Abstract
In this paper, a novel method is introduced to segment tooth from Cone Beam Computed Tomography images. Different from traditional methods, the root canal centerline identified by graph theory based energy minimization problem is applied as prior knowledge aiding for the segmentation. Besides, though we use the idea of contour tracking strategy as adopted by most published methods based on slice-by-slice basis, within a slice, the segmentation is based on the harmonic field theory, which makes our method superior to the traditional ones. Effect and efficiency of ours are proved by the experiments.
This work is partially supported by the Doctoral Innovation Fund of Hunan (CX2012B066), the Scientific Research Project in Fujian University of Technology (GY-Z160066). The authors also gratefully acknowledge the helpful comments and suggestions of the reviewers, which have improved the presentation.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
There are two major ways nowadays to capture in vivo dental information into its digital format. The first one is to use a laser scanner, which is a popular three dimensional (3D) approach and can be used to capture the geometry of crown surface accurately. However, as we know, the laser beams cannot reach the internal of object, therefore, it is unable to capture the root of tooth. As another option, Cone Beam Computed Tomography (CBCT) techniques are widely used in the clinic, since the X-ray it adopted can pass through human body and tell the differences among different organs. Actually, the CBCT images play an very important role in the computer-aided diagnosis, treatment planning and virtual surgery for dental routines such as root canal therapy, where information of the root is crucial to their successes. In order to be used by the computer, a primary step of the CBCT based approach is to distinguish tooth from the rests, which is the aim of this work. In other words, we want to find a feasible way to segment individual teeth from CBCT images. The segmentation of tooth from CBCT images is challenging according to Gao et al. [11]. Firstly, the tooth is very close to surrounding objects, such as adjacent teeth (see Fig. 1(a)) or jaw bones (seen Fig. 1(c)). Therefore, many suspicious boundaries exist and they even disappeared where adjacent teeth touch each other. Secondly, the tooth contour also suffers from topological change among different images. For example, the first molar in Fig. 1(b) can be seen as one connected region, while becomes three region in Fig. 1(c). Thirdly, the resolution of tooth is limited, because the image represents the entire head and some surroundings. As Fig. 1(d) demonstrated, the region of tooth in red rectangle is relatively small comparing to the entire region of image in blue rectangle. Fourthly, the CBCT images may be very noisy for the reason of reduced radiation dose, which is for the consideration of healthy. Lastly, efficiency is important due to large number of slices for each tooth.
1.1 Related Work
Lots of methods have been proposed for medical image segmentation. Some of them are relatively simple, such as threshold based method and region growing [17, 19]. Others may based on more some complex methodologies or procedures, such as max-flow/min-cut based graph cut [5] method and contour evaluation based Snakes [2] or Level Set [8] algorithm. But none of them can be used as general approach suitable for ever organ segmentation problems, which is the conclusion of a survey [7].
Since the CT images can be view as an entire 3D volume or a sequence of 2D images, the existed tooth segmentation methods can be classified into two categories, namely the volume based and slice based approaches. 3D mesh models (or well know as template) which represents the mean shape and prior knowledge of each teeth are generally required for the segmentation and reconstruction of volume based methods [4, 6]. In order to get the desire models, training processes are commonly needed based on some previous constructed databases, which make the method of this kind extremely complex and time costing. By contrast, slice based methods treat CT images in their very native way. Namely, they work on a slice-by-slice basis, and in order to track the segmentation boundary (or contour) in consecutive slices, the contour of segmented slices will be offered as prior knowledge for the segmentation of adjacent slices generally. Among the slice based methods, Level Set is the most adopted algorithm [10–13, 16]. The segmentation of a slice is achieved by process named contour evolution, it is a iterative procedure initialized by a given contour, and evolved under the target of minimization of a specified energy function. Therefore, the choice of the initial contour and energy function are critical to the results. Parameter tunings are generally required in order to get a excellent result for different cases [9]. Liao et al. [14, 15] adopt the thin-plate splines to connect control points specified by user interactions as the segmentation boundary of tooth in CT volume. Though user interactions can offer high level knowledge aiding for the segmentation, which are good especially for the Ground Truth generation, but too many of them will be time-costing and lead to tedious user experiences.
This paper also adopt the slice-by-slice basis, but different from the existed method, the centerline of root canal will be identified in the first place and used as prior knowledge in additional to the contour of adjacent slices. Furthermore, harmonic field theory other than algorithms such as level set is used in this paper to find the segmentation boundary. The rest of this paper will be organized as follows. In Sect. 2, a root canal centerline identification method based on graph theory is introduced. Section 3 presents the harmonic field based segmentation method. Then, Sect. 4 shows some experimental results and assessments. Finally, the Sect. 5 concludes the paper.
2 Graph Theory Based Root Canal Centerline Identification
Instead of solid object, within a tooth there exist some spaces anatomically where soft tissues, such as the blood vessels and nerve, can be found. Therefore, in the CBCT images, low intensity areas appear inside the tooth in contrast with the high intensity bone tissues as can be seen in Fig. 1.
A root canal is the tubular structure within the root of a tooth. In this paper, we treat it as an important prior knowledge based on the fact that a tooth can be located as long as its root canals are identified. In order to achieve that, centerline, which is an efficient descriptive way of root canal, will be firstly defined as a path formed by consecutive edges between two selected terminal points in the graph constructed from CT volume. Then, similar to the level set approaches, energy terms will be special designed and assigned to every edges of the graph on the condition that the centerline will possess the minimum energy values than any other path’s between the two specified terminals. Finally, the classic shortest weighted path algorithm can be used to solve the minimization problem where energy term of each edge is the weight.
2.1 Definition of the Graph and Centerline
A 3D volume can be seen as the superposition of two-dimensional (2D) CT images in the order of their Z-coordinations. The 2D pixel in images is referred to as voxel in 3D volume. A most intuitive while inefficient way of graph construction is to use every voxels as graph vertexes and the line determined by each two neighboring voxels as graph edges, because the scale of the graph will become too complicated to be processed generally. An improvement is to use a subset of entire voxels in the construction.
Actually, a skeletonization method [1] is used to remove the voxels, which definitely no belonging to the root canal region, during the graph construction. And we use the classic Dijkstra algorithm for the shortest weighted path problem solving.
A discrete version of the root canal centerline is a path of graph we constructed, which can be described by a sequence of graph vertexes \(\{x_1, x_2, ..., x_k\}\), where \(x_1\) and \(x_k\) are two vertexes closest to the terminal points of centerline. If we assign each edge a energy term \(E(x_i, x_{i+1}), (1 \le i \le k-1)\), than the energy of the path can be depicted as Eq. 1.
2.2 Designation of the Energy
In this paper, two aspects are considered in the designation of energy term as described above, including the position \(P({x_i},{x_{i + 1}})\) and tangential direction \(D({x_i},{x_{i + 1}})\) of edge \(x_i,x_{i+1}\) as described by Eq. 2.
Volume intensities and structure tensor are adopted to formulate the \(P({x_i},{x_{i + 1}})\) and \(D({x_i},{x_{i + 1}})\) for the consideration that the centerline is supposed to exist in a low intensity tubular structure. And the \(\alpha \) is set to 0.5 to balance them. In additional, every voxel intensities of input volume V is firstly reversed \(V'\) according to Eq. 3 so as to enhance the root canal region.
where M, I(x) denotes the maximum intensity of volume V and the intensity at voxel x respectively.
Then, the proposed \(P({x_i},{x_{i + 1}})\) and \(D({x_i},{x_{i + 1}})\) will meet the Eqs. 4 and 5.
where \(M, m, I(x_i)\) are the maximum, minimum intensity and the intensity at voxel \(x_i\) of the modified volume \(V'\). \(\lambda _i, e_i (i=1,2,3)\) are eigenvalue and eigenvector of structure tensor of \(x_i\) subjected to \({\lambda _1} \ge {\lambda _2} \ge {\lambda _3}\), v is the vector from \(x_i\) to \(x_{i+1}\), \(\bullet \) denotes the dot product operation.
3 Harmonic Fields Based Tooth Segmentation
Our segmentation strategy is to construct a scalar field \(\varphi \) (i.e. the harmonic field) of image pixels, which satisfies \(\varDelta \varphi = 0\) (\(\varDelta \) is the Laplacian operator) and subject to particular Dirichlet boundary conditions (namely the constrains). More details of the harmonic field can be found in our previous work [20].
3.1 Segmentation Within a Slice
If we view the Laplacian operator of each pixel together as a matrix L (i.e., the Laplacian matrix), the element \(L_{ij}\) of L can be expressed as Eq. 6.
where \(W_{ij}\) is the weighting function should be designed for different cases. Let \(I(i), I(j), L_{ij}\) denote the intensity of pixel i, j and the distance between them, then our \(W_{ij}\) meets the Eq. 7.
In order to solve the linear system \(L \varphi = 0\), the constrains should be specified. For example, pixels of two categories are preset to 0 and 1 respectively, which are the minimum and maximum scalar of the harmonic field. Generally, one category includes pixels which belonging to the aims of segmentation (e.g., the tooth in our case), others are background pixels. Therefore, pixels belonging to the identified root canal centerlines and image boundaries can be treated as the two sets of constrains respectively.
After acquiring the harmonic field by solving the linear system determined by Laplacian matrix and the constrains, one of the harmonic iso-lines can be used as the segmentation boundary.
3.2 The Contour Tracking Procedure
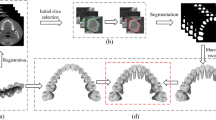
We name the entire segmentation procedure as contour tracking because the harmonic field based segmentation method described above is carried out iteratively from \(S_c\) to \(S_t\) to track the tooth contours and solve the problem. \(S_c\) and \(S_t\) are two extreme image slices closest to the crown side and tip side of tooth which include root canal centerline. User interactions are involved to assign the \(S_c\) and \(S_t\) (see Fig. 2(a) and (b)), as well as the Volume of Interest (VOI, see Fig. 2(c)) which includes the objective tooth.
During the iteration, the shape of acquired contour in current slice will guide for the extraction of contour in the next slice. Figure 3 demonstrated the entire procedure.
4 Experiments and Result
The proposed system is implemented on a personal computer (2.67 GHz Core Duo processor and 4 GB memory) based on the Visualization Toolkit [18] and using C++ for programming. The CBCT images are provided by Xiangya Hospital of Central South University.
Some experiments are carried out to evaluate the proposed method. Firstly, a Morphological Snakes (MS) algorithm [3] is adopted for the comparison. As showed in Fig. 4, our method is more accurate than the MS.
Then, the average time consumption for root canal centerline identification and contour tracking also are recorded (see Fig. 4) grouped by teeth indexes (i.e., from 11 to 17 and 21 to 27 according to the Federation Dentaire Internationale notation method).
Finally, the segmentation result of a tooth with multiple root canal is depicted in Fig. 6 to demonstrate the effect of the proposed method.
5 Conclusion and Future Works
This paper presents a novel segmentation method of tooth from CBCT images. Firstly, the root canal centerline identified by graph theory based energy minimization problem is applied as prior knowledge aiding for the segmentation. Besides, though we use the idea of contour tracking strategy as adopted by most published methods, within a slice, the segmentation is based on the harmonic field theory, which makes our method superior to the traditional ones. Effect and efficiency of ours are proved by the experiments.
As can be seen in the experiment, the major time consumption of our method is the cost for harmonic field generation. Though strategies for accelerating such as VOI extraction have been achieved in this work, further improvement is still a direction of our future works.
References
Abeysinghe, S.S., Baker, M., Chiu, W., Ju, T.: Segmentation-free skeletonization of grayscale volumes for shape understanding. In: IEEE International Conference on Shape Modeling and Applications, pp. 63–71, June 2008
Alvarez, L., Baumela, L., Henriquez, P., Marquez-Neila, P.: Morphological snakes. In: 2010 IEEE Conference on Computer Vision and Pattern Recognition, pp. 2197–2202, June 2010
Alvarez, L., Baumela, L., Marquez-Neila, P., Henriquez, P.: A real time morphological snakes algorithm. Image Process. Online 2, 1–7 (2012)
Barone, S., Paoli, A., Razionale, A.V., Savignano, R.: 3D reconstruction of individual tooth shapes by integrating dental cad templates and patient-specific anatomy. In: 2014 International Design Engineering Technical Conferences and Computers and Information in Engineering Conference, pp. 79–87 (2014)
Boykov, Y., Jolly, M.-P.: Interactive organ segmentation using graph cuts. In: Delp, S.L., DiGoia, A.M., Jaramaz, B. (eds.) MICCAI 2000. LNCS, vol. 1935, pp. 276–286. Springer, Heidelberg (2000). doi:10.1007/978-3-540-40899-4_28
Buchaillard, S.I., Ong, S., Payan, Y., Foong, K.: 3D statistical models for tooth surface reconstruction. Comput. Biol. Med. 37(10), 1461–1471 (2007)
Bulu, H., Alpkocak, A.: Comparison of 3D segmentation algorithms for medical imaging. In: Proceedings of the 26th IEEE International Symposium on Computer-Based Medical Systems, pp. 269–274 (2007)
Chung, G., Vese, L.A.: Image segmentation using a multilayer level-set approach. Comput. Vis. Sci. 12(6), 267–285 (2009)
Derraz, F., Beladgham, M., Khelif, M.: Application of active contour models in medical image segmentation. In: International Conference on Information Technology: Coding and Computing, vol. 2, pp. 675–681, April 2004
Gao, H., Chae, O.: Touching tooth segmentation from CT image sequences using coupled level set method. In: 5th International Conference on Visual Information Engineering, pp. 382–387, July 2008
Gao, H., Chae, O.: Individual tooth segmentation from CT images using level set method with shape and intensity prior. Pattern Recogn. 43(7), 2406–2417 (2010)
Hosntalab, M., Aghaeizadeh Zoroofi, R., Abbaspour Tehrani-Fard, A., Shirani, G.: Segmentation of teeth in CT volumetric dataset by panoramic projection and variational level set. Int. J. Comput. Assist. Radiol. Surg. 3(3), 257–265 (2008)
Ji, D.X., Ong, S.H., Foong, K.W.C.: A level-set based approach for anterior teeth segmentation in cone beam computed tomography images. Comput. Biol. Med. 50, 116–128 (2014)
Liao, S.-H., Tong, R.-F., Dong, J.-X.: 3D whole tooth model from CT volume using thin-plate splines. In: International Conference on Computer Supported Cooperative Work in Design, vol. 1, pp. 600–604 (2005)
Liao, S.-H., Han, W., Tong, R.-F., Dong, J.-X.: A hybrid approach to extracting tooth models from CT volumes. In: Martin, R., Bez, H., Sabin, M. (eds.) IMA 2005. LNCS, vol. 3604, pp. 308–317. Springer, Heidelberg (2005). doi:10.1007/11537908_18
Pavaloiu, I.B., Vasilateanu, A., Goga, N., Marin, I., Ilie, C., Ungar, A., Patracu, I.: 3D dental reconstruction from CBCT data. In: 2014 International Symposium on Fundamentals of Electrical Engineering, pp. 1–6, November 2014
Pohle, R., Toennies, K.D.: Segmentation of medical images using adaptive region growing (2001)
Schroeder, W., Martin, K.M., Lorensen, W.E.: The Visualization Toolkit, An Object-Oriented Approach To 3D Graphics (2006)
Verma, B., Sardana, H.K.: Real time image segmentation using watershed algorithm on FPGA. Int. J. Eng. Sci. Technol. 3(6), 4518–4522 (2011)
Zou, B.J., Liu, S.J., Liao, S.H., Ding, X., Liang, Y.: Interactive tooth partition of dental mesh base on tooth-target harmonic field. Comput. Biol. Med. 56(2015), 132–144 (2015)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Liu, SJ., Zou, Z., Liang, Y., Pan, JS. (2017). Tooth Segmentation from Cone Beam Computed Tomography Images Using the Identified Root Canal and Harmonic Fields. In: Pan, JS., Snášel, V., Sung, TW., Wang, X. (eds) Intelligent Data Analysis and Applications. ECC 2016. Advances in Intelligent Systems and Computing, vol 535. Springer, Cham. https://doi.org/10.1007/978-3-319-48499-0_13
Download citation
DOI: https://doi.org/10.1007/978-3-319-48499-0_13
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-48498-3
Online ISBN: 978-3-319-48499-0
eBook Packages: EngineeringEngineering (R0)