Abstract
Purpose
Accurate segmentation of the mandibular canal in cone beam CT data is a prerequisite for implant surgical planning. In this article, a new segmentation method based on the combination of anatomical and statistical information is presented to segment mandibular canal in CBCT scans.
Methods
Generally, embedding shape information in segmentation models is challenging. The proposed approach consists of three main steps as follows: At first, a method based on low-rank decomposition is proposed for preprocessing. Then, a conditional statistical shape model is trained, and mandibular bone is segmented with high accuracy. In the final stage, fast marching with a new speed function is utilized to find the optimal path between mandibular and mental foramen. Fast marching tries to find the darkest tunnel close to the initial segmentation of the canal, which was obtained with conditional SSM model. In this regard, localization of mandibular canal is performed more accurately.
Results
The method is applied to the identification of mandibular canal in 120 sets of CBCT images. Conditional statistical model is evaluated by calculating the compactness capacity, specificity and generalization ability measures. The capability of the proposed model is evaluated in the segmentation of mandibular bone and canal. The framework is effective in noisy scans and is able to detect canal in cases with mild bone resorption.
Conclusion
Quantitative analysis of the results shows that the method performed better than two other recent methods in the literature. Experimental results demonstrate that the proposed framework is effective and can be used in computer-guided dental implant surgery.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Cone beam computed tomography (CBCT) is an increasingly applied imaging acquisition for dental surgical planning [1] due to the lower hardware cost and accessibility compared to conventional CT. The first step in the planning of implant surgery is accurate segmentation of mandibular canal which results in safety margin around the facial nerves. These nerves give sensation to the lower lip, tongue and teeth, and if they become damaged, the recovery time would be about 3 to 6 months [2]. Localization of the canal is usually performed manually by a radiologist; however, manual segmentation becomes tedious and time-consuming due to the large amount of data to be analyzed.
Early researches in automated canal segmentation were performed on CT images. Stein et al. [3] proposed a method based on Dijkstra’s algorithm and limited Dijkstra’s search to multiple erosions and dilations to trace inside the bone. Hanssen et al. [4] improved Stein’s method by replacing Dijkstra’s algorithm with fast marching which leads to more accurate distance results. In [5], firstly the mandible was segmented by thresholding. Then, the mandibular canal was roughly segmented using image gradients and a binary mask-based line tracking method was utilized for canal localization. Rueda et al. [6] proposed a framework based on 2D active appearance models and semi-automatic landmarking to extract mandibular canal, bone and nerve. An adaptive region growing method was employed in [7], in which initial seed point inside the canal should be chosen by the user. In the recent years, some researchers attempted to segment mandibular canal in CBCT images. Localization of the mandibular canal in CBCT data is highly challenging due to the lower dose and higher noise-to-signal ratio in CBCT images comparing to conventional CT [8]. A framework based on active shape model (ASM) and Dijkstra’s algorithm was introduced in [9]. The results were promising; however, canal path near the ending point was not detected precisely. Fuzzy-connectedness approach was utilized in [10] which leads to accurate segmentation of jaw tissues including canal. The main disadvantage of fuzzy connectedness is that it is computationally inefficient and slow. A combination of 3D panoramic volume rendering algorithm and fast marching was employed in [11] to extract the whole region of the mandibular canal. The performance is highly dependent on the utilized texture features to enhance the mental foramens. The potential of active shape model (ASM) and active appearance model (AAM) was evaluated in [12] for automatic segmentation. It was reported that the accuracy of automatic segmentation of the mandibular canal by AAM and ASM methods is inadequate for use in clinical practice.
In the recent years, statistical shape models were successfully applied in many medical image segmentation tasks. Improving the accuracy of statistical shape models in segmentation tasks is still an open problem in medical image analysis. Various researches are performed to find corresponding points accurately [13]. Moreover, some researchers tried to replace principal component analysis with other dimension reduction methodologies [14]. In the last decade, many researchers utilized active shape models for automatic segmentation of the mandibular canal [9, 12]. However, they failed to achieve the accuracy high enough for safe implant surgery in CBCT images [12]. The mean interobserver variability of 1 mm is possible in clinical practice [15] and the largest error occurs in the anterior loop region due to the incomplete bony wall in combination with the unpredictable recurrent course. Identification and segmentation of mandibular canal are challenging due to several reasons. First, due to the large variation in shape and texture between mandibles of patients, building a robust statistical shape model is highly challenging. Second, multiple teeth loss results in severe bone resorption and the shape of mandible changes drastically in these patients. Third, due to the lower contrast of CBCT images compared to conventional CT, automatic segmentation is more challenging. Hence, designing an effective image enhancement and filtering method is essential for CBCT images.
In this article, a framework based on statistical shape models is developed for automatic segmentation of mandibular canal and it is applied to a dataset of CBCT images. In the proposed framework, firstly a new preprocessing algorithm based on low-rank decomposition is utilized. Then, a combination of statistical shape model and fast marching is employed for mandibular bone and canal segmentation. The rest of this paper is organized as follows. In “Material and methods” section, we introduce the proposed framework for automatic segmentation which consists of preprocessing, spatial normalization, conditional statistical shape modeling and fast marching. Then, the “Experimental results” section is reported. “Discussion and conclusion” sections are presented, respectively.
Material and methods
In this section, we introduce the proposed framework for automated segmentation of mandibular canal. Shape and position of mandibular canal, mandibular foramen and mental foramen are illustrated in Fig. 1. In CBCT images, mandibular canals often have missing edges. Moreover, the intensity of the canal is similar to the surrounding cancellous bone. Thus, it is essential to include a priori shape information in the model. This can be achieved using improved statistical shape models. Overview of the proposed method for mandibular canal segmentation is illustrated in Fig. 2.
Dataset
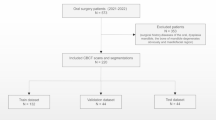
For the research presented in this paper, we collected 120 sets of CBCT images from two dedicated dental imaging centers in Tehran and Guilan provinces, Iran. In both centers, Sirona Galileos Compact 3D Cone Beam X-Ray Machine was used to acquire the high-resolution structural images. Acquisition parameters of the relevant sequences were as follows: field of view (FOV) = \(12\times 15\times 15\ \hbox {cm}^{3}\), effective dosage \({<}29\, \upmu \hbox {Sv}\) (21 mAs, 85 kV) and isotropic voxel size = 0.3 mm. The study involved 120 subjects with a mean age of \(49.7 \pm 25.2\) years. There were 68 males (56.66 %) and 52 females (43.33 %). All patients were referred to dental imaging centers to acquire CBCT images for implant surgical planning. Three-dimensional model of each subject is built from a set of 512 axial cross-sectional slices.
Image enhancement using multi-scale low-rank decomposition
CBCT images often suffer from low contrast and image enhancement techniques can improve the contrast of these images. We have utilized a combination of multi-scale modeling and low-rank matrix decomposition in [16] for image enhancement. This method was previously utilized for illumination normalization in face recognition application. Convex formulation is employed to solve the decomposition efficiently so that the multi-scale image components are incoherent. It is assumed that the 2D image matrix X with height and width of M and N, respectively, can be decomposed into different scales. In the other words, we assume that we are given a multi-scale partition \(\left\{ {Q_i } \right\} _{i=1}^L \) of an \(M\times N\) matrix, in which each block in \(Q_i \) is an order magnitude larger than the blocks in \(Q_{i-1} \). In other to transform between data matrix and block matrices, a block reshape operator \(R_A (Y)\) is defined to extract a block A from the matrix Y and then it is reshaped into an \(m_i \times n_i \) matrix. Given an \(M\times N\) input matrix X and the corresponding multi-scale partition, the following multi-scale low-rank modeling is proposed in [16]:
where \(U_A ,S_A \) and \(V_A \) form singular value decomposition (SVD) of \(R_A (Y_i )\). Given the data matrix X, the goal is to recover \(\left\{ {Y_i } \right\} _{i=1}^L \) from X. This can be achieved using convex programming, and multi-scale low-rank decomposition problem is formulated as follows:
where \(\left\| . \right\| _{(i)} \) is the block-wise nuclear norm for the i-th scale as \(\left\| . \right\| _{(i)} =\mathop {\sum }\nolimits _{A\in Q_i } {\left\| {R_A (.)} \right\| }_{\mathrm{nuc}}\). \(R_A (Y)\) is a block reshape operator which extracts a block matrix A from the full matrix X. This notation is considered to easily transform between the data matrix and the block matrices. Nuclear norm is the sum of the singular values of a matrix. The main characteristic of nuclear norm is that it is the tightest convex lower approximation to the rank function. The results of filtering with different scale numbers are illustrated in Fig. 3. Quantitative evaluation of filtering using different metrics is illustrated in Table 1. Furthermore, conventional median filter is included in this table. PSNRFootnote 1 values of different scales are approximately the same. In addition to PSNR, Root Mean Square Error (RMSE) [17] and Structural Similarity index (SSIM) [18] are quantitative measures which are utilized to choose the best result. PSNR and RMSE are slightly biased toward over smoothed results, i.e. an algorithm which filters not only the noise but also a part of the textures will get a good score. Structural similarity index [18] is a quality reconstruction metric that considers the similarity of the edges (high-frequency content) between the denoised image and the ground truth. To get a good SSIM score, the filtering method should remove the noise and preserve the edges and textures of the objects. Considering these criteria, the second scale gives the best result. Hence, it is the ideal filter for this application. The main advantage of this method is that irregular patterns are prohibited due to the low-rank decomposition. Hence, instead of global smoothing, local processing is done. The proposed filtering technique gives the best enhancement in uniform regions, while the edges are preserved.
Building statistical shape model
Statistical shape models (SSM) are established as a robust tool for 3D segmentation of medical images. The process of building SSM can be divided into two phases: learning and segmentation phase. Block diagram of the steps performed in this paper for building and testing SSM is depicted in Fig. 4. Mandibles differ in size and shape; hence, normalization preprocessing step is essential for studying the shape. For this purpose, we build a reference mandible surface, firstly. The mandible segmentations are converted to triangulated surface using marching cubes [19]. Mandible shapes are defined by vectors containing the coordinates of a set of landmark points which correspond to different mandible instances and that are typically located on the boundaries of the mandible. To construct the reference shape, the average shape of 84 training mandibles is calculated from corresponding surface meshes. The shape correspondences between the individual and average mandible shape is determined using pair-wise surface registration which is performed by nonrigid registration [20]. In the registration process, we use free-form deformation as the transformation model, the sum of squared difference as the similarity metric and the gradient descent algorithm for optimization. The point positions are optimized to minimize the model variance and obtain the most compact shape model. Training mandibles are transferred to the reference mandible space using the obtained transformation functions. Conditional statistical shape model [21] is utilized to embed the information about the position of the mandibular and mental foramen which are the starting and ending points of mandibular canal. This would avoid treating all regions of shape equally. At first, learning phase is explained. In order to model relations between shapes, let Y and Z be the shape of mandible and the combined shape of the mandibular and mental foramen. The conditional distribution of shape Y given a known shape \(Z=Z_0 \) is formulated using Gaussian conditional density as following:
with
where \(\Sigma _{ij} \) is the joint covariance, and \({\upmu } _Y \) and \({\upmu } _Z \) are the mean shapes of Y and Z in the training set. \(\Sigma _{YY} ,\Sigma _{YZ} ,\Sigma _{ZY} ,\Sigma _{ZZ} \) are the joint covariance matrix defined as follows:
Ridge regression is employed to calculate \(\Sigma _{ZZ}^{-1} \), i.e. is replaced by \((\Sigma _Z Z+\gamma I)^{-1}\). The average shape and model deformation of conventional SSM are constant with subjects, whereas in conditional SSM, the average shape fits the patient-specific shape and the model deformation is restricted. The segmentation phase includes the following steps:
-
1.
Automatic localization of the mandible coordinate system
-
2.
Initial rough segmentation of the mandibular bone region by thresholding
-
3.
Fitting SSM using Levenberg–Marquardt algorithm
-
4.
Refinement of mandibular canal using fast marching which will be explained in the upcoming section.
Step (1) is performed by a previously reported method [22] which consists of spatial normalization, localization of anatomical landmarks using the statistical landmark model and refinement of the anatomical coordinate using the average surface image. In step (2), thresholding is performed and the largest connected component is extracted. The threshold value is automatically learned from the crossvalidation within the training dataset. The threshold value of 210 gives the lowest surface distance in this task. The procedure of fitting SSM to a point cloud is done using Levenberg–Marquardt algorithm through the following cost function as in [23]:
where b is the shape parameter vector, S(b) is the shape instance defined by parameter b, and E is a cloud of edge points. \(N_E \), \(N_q \), D and M are the number of edge points, the number of vertices in S(b), the distance metric and the number of modes, respectively. The first two terms represent fitness between a shape instance and the detected edge points, and the last term represents a penalty to avoid a shape go far from the mean. Due to the importance of detecting edge points correctly, we proposed a filtering method in “Image enhancement using multi-scale low-rank decomposition” section. In Eq. (6), we use the same notation, D(x, A) for two similar distance metrics: point-to-surface mesh distance and point-to-point cloud distance. Both metrics measure the shortest distance between x and A as follows:
If the argument A is a surface mesh such as the first term in Eq. (6), \(p\in A\) indicates any point on the surface mesh including every point on a triangle consisting of connected three vertices. If A is a point cloud such as the second term in Eq. (6), \(p\in A\) simply indicates a point in the point cloud.
Hence, embedding the information about the shape of the mandibular and mental foramen leads to more flexibility in modeling shape variations. In summary, the conditional SSM is fitted to the boundary edge points of the roughly segmented mandible using simple thresholding of CBCT images to obtain initial parameter settings for subsequent segmentation procedures. Edge detection is done based on intensity profile analysis, and the perpendicular direction at each surface point is estimated in each iteration.
Fast marching
Early researchers utilized Dijkstra’s algorithm for finding the shortest path on the graph of mandible [3, 9], however since the boundary of mandibular bone is not always present or visible in CBCT datasets, Dijkstra’s algorithm often shortcuts outside the mandibular canal. Fast marching is more recent approach for optimal path problem which gives more accurate distance results for image volumes. Fast marching [24, 25] is an efficient iterative algorithm for numerical approximation of fronts propagating in \( \mathbb {R}^{n}\)space. A propagating front is defined as a closed hypersurface, each point of which moves with speed function F in the direction of the surface normal. Suppose that \(S(t)\subset \mathbb {R}^{n}\) is the propagating interface in \(\mathbb {R}^{n}\) space. The evolution of the front can be modeled using Eikonal equation:
where T is the arrival time function, and F is the speed function. In fast marching, one of the most critical parameters is speed function. Since the canal has a low intensity, we consider a speed function in which the speed is inversely related to the intensity. Thus, the shortest path between mental and mandibular foramen will be mandibular canal. The segmented bone surface from the previous stage is utilized to select the background region, and we set all those pixels to a high pixel value. The curved pixel length is calculated which is equal to the length of the canal (\(L_\mathrm{canal})\). Then, we warp the local neighborhood of the canal to a small volume \(I_L \) of dimensions \(4\,\mathrm{mm}\times 4\,\mathrm{mm}\times L_\mathrm{canal}\). In this regard, the curved canal will be a straight line in \(I_L \). After these steps, uniformly distributed normal planes along the channel are estimated and the local neighborhood of the canal is warped to a small volume. The intensities of the warped volume, \(I_\mathrm{w} \) are converted to a speed map F as follows:
where \(I_\mathrm{w} \) and \(H_g \) are the warped volume and Gaussian kernel with \(\sigma =1.5\), respectively. The first speed term involves the smoothed intensity by Gaussian kernel, while the second speed term involves the local gradient. The source and sink are considered as mental and mandibular foramen. Fast marching is performed using speed map F, and the shortest path is detected using Runge–Kutta algorithm [26]. Then, the shortest path is warped back to the original volume.
Experimental results
The proposed algorithm is implemented on MATLAB 8.1 environment [27] and C++ platform [28] (MS Visual Studio 2013), a personal computer with a P4 (3 GHz) processor and 8 GB memory. Image enhancement algorithm which is explained in “Image enhancement using multi-scale low-rank decomposition” section is built in MATLAB and the filtered images are fed into the C++ program for building SSM. Estimated time for segmenting each dataset with 512 slices by our algorithm is less than 5 min. We asked two radiologists with at least 10 years of experience for manual segmentation which is used as ground truth. For manual segmentation, an expert should spend at least 1 h to segment 512 slices. The dataset is divided into training and test sets. We have considered 70 % of the data for training and the rest for testing, which is considered as a common rule of thumb in machine learning [29, 30]. Splitting the dataset into the training and testing set is performed randomly. Furthermore, the ground truth segmentation of mandibular canal is provided by two radiologists for each case in the dataset.
The first step is preprocessing which is performed using low-rank decomposition with different scales. We compared the filtering result using the proposed methodology with conventional median filter and diffusion filtering previously proposed by Kroon for CBCT images [31]. Decomposition using different scales such as 4, 8 and 16 are illustrated in Fig. 3. The best performance is achieved using the block size of 8. By comparing the area identified by a red bounding box in each subfigure, it can be observed that some details are lost in diffusion filtering. However, these details are preserved in low-rank decomposition. Quantitative comparison of different filtering methods is reported in Table 1. After preprocessing, conditional SSM is trained using 84 training sets of CBCT images. The standard measures such as compactness capacity, generalization ability and specificity are employed to compare conditional SSM model and conventional SSM model by Cootes et al. [32]. The evaluation metrics are explained thoroughly in “Appendix A.” Figure 5 shows the reconstruction error for conditional and conventional SSM as a function of the number of variation modes. For a constant number of modes, the reconstruction error is higher for conventional SSM. The generalization ability of conditional SSM is better than conventional SSM. The specificity and compactness for conditional and conventional SSM distributions are illustrated in Fig. 6. As it is evident from this figure, the error made by specificity measure is lower for conditional SSM. C(Conditional_SSM) is slightly larger than C(Conventional_SSM), but considering the error for each M, we can say that these two methods offer similar compactness level or conditional SSM is a bit worse than conventional SSM.
Figure 7 visualizes sample results of mandible segmentation obtained using the combination of conditional SSM and fast marching. Figure 8 shows accuracy levels and box plots for mandible segmentation results obtained using our method and two other automatic methods in [9, 12] according to the ASSD and Dice criteria for all test dataset. Moreover, Table 2 summarizes the mean, standard deviation and median values of the presented results in Fig. 8. The distance values between manual and automatic segmented mandibular canal (for both right and left nerve) are reported in Table 3. From the quantitative results, it can be concluded that our method can segment mandibular canal with a good level of accuracy and perform better than the methods previously proposed in [9, 12].
Discussion
As we mentioned earlier, segmentation of mandibular canal is a challenging and time-consuming task. The goal of this research was to propose a framework based on statistical shape model for automatic segmentation of the canal. To this aim, we first developed a filtering approach based on low-rank decomposition for CBCT images. The high accuracy of this preprocessing step is essential for fitting SSM using Levenberg–Marquardt algorithm since the accuracy of fitting is dependent on the efficiency of edge points. Then, we segmented mandible in a patient dataset and then considered it as the input information for the canal localization procedure. Fast marching tries to find the darkest tunnel close to the initial segmentation of the canal found, which was obtained by conditional SSM model. Quantitative evaluation of the conditional statistical model was performed by compactness capacity, specificity and generalization ability measures. The overall performance of conditional SSM is superior to conventional SSM based on Figs. 5 and 6. Moreover, a combination of conditional SSM and fast marching was utilized for automatic detection of mandibular canal. Although the error of many methods is inadequate, especially near the canal ending and starting point, adding condition points’ information led to higher accuracy of the method. Figure 9 represents the efficiency of our method in a noisy environment.
In order to compare our proposed methodology with the previous works, we implemented Kainmueller and Kroon’s methods [9, 12] on our dataset. Kroon utilized statistical shape models to localize mandibular canal. In order to enhance CBCT images, he proposed coherence diffusion filtering. There are various schemes such as optimized, standard and nonnegative for solving discretized diffusion filtering equation. The performance of diffusion filtering in various schemes was previously evaluated in [33] and optimized scheme outperformed other schemes in terms of SSIM index. In this article, a new filtering method based on multi-scale low-rank decomposition is proposed. Quantitative comparison of diffusion filtering and the proposed method based on low-rank decomposition is reported in Table 1. In Kainmueller’s paper, active shape model was constructed using 106 datasets and canal segmentation is performed by
a Dijkstra’s algorithm based optimization. It was reported that the right nerve and the left nerve could be detected with an average error of 1.0 and 1.20 mm, respectively. Kroon failed in achieving the desired accuracy for clinical practice. Comparative results for mandibular bone segmentation are reported in Table 2. Based on Dice’s coefficient and ASSD (mm), it can be concluded that the proposed method outperforms Kainmueller and Kroon’s approaches. Moreover, comparative metric results for canal detection are summarized in Table 4. The average mean curve distance to the respective gold standard was utilized as the evaluation metric in Kainmueller’s paper. Comparative performance of the methods based on this metric is reported in Table 5. Hence, it can be concluded that the proposed methodology has higher generalization ability, as well as robustness to unusual mandible shapes.
In the previous methods proposed for canal detection, the error in the mandibular and mental foramen region is more than 1 mm which is not sufficient in the clinical practice. In Fig. 10, the Euclidian distance error between the mandibular canal annotation from the proposed method and expert annotation is illustrated. As it can be seen, the mean error in the nerve entry and exit points is less than 1 mm and standard deviation is small. This is one of the main advantages of the proposed method in this article.
Figure 11 is related to the low accuracy level of our method due to the severe bone resorption. In some cases, such as bone loss resulting from missing teeth or cases with impacted tooth, there are large variations in mandible shape. The most common cause of bone loss is tooth loss, especially multiple teeth. When multiple teeth in an area are missing for a long term, facial drooping will occur. One possible way to improve the accuracy in these cases is increasing the number of condition points. However, in this regard, the accuracy of selecting condition points will affect the whole process.
The main challenging part of building statistical shape models is finding corresponding points. Various methods can be utilized to perform this step such as spherical harmonic basis functions [34] and minimum description length (MDL) [35]. However, these methods are mainly suitable for closed surface objects or manual initialization by anatomical landmarks is essential [13]. When the 3D shapes are not topologically equivalent to a sphere, the accuracy of registration methods will decrease significantly. The shape of mandible does not resemble a sphere, and mapping to a sphere is not accurate. Furthermore, this shape is not a closed surface since the right and left mandibular canals are removed from the mandible. In the future work, we will attempt to seek and develop more efficient methods to find the corresponding points. We are aiming to utilize the potential of Lie groups and Lie Algebras theory [36] in this research.
Conclusion
Accurate localization of mandibular canal is essential in dental implant surgery. The main challenges are large variation in shapes and texture between mandibles, the high level of noise and low contrast in CBCT images and small dimension of the canal. Many researchers attempted to utilize statistical shape models for automatic segmentation of the mandibular canal. However, the accuracy of automatic segmentation is inadequate for use in clinical practice. In this article, we presented an accurate and effective framework which is able to segment mandibular canal automatically in CBCT images. From the methodological viewpoint, a particular aspect which differentiates the proposed method from existing methods is the combination of anatomical and statistical information including mental and mandibular foramen position. The proposed framework based on conditional SSM and fast marching leads to more accurate detection of the canal. A priori information about shape makes the mandibular canal segmentation more robust. Based on the quantitative results, we can conclude that the proposed segmentation framework outperforms two other methods in the literature. Due to the variability between the shape of mandibular bone in male and female subjects, future work could be addressed to employ different statistical shape models for male and female subjects and investigate the efficiency of our method for difficult datasets with severe bone resorption.
Notes
Peak Signal-to-Noise Ratio.
References
Roberts JA, Drage NA, Davies J, Thomas DW (2014) Effective dose from cone beam CT examinations in dentistry. Br J Radiol 82(973):35–40
Robinson PP (1988) Observations on the recovery of sensation following inferior alveolar nerve injuries. Br J Oral Maxillofac Surg 26:177–189
Stein W, Hassfeld S, Muhling J (1998) Tracing of thin tubular structures in computer tomographic data. Comput Aided Surg 3:83–88
Hanssen N, Burgielski Z, Jansen T, Liévin M, Ritter L, von Rymon-Lipinski B, Keeve E (2004) Nerves-level sets for interactive 3D segmentation of nerve channels. In: IEEE international symposium on biomedical imaging nano macro 2004. IEEE, pp 201–204
Kondo T, Ong SH, Foong KW (2004) Computer-based extraction of the inferior alveolar nerve canal in 3-D space. Comput Methods Programs Biomed 76:181–191
Rueda S, Gil JA, Pichery R, Alcañiz M (2006) Automatic segmentation of jaw tissues in CT using active appearance models and semi-automatic landmarking. In: Larsen R, Nielsen M, Sporring J (eds) Medical image computing and computer-assisted intervention 2006. Springer, Heidelberg, pp 167–174
Yau HT, Lin YK, Tsou LS, Lee CY (2008) An adaptive region growing method to segment inferior alveolar nerve canal from 3D medical images for dental implant surgery. Comput Aided Des Appl 5:743–752
Orth RC, Wallace MJ, Kuo MD (2008) C-arm cone-beam CT: general principles and technical considerations for use in interventional radiology. J Vasc Interv Radiol 19:814–820
Kainmueller D, Lamecker H, Seim H, Zinser M, Zachow S (2009) Automatic extraction of mandibular nerve and bone from cone-beam CT data. In: Yang G-Z, Hawkes D, Rueckert D, Noble A, Taylor C (eds) Medical image computing and computer-assisted intervention 2009. Springer, Heidelberg, pp 76–83
Lloréns R, Naranjo V, López F, Alcañiz M (2012) Jaw tissues segmentation in dental 3D CT images using fuzzy-connectedness and morphological processing. Comput Methods Programs Biomed 108:832–843
Kim G, Lee J, Lee H, Seo J, Koo Y-M, Shin Y-G, Kim B (2011) Automatic extraction of inferior alveolar nerve canal using feature-enhancing panoramic volume rendering. IEEE Trans Biomed Eng 58:253–264
Gerlach NL, Meijer GJ, Kroon D-J, Bronkhorst EM, Bergé SJ, Maal TJJ (2014) Evaluation of the potential of automatic segmentation of the mandibular canal using cone-beam computed tomography. Br J Oral Maxillofac Surg 52:838–844
Styner MA, Rajamani KT, Nolte L-P, Zsemlye G, Székely G, Taylor CJ, Davies RH (2003) Evaluation of 3D correspondence methods for model building. In: Biennial international conference on information processing in medical imaging. Springer, pp 63–75
Roohi SF, Zoroofi RA (2013) 4D statistical shape modeling of the left ventricle in cardiac MR images. Int J Comput Assist Radiol Surg 8:335–351
Gerlach NL, Meijer GJ, Maal TJ, Mulder J, Rangel FA, Borstlap WA, Bergé SJ (2010) Reproducibility of 3 different tracing methods based on cone beam computed tomography in determining the anatomical position of the mandibular canal. J Oral Maxillofac Surg 68:811–817
Ong F, Lustig M (2016) Beyond low rank + sparse: multi-scale low rank matrix decomposition. IEEE J Sel Top Sign Proces 10(4):672–687
Wackerly D, Mendenhall W, Scheaffer RL (2008) Mathematical statistics with applications, 7th edn. Thomson Brooks/Cole, Scarborough
Wang Z, Bovik AC, Sheikh HR, Simoncelli EP (2004) Image quality assessment: from error visibility to structural similarity. IEEE Trans Image Process 13:600–612
Lorensen WE, Cline HE (1987) Marching cubes: a high resolution 3D surface construction algorithm. In: Stone MC (ed) ACM siggraph computer graphics. ACM, New York, pp 163–169
Rueckert D, Sonoda LI, Hayes C, Hill DL, Leach MO, Hawkes DJ (1999) Nonrigid registration using free-form deformations: application to breast MR images. IEEE Trans Med Imaging 18:712–721
Yokota F, Okada T, Takao M, Sugano N, Tada Y, Tomiyama N, Sato Y (2013) Automated CT segmentation of diseased hip using hierarchical and conditional statistical shape models. In: Mori K, Sakuma I, Sato Y, Barillot C, Navab V (eds) Medical image computing and computer-assisted intervention 2013. Springer, Heidelberg, pp 190–197
Yokota F, Okada T, Takao M, Sugano N, Tomiyama N, Sato Y, Tada Y (2012) Automated localization of pelvic anatomical coordinate system from 3D CT data of the hip using statistical atlas. Med Imaging Technol 30:43–52
Lamecker H, Lange T, Seebaß M (2004) Segmentation of the liver using a 3D statistical shape model. Konrad-Zuse-Zentrum für Informationstechnik, Apr 2
Sethian JA (1996) A fast marching level set method for monotonically advancing fronts. Proc Natl Acad Sci 93:1591–1595
Sethian JA (1999) Fast marching methods. SIAM Rev 41:199–235
Atkinson KE (2008) An introduction to numerical analysis. Wiley, London
MATLAB R2015a—Die Sprache für technische Berechnungen—MathWorks Deutschland (n.d.). http://de.mathworks.com/products/matlab/
Visual Studio 2013—Microsoft Developer Tools (n.d.). https://www.visualstudio.com/
Atkinson KE (2008) An introduction to numerical analysis. John Wiley & Sons
Training and Testing Data Sets (n.d.). https://msdn.microsoft.com/en-us/library/bb895173.aspx (2016)
Kroon D-J, Slump CH, Maal TJ (2010) Optimized anisotropic rotational invariant diffusion scheme on cone-beam CT. In: Jiang T, Navab N, Pluim JPW, Viergever MA (eds) Medical image computing and computer-assisted intervention 2010. Springer, Heidelberg, pp 221–228
Cootes TF, Edwards GJ, Taylor CJ (1998) Active appearance models. In: Computer vision—ECCV’98. Springer, pp 484–498
Abdolali F, Zoroofi RA, Otake Y, Sato Y (2016) Automatic segmentation of maxillofacial cysts in cone beam CT images. Comput Biol Med 72:108–119
Brechbühler C, Gerig G, Kübler O (1995) Parametrization of closed surfaces for 3-D shape description. Comput Vis Image Underst 61:154–170
Rissanen J (1978) Modeling by shortest data description. Automatica 14:465–471
Varadarajan VS (2013) Lie groups, Lie algebras, and their representations. Springer, Berlin
Davies R, Taylor C et al (2008) Statistical models of shape: optimisation and evaluation. Springer, Berlin
Kohavi R et al (1995) A study of cross-validation and bootstrap for accuracy estimation and model selection. In: Ijcai, pp 1137–1145
Dice LR (1945) Measures of the amount of ecologic association between species. Ecology 26:297–302
Heimann T, Van Ginneken B, Styner MA, Arzhaeva Y, Aurich V, Bauer C, Beck A, Becker C, Beichel R, Bekes G et al (2009) Comparison and evaluation of methods for liver segmentation from CT datasets. IEEE Trans Med Imaging 28:1251–1265
Acknowledgments
This work is partly supported by MEXT/JSPS Grant-in-Aid for Scientific Research Nos. 26108004 and 25242051.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Human and animal rights
For this type of study formal consent is not required because this study is a retrospective study.
Informed consent
Written informed consent was not required for this study because this study is a retrospective study.
Appendix A: Performance evaluation metrics
Appendix A: Performance evaluation metrics
To investigate the statistical behavior of conditional SSM, we utilize compactness, specificity and generalization measures. Compactness is an estimation of parameters required to generate a valid instance of the modeled object [37]. The compactness of the shape model is calculated as the cumulated variance for the first \(i=1,\ldots ,M\) modes:
where \(C(\tau )\) and \({\lambda }_i \) are the compactness capacity and i-th largest eigenvalue, respectively. Generalization of a model measures the ability to represent unseen instances of the object class modeled [37], and it is defined as follows:
where \(N_s \) is the number of training data, \(t_k \) is the training sample that is eliminated in leave-one-out procedure, and \(r_k \) is the reconstructed shape using \({\tau }\) parameters. The specificity of a shape model is described as how much it can represent valid instances of the modeled class of object [37] and it is formulated as following:
where \(s_k (\uptau )\) is an arbitrary sample constructed by \(\uptau \) parameters, \(N_r \) is the number of data, and \({t}^{\prime }_k \) is the closest sample in training datasets to \(s_k (\uptau )\). Leave-one-out crossvalidation [38] is employed to compare the segmented mandible bone with the gold standard. To create the gold standard dataset, each mandible and the mandibular canal was manually segmented by two radiologists in a slice-by-slice fashion. In order to compare the automatic segmentation results with the gold standard, two criteria are employed: (1) Dice’s coefficient [39] and (2) average symmetric surface distance [40]. Dice’s coefficient measures the overlap between the automatic segmentation result and reference manual annotations. This similarity measure is defined as following:
where A and M are segmentation results obtained by automatic segmentation and gold standard, respectively. This criterion is one of the most well-known methods in evaluating different segmentation methods.
Average symmetric surface distance (ASSD) [40] is defined as the space between two segmentations A and M in millimeters. If we assume that \(S_\mathrm{A}\) and \(S_{\mathrm{M}}\) are surface voxels of A and M, the Euclidean distance for each surface voxel of \(S_{\mathrm{A}}\) to the closest surface voxel of \(S_{\mathrm{M} }\) is calculated. To preserve symmetry, the same process is applied for the surface voxels of \(S_{\mathrm{M}}\) to \(S_{\mathrm{A}}\). Therefore, ASSD is expressed as the average of all stored distances as follows:
where d is the shortest distance of voxel v to surface S, \(\left\| . \right\| \) and \(\left| . \right| \) represent vector norm and number of vertices, respectively. ASSD provides a volumetric-based evaluation criterion for the assessment of segmentation result.
Rights and permissions
About this article
Cite this article
Abdolali, F., Zoroofi, R.A., Abdolali, M. et al. Automatic segmentation of mandibular canal in cone beam CT images using conditional statistical shape model and fast marching. Int J CARS 12, 581–593 (2017). https://doi.org/10.1007/s11548-016-1484-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-016-1484-2