Abstract
Purpose
In both structural and functional MRI, there is a need for accurate and reliable automatic segmentation of brain regions. Inconsistent segmentation reduces sensitivity and may bias results in clinical studies. The current study compares the performance of publicly available segmentation tools and their impact on diffusion quantification, emphasizing the importance of using recently developed segmentation algorithms and imaging techniques.
Methods
Four publicly available, automatic segmentation methods (volBrain, FSL, FreeSurfer and SPM) are compared to manual segmentation of the thalamus and hippocampus imaged with a recently proposed T1-weighted MRI sequence (MP2RAGE). We evaluate morphometric accuracy on 22 healthy subjects and impact on diffusivity measurements obtained from aligned diffusion-weighted images on a subset of 10 subjects.
Results
Compared to manual segmentation, the highest Dice similarity index of the thalamus is obtained with volBrain using a local library (\(M=0.913\), \(\hbox {SD}=0.014\)) followed by volBrain using an external library (\(M=0.868\), \(\hbox {SD}=0.024\)), FSL (\({M}=0.806\), \(\mathrm{SD}=0.034\)), FreeSurfer (\({M}=0.798\), \(\mathrm{SD}=0.049\)) and SPM (\({M}=0.787\), \(\mathrm{SD}=0.031\)). The same order is found for hippocampus with volBrain local (\({M}=0.892\), \(\mathrm{SD}=0.016\)), volBrain external (\({M}=0.859\), \(\mathrm{SD}=0.014\)), FSL (\({M}=0.808\), \(\mathrm{SD}=0.017\)), FreeSurfer (\({M}=0.771\), \(\mathrm{SD}=0.023\)) and SPM (\({M}=0.735\), \(\mathrm{SD}=0.038\)). For diffusivity measurements, volBrain provides values closest to those obtained from manual segmentations. volBrain is the only method where FA values do not differ significantly from manual segmentation of the thalamus.
Conclusions
Overall we find that volBrain is superior in thalamus and hippocampus segmentation compared to FSL, FreeSurfer and SPM. Furthermore, the choice of segmentation technique and training library affects quantitative results from diffusivity measures in thalamus and hippocampus.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The extensive use of magnetic resonance imaging (MRI) to investigate pathology in the brain entails identification of specific regions of interest (ROI) for quantitative analysis. Accurate manual tracing of deep brain structures, such as the thalamus and hippocampus, demands a high level of tracer expertise and preferably standardized segmentation protocols. Introducing automatic or semi-automatic techniques into post-processing pipelines accelerates data analysis and offers reproducible and consistent decisions across datasets in large studies, which is crucial for obtaining reliable results [1].
Several software solutions for automatic segmentation are publicly available. Frequently used softwares in clinical research include “Oxford Centre for Functional MRI of the Brain” (FMRIB) Software Library (FSL) (http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/), FreeSurfer (http://surfer.nmr.mgh.harvard.edu) and Statistical Parametric Mapping (SPM) (http://www.fil.ion.ucl.ac.uk).
The segmentation techniques applied in FSL, FreeSurfer and SPM are model-based methods. In highly variable data, such as MRI of the brain, it may be difficult for the segmentation tools to model the ROIs with sufficient accuracy, even when the techniques are trained on representative datasets. To address this, multi-atlas label fusion has been suggested and has demonstrated excellent segmentation abilities [2–4]. Label fusion relies on a representative image library with corresponding validated structure segmentations (atlases). Recently, multi-atlas segmentation techniques, such as patch-based segmentation, have become popular [5, 6]. Patch-based methods have the advantage of requiring a smaller training library compared to regular label fusion and is therefore relatively easy to implement in a local setting [5, 7]. Even though these improvements in segmentation algorithms have demonstrated highly accurate morphometric results,Footnote 1 most of the novel approaches are still not publicly available and therefore less used in clinical research. Moreover, the impact of segmentation accuracy on quantification of parameters from other imaging modalities, such as diffusion and perfusion MRI, is not well studied.
Quantitative diffusion tensor imaging (DTI) is widely used to investigate microstructural changes in tissue. In diseases that cause subtle microstructural changes, such as mild traumatic brain injury (mTBI), there is a need for sensitive biomarkers in clinically relevant areas of the brain. Thalamus and hippocampus are two deep brain structures, where previous DTI studies have shown microstructural changes linked to cognitive impairment [8, 9], stress [10] and headache [11]. Segmentation directly on the DTI maps is prone to inconsistency and bias, as DTI provides limited anatomical information. Unbiased and automatic studies rely on accurate T1-weighted (T1w) segmentation and co-registration for obtaining quantitative measurements within relevant brain regions. Thus, it is highly relevant to investigate the impact of automatic segmentation accuracy on these quantitative measures.
Patch-based segmentation methods [5] perform well on conventional T1w images, such as Magnetization Prepared Rapid Acquisition Gradient Echo (MPRAGE) [12]. To the best of our knowledge, the accuracy of different automated segmentation methods has not yet been compared using T1w images from the recently proposed MP2RAGE sequence, which significantly reduces the intensity bias and provides superior grey matter (GM) to white matter (WM) contrast [13].
In this study, we compared the performance of a multi-atlas, patch-based segmentation method, as implemented in the online software platform volBrain (with two different training libraries), to three widely applied methods implemented in FSL, FreeSurfer and SPM. We used manual segmentation as the gold standard and measured the segmentation accuracy of thalamus and hippocampus when imaged with MP2RAGE. Additionally, we applied the segmented masks of thalamus and hippocampus on co-registered fractional anisotropy (FA) and mean diffusivity (MD) maps for the purpose of evaluating the effect on the quantification of these diffusivity metrics.
Material and methods
Participants
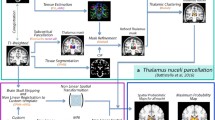
Twenty-two healthy subjects were included in the study (age range 19–40 years, 12 females). MP2RAGE images were acquired from all subjects and DTI images in 10 subjects. All subjects were scanned on a Siemens Magnetom Skyra 3T MRI system with a 32-channel head coil. MP2RAGE parameters were acquired with \(\hbox {TR}=5\,\hbox {s}, \hbox {TI}_{1}=0.7\,\hbox {s}\), \(\mathrm{TI}_{2}=2.5\,\hbox {s}\), \(\alpha _{1}=4^{\circ }, \alpha _{2}=5^{\circ }\), a 3D sequence imaged at isotropic \(1\,\hbox {mm}^{3}\) resolution (acquisition matrix: 240 \(\times \) 256, 176 sagittal slices) and turbo factor of 176 as defined by Marques et al. [13]. DTI was acquired with 32 directions, 5 B0 acquisitions, \(\hbox {TR=}10.9\,\hbox {s}\), \(\hbox {TE}=0.079\), \(\hbox {TI}= 2.1\,\hbox {s}\), imaged at isotropic \(2.3\,\hbox {mm}^{3}\) resolution (acquisition matrix: 96 \(\times \) 96, 38 axial slices), and inversion recovery-based CSF suppression to reduce partial volume effects. Figure 1 provides an overview of the methods and comparisons.
Pre-processing
MP2RAGE images were calculated as the ratio of two MPRAGE images, acquired with different inversion times [13]. As reported by Fujimoto et al. [14], the amplified background noise in MP2RAGE images may introduce errors in FreeSurfer segmentations. To deal with this, we used a slightly different approach than Fujimoto and colleagues. We classified the intensities of the proton density-weighted image acquired during the second inversion recovery into four classes using a Fuzzy C-means algorithm [15]. This robustly separates the background (1 class) from foreground (3 classes). To regain a “natural” background noise, we added the background noise of the second inversion recovery to the combined (flat) image. The modified image was used as input to all segmentation pipelines, and no errors were detected. Despite inherent intensity normalization of the MP2RAGE images, all segmentation tools were run with intensity non-uniformity correction, as this was the default setting.
Diffusion data were eddy current and motion corrected using FSL, and EPI distortion correction was performed with ExploreDTI toolbox [16]. During pre-processing, the diffusion data were resampled to the space of the undistorted MP2RAGE image and FA and MD values were subsequently calculated with the ExploreDTI pipeline.
Manual segmentation of ROIs
Thalamus and hippocampus from the 22 MP2RAGE images were manually segmented by an experienced neuroradiologist (EN: 7 years of experience in neuroradiology) and a trained assistant (TA) using ITK-SNAP (www.itk-snap.org) [17]. The thalami were first manually traced by EN in the axial plane using anatomical landmarks. Next an initial training phase of TA using the protocol outlined by Power et al. [18] and supervised by EN was established. TA then adjusted the thalami in all three principal planes according to the protocol. Bilateral thalamus segmentation took 35–40 min per subject. The hippocampi were manually traced according to the EADC-ADNI segmentation protocol [19] by TA, initially supervised by EN. Segmentation of both hippocampi took 25–35 min per subject. As done in the EADC-ADNI protocol [19], all manual segmentations were performed in MNI space, where similar appearance of the nuclei is expected to improve tracing consistency and speed when using the segmentation protocols. The final segmentations were transformed back to scanner native space for comparison. Manual intra-operator reliability on hippocampus was tested 10 months after initial segmentations by TA on the ten subjects with DTI. A 1-h training session, reading the protocol and training on a separate subject were performed before segmentation of the ten subjects was carried out.
Automatic segmentation
The following provides a brief overview of the four processing methods volBrain, FSL, FreeSurfer and SPM with the applied settings.
volBrain
volBrain (http://volbrain.upv.es), which is an open-access platform, is based on an advanced pipeline providing automatic segmentations of several brain structures [20]. The version of volBrain used in the current setting involves an updated version of a recent patch-based method [5]. We tested the segmentation method using two different libraries: (1) the default volBrain library (external) consisting of 50 conventional T1w images (MPRAGE and SPGR), and (2) our own manually segmented library (local) of 22 MP2RAGE images in a leave-one-out fashion. In both cases, the images were flipped across the mid-sagittal plane to artificially increase the library size as done in related work [7].
FSL
FSL is freely available for download online (http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/). The FMRIB’s Integrated Registration & Segmentation Tool (FIRST), from FSL v5.0, was used to segment subcortical structures [21]. FSL FIRST is a model-based segmentation tool that uses training data from manually segmented images. In the current pipeline, we used the default setting of FIRST, which applies empirically optimized settings (numbers of modes and shape/boundary correction) for each structure (see http://fsl.fmrib.ox.ac.uk/fsl/fsl-4.1.9/first/index.html).
FreeSurfer
FreeSurfer image analysis suite (version 5.3), which is documented and freely available for download online (http://surfer.nmr.mgh.harvard.edu), was used in a default mode in the current setting. The segmentation maps are created using spatial intensity gradients across tissue classes and are therefore not simply reliant on absolute signal intensity [22].
SPM
SPM is a MATLAB-based (MathWorks Inc.) freely available software, which can be downloaded online (http://www.fil.ion.ucl.ac.uk). Segmentation was performed with SPM12 and MATLAB R2015b by combining the unified segmentation tool with the neuroinformatics SPM template [23], which consists of multiple structures of the brain segmented in Montreal Neurological Institute (MNI) space.
ROI volumes of the thalamus and hippocampus. From left manual, volBrain local, volBrain external, FSL, FreeSurfer and SPM. Boxes indicate 25 and 75 % percentile and the bold line the median. Whiskers indicate the most extreme point within 1.5 times the interquartile range. Data points outside this range are plotted individually. Horizontal bars indicate non-significant test for difference in volume. The remaining comparisons showed significant differences in volume
DSI, FPR and FNR for segmentations of the thalamus and the hippocampus using volBrain local (vBlocal), volBrain external (vBext), FSL, FreeSurfer and SPM compared to the manual “gold standard”. Boxes indicate 25 and 75 % percentile and the bold line the median. Whiskers indicate the most extreme point within 1.5 times the interquartile range. Data points outside this range are plotted individually. Horizontal bars indicate non-significant test for difference in DSI, FPR and FNR. The remaining comparisons showed significant differences
Data and statistical analysis
The segmentations obtained from the four automatic methods were compared to the manual segmentations using volume, Dice similarity index (DSI), false positive rate (FPR), false negative rate (FNR), and Hausdorff distance estimated bilaterally. We report mean (M) and standard deviations (SD) and visualize data with boxplots.
DSI is defined as \(\frac{2{C}}{{A}+{B}}\) and is the quotient of similarity ranging from zero to one. A and B are the number of voxels in segmentation A and segmentation B, respectively, and C is the number of voxels shared by the two segmentations. FPR and FNR were calculated, respectively, as number of false positive and false negative voxels as percentage of the total manually segmented number of voxels. Hausdorff distance, h, indicates the maximum distance error and is defined as the maximum distance, d, from the surface of segmentation A to the nearest point in the surface of segmentation B: \({h}({A,B})= \max _{{a}\in {A}}\{\min _{{b}\in {B}}\{{d}({a},{b})\}\}\). Intra-rater reliability, volume, DSI, FPR, FNR and Hausdorff distance were analysed with two-way ANOVA and post-estimation was carried out, with a significance level of 0.05. Finally, FA and MD values were analysed using two-way ANOVA. Post hoc analyses of FA and MD were carried out with a primary analysis of the automatic segmentations against the manual segmentation and secondary between the automatic methods. Results are presented at a significance level of 0.05, and in addition, diffusivity results were reported with correction for multiple comparisons (60 tests on diffusion metrics were carried out, which yields a Bonferroni-corrected threshold of \(p=0.0008\)).
Results
Intra-operator reliability of manual segmentation
The 10-month intra-operator reliability test of hippocampus manual segmentation resulted in a mean volume difference of 3.1 % (\(\hbox {SD}=4.9\,\%\)), which was not significantly different (\(p>0.05\)). Mean DSI was 0.913 (\(\hbox {SD}=0.010\)), and mean FPR and FNR were, respectively, 10.4 (\(\hbox {SD}=3.4\,\%\)) and 7.3 % (\(\hbox {SD}=1.6\,\%\)). Intra-operator Hausdorff distances ranged from 2.2 to 4.9 mm. For DTI metrics, the overall model was significantly different for both FA (\(p<0.001\)) and MD (\(p<0.001\)). Post-estimation revealed a FA mean difference of 0.003 (\(\hbox {SD}=0.002\)) which was significantly different (\(p=0.003\)). MD obtained a mean difference of \(0.006\times 10^{-3}(\hbox {SD}=0.006\times 10^{-3})\), which was significantly different (\(p=0.018\)). If Bonferroni corrected, there is no significant difference between the manual segmentations.
Hausdorff distance of the automatic segmentations of hippocampus and thalamus compared to the manual segmentation. Boxes indicate 25 and 75 % percentile and the bold line the median. Whiskers indicate the most extreme point within 1.5 times the interquartile range. Data points outside this range are plotted individually. The horizontal bar indicates a non-significant test for difference in Hausdorff distance. The remaining comparisons showed significant differences
Thalamus and hippocampus volumes
Figure 2 shows the volumes of thalamus and hippocampus for each of the segmentation methods. Overall the model was significantly different in both ROIs (\(p<0.001\)). There was no significant difference (\(p>0.05\)) in manual versus volBrain local, manual versus volBrain external and volBrain local versus volBrain external in thalamus, but all other comparisons for thalamus were significantly different (\(p<0.05\)). The hippocampus segmentations showed significantly higher volumes of volBrain external, FSL, FreeSurfer and SPM compared to the manual and volBrain local, and only FSL versus FreeSurfer and volBrain external versus SPM were not significantly different (\(p>0.05\)) from each other.
Manual versus automatic segmentation
Comparison of manual and automatic segmentation methods showed a substantial variation in DSI across the methods (see Fig. 3), and the overall model was significantly different (\(p<0.001\)) for both thalamus and hippocampus DSI, FPR and FNR. To maintain overview, only non-significant (\(p>0.05\)) p values are marked in Fig. 3. All other p values are significant (\(p<0.001\)).
DSI of the thalamus was significantly higher for volBrain local (\({M}=0.913\), \(\mathrm{SD}=0.014\)) and volBrain external (\({M}=0.868\), \(\mathrm{SD}=0.024\)) compared to FSL (\({M}=0.806\), \(\mathrm{SD}=0.034\)), FreeSurfer (\({M}=0.798\), \(\mathrm{SD}=0.049\)) and SPM (\({M}=0.787\), \(\mathrm{SD}=0.031\)). FreeSurfer was not significantly different from FSL or SPM. FPR in the thalamus when segmented with FSL (\(M=41\,\%\)) and SPM (\(M=42\,\%\)) was significantly higher than the other segmentation methods. Over-segmentations are exemplified in Fig. 5a, b, where the significantly lower FPR of volBrain local (\(M=9\,\%\)) and external (\(M=14\,\%\)) also can be observed. FreeSurfer FPR was significantly higher than volBrain and significantly lower than FSL and SPM. The mean FNR of the four methods ranged from 5 to 13 % all being significantly different, except volBrain local versus SPM, volBrain external versus FreeSurfer and FSL versus SPM.
The DSI of the hippocampus demonstrated significantly different values between all methods, with volBrain local (\({M}=0.892\), \(\mathrm{SD}=0.016\)) showing the best performance, followed by volBrain external (\({M}=0.859\), \(\mathrm{SD}=0.014\)), FSL (\({M}=0.808\), \(\mathrm{SD}=0.017\)), FreeSurfer (\({M}=0.771\), \(\mathrm{SD}=0.023\)) and SPM (\({M}=0.735\), \(\mathrm{SD}=0.038\)). A similar pattern was observed for FPRs, with volBrain local performing best (\({M}=9\,\%\)) followed by volBrain external (\({M}=26\,\%\)), FSL (\({M}=36\,\%\)), SPM (\({M}=40\,\%\)) and FreeSurfer performing worst (\({M}=41\,\%\)). FSL versus SPM and FreeSurfer versus SPM were the only methods which were not significantly different in FPR. Mean FNR ranged from 5 to 19 %, and all methods were significantly different, except volBrain local versus FreeSurfer.
Examples of manual and automatic segmentations of thalamus and hippocampus presented in a the subject where volBrain local had the best performance, and b where volBrain local had the worst performance. Upper two rows thalamus in an axial view, overlaid on native T1 and co-registered FA images. Third row 3D reconstructions of thalamus. The lower three rows contain similar visualizations for hippocampus segmentations. Left to right manual, volBrain local, volBrain external, FSL, FreeSurfer and SPM methods. Green areas indicate overlap between automatic methods and manual segmentation. Red indicates areas, which are included in the automatic, but not the manual method (false positives). Blue indicates areas, which are included by the manual, but not the automatic method (false negatives)
In terms of Hausdorff distance, the overall model was significantly different in both thalamus and hippocampus (\(p<0.001\)). Figure 4 shows the Hausdorff distances for the automatic hippocampus and thalamus segmentations with low distances indicating good performance. Post-estimation showed that all methods had significantly different Hausdorff distances (\(p<0.05\)) except volBrain external versus FSL in thalamus. The best performance was seen with volBrain local, and the highest Hausdorff distances were measured with FreeSurfer in both thalamus and hippocampus.
Visual inspection of ROIs
Examples of manual segmentations and the corresponding automatic segmentations of the thalamus and hippocampus, overlaid on the T1w image and the FA map, are shown in Fig. 5
As illustrated, FreeSurfer, FSL and SPM generally over-segment the thalamus, especially the non-thalamic tissue near the border of the internal capsule (IC). volBrain external over-segments to a lesser extent, and volBrain local demonstrated only subtle over-segmentation at the inferior and lateral border of the thalamus. The same pattern of over-segmentation is found in the hippocampus with more extensive over-segmentation by FSL, FreeSurfer and SPM, but also slightly by volBrain external, compared to the manual (Fig. 5a, b). The over-segmentation of FSL, FreeSurfer and SPM in the hippocampus is mainly restricted to the superior and the rostral part of the hippocampus in the transition to thalamus and fornix.
Diffusivity results: Thalamus
The model was overall significantly different in the diffusivity measurements for both FA (\(p<0.001\)) and MD (\(p<0.001\)) in thalamus. Figure 6 shows mean FA and MD values in thalamus extracted from the six different segmentations and how MD values of all segmentations consistently change based on the segmentation method used, while FA values change less consistently. The volBrain local method provided the most accurate measurements compared to the manual segmentation.
Diffusivity results: Hippocampus
The model also provided overall significantly different results for FA (\(p<0.001\)) and MD (\(p<0.001\)) in hippocampus. Diffusivity results of the hippocampus are shown in Fig. 7. The figure illustrates the same consistent increase or decrease in MD between methods and subjects, but with different offsets and variation compared to the manual segmentation. FA showed a less consistent pattern. All automatic methods were significantly different from the manual values. When corrected for multiple comparisons, FA values for all methods stayed significantly different from the manual segmentation, except volBrain local and SPM, and for MD, all methods stayed significantly different except volBrain local.
Post hoc analysis on diffusivity parameters between manual and automatic segmentation
Post hoc analysis for thalamus and hippocampus is reported in Table 1. The post hoc analysis of thalamus revealed that only volBrain local was not significantly different from the manual segmentation and obtained the lowest mean difference of \(M=-0.3\,\%\) in FA and \(M=-0.1\,\%\) in MD. The other methods obtained a higher mean difference, ranging from \(M=3\,\%\) to \(M=9\,\%\) in FA and \(M=1\)–3 % in MD.
All methods obtained significantly different diffusivity parameters in the hippocampus when compared to the manual segmentation. The volBrain local demonstrated the most accurate result in the hippocampus, with a mean difference of \(M=-1\,\%\) of FA and \(M=-0.5\,\%\) of MD. If corrected for multiple comparisons, volBrain local FA and MD were not significantly different from the manual and neither was the SPM result of FA.
Post hoc analysis on diffusivity parameters between the automatic segmentation methods
Between-method comparison revealed more variable results. For an overview, see Table 2 with indication of corrected and un-corrected p values for both thalamus and hippocampus. All methods, except FSL, FreeSurfer and SPM, were significantly different from each other, when measuring FA in the thalamus. When measuring MD in the thalamus, all five methods yielded significantly different results. For hippocampus FA measurements, only volBrain local stood out as different from all the other methods. Furthermore, volBrain external was significantly different from FreeSurfer, while FSL versus FreeSurfer and SPM were also significantly different. For hippocampus MD, all methods were significantly different, except volBrain external versus FSL and FreeSurfer versus SPM.
Discussion
In this study, we evaluated the performance of a recent patch-based segmentation method [5] as implemented in volBrain [20] and three widely used conventional methods as implemented in FSL [21], FreeSurfer [22] and SPM [23]. Using MP2RAGE images, we tested the algorithms on two often investigated deep brain structures: the thalamus and the hippocampus. We found that the patch-based segmentation had the best overall accuracy. FreeSurfer, FSL and SPM all over-segmented the thalamus including non-thalamic tissue near the border of the IC and under-segmented in regions of the medial and lateral geniculate of the thalamus. In the segmentation of hippocampus, volBrain performed best followed by FSL, FreeSurfer and SPM. Moreover, we demonstrated that volBrain, based on a local library, was the only method, in which the diffusivity metrics of the thalamus did not differ significantly from the metrics obtained based on manual segmentation (Table 1). Analysis of hippocampus revealed that volBrain and SPM (although reporting low DSI) were not significantly different (Bonferroni corrected) from the manual method in terms of FA, and for MD only, volBrain local was not significantly different. This demonstrates that segmentation accuracy impacts the obtained diffusivity results, and less accurate methods, such as FSL, FreeSurfer and SPM, do not produce consistent diffusivity results.
The accuracy of the patch-based segmentation method in our study is comparable to previous results on hippocampus segmentations using MPRAGE images [5, 6]. A study by Patenaude et al. [21], using conventional T1w images and a leave-one-out comparison on its own library, found higher DSIs using FSL than found here. Patenaude and colleagues reported a mean DSI of 0.887 and 0.840 for the thalamus and hippocampus, respectively. This difference may reflect the importance of using coherent labelling protocols and similar imaging parameters within the template library. Patenaude et al. did, however, not reach the accuracy of the volBrain local segmentation in our study with DSI of 0.913 and 0.892, respectively. To compare the performance of the volBrain method with a training library different from MP2RAGE, we applied volBrain with an external training library consisting of MPRAGE and SPGR images. We found that volBrain still performed better than FSL, FreeSurfer and SPM (Fig. 3). The results of the intra-reliability test on hippocampus further emphasize the advantage of automatic segmentation. We found a mean DSI of 0.915, which is consistent with the previous findings by Frisoni et al. [24] of DSI \(=\) 0.89. This result is at an accuracy level of volBrain local. However, in contrast to manual segmentations, automatic methods are deterministic and yield consistent errors. Thus, automatic segmentation methods are more robust in a longitudinal setting.
FSL, FreeSurfer and SPM over-segmented the structures with FPRs in the range of 15–42 %. This resulted in consistent inclusion of white matter in the segmented regions of thalamus and hippocampus (both grey matter structures) as qualitatively verified using FA maps (see Fig. 5a, b). Regarding the volBrain method, no systematic over- or underestimation for thalamus was observed with neither local nor external libraries (FPR out-balanced FNR). Patches can capture texture similarities [5], and this is perhaps why the patch-based method attains consistently high accuracy on both thalamus and hippocampus. volBrain local was unbiased for hippocampus, while volBrain external slightly over-segmented hippocampus (FPR \(M = 26\,\%\)). This was unexpected, because the two libraries were constructed using hippocampus masks segmented based on the same protocol (the EADC protocol), while the protocols differed for the thalamus libraries. For hippocampus, this may be explained by different interpretations (different operators) of the EADC-ADNI protocol in the segmentation procedure of the hippocampus or by differences in contrast to the T1w images in the training libraries (MP2RAGE versus MPRAGE/SPGR).
The Hausdorff distance showed a stepwise increase between the manual and automatic methods, with mean values in the range 2–6 mm, the lowest being volBrain local followed by volBrain external, FSL, SPM and FreeSurfer (see Fig. 4). When considering the obtained FPR and FNR, the Hausdorff distance most likely reflects a maximum over-segmentation. However, evaluating the examples in Fig. 5a, b where the geniculate bodies of the thalamus are excluded (except for volBrain local), the distance may be due to under-segmentation in this specific region.
The intra-operator reliability test of manual hippocampus segmentation showed a consistent segmentation and no significant difference between volumes segmented with a time interval of 10 months. Our intra-operator DSI (\({M}=0.913\), \(\mathrm{SD}=0.010\)) is in line with previous reports of manual hippocampus segmentation reliability (\({M}=0.89\), \(\mathrm{SD}=0.01\)) [24]. The DSI of repeated tracings reveal that manual segmentation of hippocampus has the same level of accuracy as between manual segmentation and the volBrain local method. The volBrain local method though has the advantage of being more consistent, faster and less costly when the library has been established [25].
The obtained segmentation accuracies are partly reflected in the analysis of the diffusivity metrics. The volBrain local method was the only method not yielding significantly different FA and MD results in the thalamus compared to the results obtained by manual segmentation with a mean difference of −1 and −0.1 % in FA and MD, respectively. The other methods yielded mean differences between 1 and 9 % and were all significantly different in FA and MD compared to the values obtained with manual segmentation. This can be explained by the over-segmentation expanding into IC and the ventricular cerebral spinal fluid (CSF) (Fig. 5a, b). In hippocampus, the manual method was significantly different in FA compared to all methods (\(p<0.03\)). When correcting for multiple comparisons, FA values in both volBrain local and SPM and MD in volBrain local was not significantly different from manual measurements. The finding of SPM not being significantly different from the manual method, despite the inaccuracy of the SPM segmentation, can be explained by the segmentation expanding into both WM and CSF, which on average blur the FA differences as WM and CSF, respectively, represent higher and lower FA values. Higher FA values of volBrain external, FSL and FreeSurfer in the hippocampus can be explained by over-segmentation into areas at the transition to the thalamus and fornix. The difference between volBrain local and the other methods in the hippocampus segmentation is furthermore confirmed by the post hoc analysis (Table 2), which shows that both volBrain external, FSL, FreeSurfer and SPM all significantly differ in FA from volBrain local estimates, but not from each other, if corrected for multiple comparisons. This is visualized in Fig. 7 by the offsets between volBrain local and volBrain external, FSL, FreeSurfer and SPM. The FA and MD results of the intra-operator reliability test in hippocampus showed a significant difference between the two segmentations. This difference is similar to that of the best automatic segmentation (volBrain local). In both cases, the difference seems to be systematic (Fig. 7). However, this bias will be removed if using automatic methods in a longitudinal setting, as automatic methods are consistent and not prone to changing interpretations of the segmentation protocol. Finally, it should be noted that Bonferroni correction removed the significant differences between the manual segmentations.
Although the mean difference of FA and MD varies, all segmentation methods yielded consistent inter-subject differences compared to the manual approach (Figs. 6, 7). This was most pronounced for MD results. Group comparisons may therefore relatively yield similar results, using the same method within the same study. However, in diseases and disorders with subtle structural changes where the influence of segmentation errors could blur the findings and result in reduced sensitivity, it is crucial to use the most accurate method to detect pathological changes. A study by Barbagallo et al. [26] found a significant difference in MD in the thalamus between amyotrophic lateral sclerosis (ALS) patients and controls (\(0.06 \times 10^{-3}\), \(p=0.019\)) using FSL FIRST, but the FA difference of 0.01 was not significant (\(p>0.025\)). We speculate that such a result might have been significant if a more accurate segmentation method had been used. We found that volBrain local obtained the most accurate measurements compared to the manual segmentation (FA mean difference \(= -\)0.001), and all the other methods obtained a mean difference of FA higher than the 0.01 level obtained between groups in the study by Barbegello and colleagues. Although it is not directly comparable, the impact of using different methods (more or less accurate) in clinical studies should be investigated further.
The variation of our diffusivity measurements was considerably smaller compared to those reported in the study by Barbagello et al. The volBrain local method obtained SDs of FA values in the thalamus and hippocampus of, respectively, 0.008 and 0.007 and SDs of MD values of, respectively, 0.014 (\(\times 10^{-3}\)) and 0.017 (\(\times 10^{-3})\). In the Barbagello study, the corresponding SDs were 0.02 and 0.01 for FA and 0.05 \((\times 10^{-3})\) and 0.07 \((\times 10^{-3})\) for MD. This could be due to the reliability of the MP2RAGE images, as pointed out in a recent study [27]. The MP2RAGE sequence is less influenced by B1 as well as M0 and T2*, improving the image contrast and sharpness, which makes it easier to discriminate between grey- and white matter structures [13]. Another reason for the higher FA and MD variation in the Barbagello study could be due to the ALS pathology.
In the present study, we executed the FSL pipeline with the empirical optimized default settings. Experimentation on optimizing the FSL modes and boundary correction adapted to the MP2RAGE images may improve the final segmentation of FSL. The influence of adapting the FSL segmentation tool, FIRST, to MP2RAGE remains to be investigated. However, Patenaude et al. [21] used an adapted and optimized setting to their T1w images (no specification of the T1 sequence) and did still not reach the DSI levels of the patch-based method in volBrain local. Furthermore, it was not possible to use the same template library in volBrain and FSL, which would have been optimal for comparing the methods. Thus volBrain local has an advantage because it uses consistent training and testing data. This is similar to the Patenaude study [21], which showed good performance when using customized local settings. Patch-based methods have the advantage of requiring a relatively small library, which makes it feasible to implement and optimize locally.
State-of-the-art segmentation methods, like patch-based methods [5], together with high image quality, as in MP2RAGE data, may lead to higher sensitivity in future studies of morphometry and of microstructural changes. It could also be of interest to apply the presented methods retrospectively to clinical studies and evaluate whether statistical power and conclusions might be altered.
In conclusion, we have shown the potential of a recent, automatic patch-based segmentation method, volBrain, to provide more accurate thalamus and hippocampus segmentations in MP2RAGE images compared to conventional approaches. We have furthermore demonstrated that FA and MD values, extracted from co-registered DTI, deviated less from the reference of the manual segmentation, when using patch-based methods compared to the segmentations of FSL, FreeSurfer and SPM. We have illustrated under- and particularly over-segmentations on T1w images and FA maps, especially for FSL, FreeSurfer and SPM. Finally, we propose that MP2RAGE images are more suitable for thalamus and hippocampus segmentation compared to conventional T1w images.
Notes
See, for example, the recent MICCAI workshop on Multi-Atlas Labeling, https://masi.vuse.vanderbilt.edu/workshop2012/images/c/c8/MICCAI_2012_Workshop_v2.pdf.
References
Mulder ER, de Jong RA, Knol DL, van Schijndel RA, Cover KS, Visser PJ, Barkhof F, Vrenken H (2014) Hippocampal volume change measurement: quantitative assessment of the reproducibility of expert manual outlining and the automated methods FreeSurfer and FIRST. Neuroimage 92:169–181
Heckemann RA, Hajnal JV, Aljabar P, Rueckert D, Hammers A (2006) Automatic anatomical brain MRI segmentation combining label propagation and decision fusion. Neuroimage 33(1):115–126
Rohlfing T, Brandt R, Menzel R, Maurer CR (2004) Evaluation of atlas selection strategies for atlas-based image segmentation with application to confocal microscopy images of bee brains. Neuroimage 21(4):1428–1442
Aljabar P, Heckemann RA, Hammers A, Hajnal JV, Rueckert D (2009) Multi-atlas based segmentation of brain images: atlas selection and its effect on accuracy. Neuroimage 46(3):726–738
Coupé P, Manjón JV, Fonov V, Pruessner J, Robles M, Collins DL (2011) Patch-based segmentation using expert priors: application to hippocampus and ventricle segmentation. Neuroimage 54(2):940–954
Tong T, Wolz R, Coupé P, Hajnal JV, Rueckert D (2013) Segmentation of MR images via discriminative dictionary learning and sparse coding: application to hippocampus labeling. Neuroimage 76:11–23
Eskildsen SF, Coupé P, Fonov V, Manjón JV, Leung KK, Guizard N, Wassef SN, Østergaard LR, Collins DL (2012) BEaST: brain extraction based on nonlocal segmentation technique. Neuroimage 59(3):2362–2373
Falangola MF, Jensen JH, Tabesh A, Hu C, Deardorff RL, Babb JS, Ferris S, Helpern JA (2013) Non-Gaussian diffusion MRI assessment of brain microstructure in mild cognitive impairment and Alzheimer’s disease. Magn Reson Imaging 31(6):840–846
Mitchell AS, Sherman SM, Sommer MA, Mair RG, Vertes RP, Chudasama Y (2014) Advances in understanding mechanisms of thalamic relays in cognition and behavior. J Neurosci 34(46):15340–15346
Vestergaard-Poulsen P, Wegener G, Hansen B, Bjarkam CR, Blackband SJ, Nielsen NC, Jespersen SN (2011) Diffusion-weighted MRI and quantitative biophysical modeling of hippocampal neurite loss in chronic stress. PLoS ONE 6(7):e20653
Granziera C, Daducci A, Romascano D, Roche A, Helms G, Krueger G, Hadjikhani N (2014) Structural abnormalities in the thalamus of migraineurs with aura: a multiparametric study at 3 T. Hum Brain Mapp 35(4):1461–1468
Coupé P, Eskildsen SF, Manjón JV, Fonov VS, Collins DL (2012) Simultaneous segmentation and grading of anatomical structures for patient’s classification: application to Alzheimer’s disease. Neuroimage 59(4):3736–3747
Marques JP, Kober T, Krueger G, van der Zwaag W, Van de Moortele PFF, Gruetter R (2010) MP2RAGE, a self bias-field corrected sequence for improved segmentation and T1-mapping at high field. Neuroimage 49(2):1271–1281
Fujimoto K, Polimeni JR, van der Kouwe AJW, Reuter M, Kober T, Benner T, Fischl B, Wald LL (2014) Quantitative comparison of cortical surface reconstructions from MP2RAGE and multi-echo MPRAGE data at 3 and 7 T. Neuroimage 90:60–73
Dudo RO, Hart PE, Stork D (2001) Pattern classification, 2nd edn. Wiley, Hoboken
Leemans A, Jeurissen B, Sijbers J, Jones D (2009) ExploreDTI: a graphical toolbox for processing, analyzing, and visualizing diffusion MR data. In: Proceedings 17th scientific meeting, international society for magnetic resonance in medicine, vol 17, no 2, p 3537
Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G (2006) User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 31(3):1116–1128
Power BD, Wilkes FA, Hunter-Dickson M, van Westen D, Santillo AF, Walterfang M, Nilsson C, Velakoulis D, Looi JCL (2015) Validation of a protocol for manual segmentation of the thalamus on magnetic resonance imaging scans. Psychiatry Res 232(1):98–105
Boccardi M, Bocchetta M, Apostolova LG, Barnes J, Bartzokis G, Corbetta G,DeCarliC, deToledo-Morrell L, Firbank M, Ganzola R, Gerritsen L, Henneman W, Killiany RJ, Malykhin N, Pasqualetti P, Pruessner JC, Redolfi A, Robitaille N, Soininen H, Tolomeo D, Wang L, Watson C, Wolf H, Duvernoy H, Duchesne S, Jack CR, Frisoni GB (2014) Delphi definition of the EADC-ADNI harmonized protocol for hippocampal segmentation on magnetic resonance. Alzheimers Dement 11(2):126–138
Manjón JV, Coupé P (2015) volBrain: an online MRI brain volumetry system. Hum Brain Mapp 15:2015
Patenaude B, Smith SM, Kennedy DN, Jenkinson M (2011) A Bayesian model of shape and appearance for subcortical brain segmentation. Neuroimage 56(3):907–922
Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, Van Der Kouwe A, Killiany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale AM (2002) Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron 33(3):341–355
Ashburner J, Friston KJ (2005) Unified segmentation. Neuroimage 26(3):839–851
Frisoni GB, Jack CR, Bocchetta M, Bauer C, Frederiksen KS, Liu Y, Preboske G, Swihart T, Blair M, Cavedo E, Grothe MJ, Lanfredi M, Martinez O, Nishikawa M, Portegies M, Stoub T, Ward C, Apostolova LG, Ganzola R, Wolf D, Barkhof F, Bartzokis G, DeCarli C, Csernansky JG, Detoledo-Morrell L, Geerlings MI, Kaye J, Killiany RJ, Lehericy S, Matsuda H, O’Brien J, Silbert LC, Scheltens P, Soininen H, Teipel S, Waldemar G, Fellgiebel A, Barnes J, Firbank M, Gerritsen L, Henneman W, Malykhin N, Pruessner JC, Wang L, Watsonl C, Wolf H, Deleon M, Pantel J, Ferrari C, Bosco P, Pasqualetti P, Duchesne S, Duvernoy H, Boccardi M, Albert MS, Bennet D, Camicioli R, Collins DL, Dubois B, Hampel H, Denheijer T, Hock C, Jagust W, Launer L, Maller JJ, Mueller S, Sachdev P, Simmons A, Thompson PM, Visser PJ, Wahlund LO, Weiner MW, Winblad B (2015) The EADC-ADNI harmonized protocol for manual hippocampal segmentation on magnetic resonance: evidence of validity. Alzheimer’s Dement 11(2):111–125
Næss-Schmidt ET, Tietze A, Mikkelsen IK, Petersen M, Blicher JU, Coupé P, Manjón JV, Eskildsen SF (2015) Patch-based segmentation from MP2RAGE images: comparison to conventional techniques. In: Wu G, Coupé P, Zhan Y, Munsell B, Rueckert D (eds) First international workshop, patch-techniques in medical imaging. Lecture notes in computer science, held in conjunction with MICCAI 2015, vol 9467. Munich, Germany, pp.180–187
Barbagallo G, Nicoletti G, Cherubini A, Trotta M, Tallarico T, Chiriaco C, Nisticò R, Salvino D, Bono F, Valentino P, Quattrone A (2014) Diffusion tensor MRI changes in gray structures of the frontal-subcortical circuits in amyotrophic lateral sclerosis. Neurol Sci 35(6):911–918
Okubo G, Okada T, Yamamoto A, KanagakiM, Fushimi Y, Okada T, Murata K, Togashi K (2015) MP2RAGE for deep gray matter measurement of the brain: a comparative study with MPRAGE. J Magn Reson Imaging 43(1):55–62
Acknowledgments
This work was funded in part by MINDLab UNIK initiative at Aarhus University, funded by the Danish Ministry of Science, Technology and Innovation, Grant Agreement Number 09-065250, partly by the Spanish grant TIN2013-43457-R from the Ministerio de Economia competitividad and with financial support from the French State, managed by the French National Research Agency (ANR) in the frame of the Investments for the future Programme IdEx Bordeaux (ANR-10-IDEX-03-02) by funding HL-DTI grant, Cluster of excellence CPU, LaBEX TRAIL (HR-DTI ANR-10-LABX-57) and the CNRS multidisciplinary project “Défi ImagIn”.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in the study involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. The study was a retrospective study. For this type of study, formal consent is not required.
Rights and permissions
About this article
Cite this article
Næss-Schmidt, E., Tietze, A., Blicher, J.U. et al. Automatic thalamus and hippocampus segmentation from MP2RAGE: comparison of publicly available methods and implications for DTI quantification. Int J CARS 11, 1979–1991 (2016). https://doi.org/10.1007/s11548-016-1433-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-016-1433-0