagrangian method for total variation video restoration. IEEE Trans Image Process 20(11):3097–3111
Goldstein T, Osher S (2009) The split Bregman method for L1-regularized problems. SIAM J Imaging Sci 2(2):323–343
Fong D, Saunders M (2011) LSMR: an iterative algorithm for sparse least-squares problems. SIAM J Sci Comput 33(5):2950–2971
Klambauer G, Unterthiner T, Mayr A, Hochreiter S (2017) Self-normalizing neural networks. arXiv:1706.02515v5
Everingham M, Gool LV, Williams CKI, Winn J, Zisserman A (2010) The PASCAL visual object classes (VOC) challenge. Int J Comput Vis 88:303–338
Acknowledgements
This work was accomplished with funding support from the North American Spine Society 2017 young investigator award.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
This article does not contain patient data.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 1 (MP4 51745 kb)
Rights and permissions
About this article
Cite this article
Mwikirize, C., Nosher, J.L. & Hacihaliloglu, I. Learning needle tip localization from digital subtraction in 2D ultrasound. Int J CARS 14, 1017–1026 (2019). https://doi.org/10.1007/s11548-019-01951-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-019-01951-z