Abstract
Quantitative thickness computation of knee cartilage in ultrasound images requires segmentation of a monotonous hypoechoic band between the soft tissue-cartilage interface and the cartilage-bone interface. Speckle noise and intensity bias captured in the ultrasound images often complicates the segmentation task. This paper presents knee cartilage segmentation using locally statistical level set method (LSLSM) and thickness computation using normal distance. Comparison on several level set methods in the attempt of segmenting the knee cartilage shows that LSLSM yields a more satisfactory result. When LSLSM was applied to 80 datasets, the qualitative segmentation assessment indicates a substantial agreement with Cohen’s κ coefficient of 0.73. The quantitative validation metrics of Dice similarity coefficient and Hausdorff distance have average values of 0.91 ± 0.01 and 6.21 ± 0.59 pixels, respectively. These satisfactory segmentation results are making the true thickness between two interfaces of the cartilage possible to be computed based on the segmented images. The measured cartilage thickness ranged from 1.35 to 2.42 mm with an average value of 1.97 ± 0.11 mm, reflecting the robustness of the segmentation algorithm to various cartilage thickness. These results indicate a potential application of the methods described for assessment of cartilage degeneration where changes in the cartilage thickness can be quantified over time by comparing the true thickness at a certain time interval.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Osteoarthritis (OA) in the knee joint is a prevalent disease occurred among the elderly [12]. Degenerative change in the cartilage is one of the primary features of this disease [13]. X-rays, that are useful to depict bony structures, can provide an assessment of joint space width (JSW) for OA screening and diagnosis [21]. However, JSW assessment only provides an indirect measurement of the cartilage thickness through the distance between femur and tibia that demonstrates weak sensitivity to change [3]. Magnetic resonance imaging (MRI) allows a precise depiction of the entire joint structures, including the cartilage and its pathological changes [9]. In recent years, there have been considerable developments for cartilage segmentation to assess the cartilage thickness in three-dimensional (3-D) MRI images [6,7,8, 20, 22, 23, 29]. However, routine clinical use of MRI is limited due to high cost and low availability of the equipment [19].
Among other medical imaging modalities, ultrasound is considered to be non-invasive, radiation-free, portable, real-time, and cost-effective. As an affordable and widely accessible diagnostic tool compared to MRI, ultrasound imaging offers an excellent alternative to help diagnose the presence of the knee OA disease [17]. While MRI provides a comprehensive assessment of intra-articular structures, ultrasound imaging has a complementary role in the evaluation of extra-articular structures [13]. Although the visualization of deeper articular structure and subchondral bone is prevented by the nature of the sound, it has demonstrated its ability in depicting more appearance features of the knee OA than X-rays and computed tomography [1]. It has been used to measure the thickness and detect the degenerative change in the cartilage [2], in patients with knee pain [13], osteoarthritis and rheumatoid arthritis [11] where the cartilage thickness was measured manually by drawing a perpendicular line between hyperechoic lines of the soft tissue-cartilage interface and of the cartilage-bone interface (see Fig. 1) [13, 19].
Segmentation is a necessary task that has a significant influence on the accuracy of the thickness computation [8]. As in Fig. 1, the femoral condylar cartilage in ultrasound images is depicted as a monotonous hypoechoic band between the adjacent tissues and the smooth bony surface [13]. Hence, the goal in segmenting knee cartilage from the ultrasound images is to locate the boundaries between the two interfaces. Segmenting the cartilage from surrounding tissues is a challenging task due to the boundary between different tissues is not sufficiently distinct.
Due to physical constraint in the ultrasound image acquisition, speckle noise and intensity inhomogeneity occur in the ultrasound images, which tend to adversely affect the image contrast (see Fig. 1). While speckle noise appears as dense, bright and dark granular objects in close proximity throughout the image, intensity inhomogeneity causes a slowly changing intensity contrast where the same tissue region may exhibit various contrasts at several locations and the intensity distributions between different tissues are overlapped significantly. When spatial intensity variation caused by intensity inhomogeneity is considered and not the one caused by speckle, the problem of intensity inhomogeneity can be addressed similar to the intensity bias correction in MRI images [27]. It is applied retrospectively in the acquired images and also often combined with the segmentation methods, for which, several level set methods for joint segmentation and intensity bias correction have been proposed [15, 18, 26, 32]. Although developed for other imaging modality, these methods are suitable and can be adapted as a technique for locating the cartilage boundary in the ultrasound images. It has been explained in [27] that the underlying assumption of the multiplicative noise model is related to the ultrasound physics of image formation. While intensity-based segmentation techniques are generally insensitive to noise, the use of local image intensity and intensity bias estimation help to cope with intensity inhomogeneity.
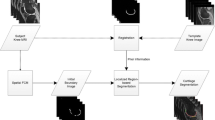
This paper presents segmentation and thickness computational approaches to the knee cartilage in two dimensional (2-D) ultrasound images as an initial step for assessment of the cartilage degeneration. A summary on the methodologies to obtain short-axis knee cartilage of the 2-D ultrasound images is given. The locally statistical level set method (LSLSM) applied in locating the cartilage boundary in a noisy and bias-corrupted image is explained. To ensure the thickness computation is performed in the cartilage region only, the cartilage region is then extracted where its surrounding tissue regions are removed from the final segmentation results. The normal distance as a technique for computing the cartilage thickness on the isolated cartilage region is described. Segmentation result and computational time of LSLSM are compared to other relevant level set methods in the attempt of segmenting the knee cartilage. Next, qualitative and quantitative evaluation of the segmentation performance of different level set methods over 80 data sets is illustrated using Cohen’s κ statistics, Dice similarity coefficient and Hausdorff distance metrics. The mean, standard deviation, and intraclass correlation coefficient are computed to determine the accuracy and reproducibility of the thickness computation on a set of segmented cartilage images. Bland-Altman plot is used to measure agreement between thickness measurement obtained manually and using normal distance. The methods described can be used to assess cartilage degeneration where changes in the cartilage thickness are quantified over time by comparing the true thickness at a certain time interval.
2 Materials and methods
2.1 Image acquisition
An ultrasound image acquisition protocol to capture the cartilage of the knee joint is described here. The Toshiba Aplio MX ultrasound machine is utilized with a 2-D linear array, 8-12 MHz multifrequency transducer (PLT-805AT). In order to obtain short-axis views of the femoral condylar cartilage on the trochlear notch, the subject were scanned in vivo in the supine position with the knee fully flexed (120∘). The transducer was placed transversely to the leg and perpendicular to the bone surface, just above the superior margin of the patella [13, 19]. Ten asymptomatic subjects (male, age range: 23-27 years, mean age: 24.75 ± 2.18) were recruited. The written consent was obtained prior to the ultrasound scanning. Four different scans of the cartilage were obtained from both left and right knee joints with repositioning of the ultrasound probe between acquisitions. The images were stored in DICOM format at a resolution of 0.1316 × 0.1316 mm. The musculoskeletal sonography was performed by a professional sonographer. The study received approval from the University of Malaya Medical Ethics Committee (MECID No. 20147-396).
2.2 Locally statistical level set method
Let I denote a given image defined on the image domain Ω. In the level set methods, a contour is represented by the zero level set of the level set function. Let ϕ : Ω →R be a level set function that takes positive and negative signs inside and outside the contour, which represent an image partition of the image domain Ω into two disjoint regions, respectively
The two regions Ω1 and Ω2 are accordingly represented with their membership functions defined by M 1(ϕ) = H(ϕ) and M 2(ϕ) = 1 − H(ϕ). The regularized Heaviside function H 𝜖 (ϕ) and its derivative, the smoothed Dirac delta function δ 𝜖 (ϕ) with 𝜖 = 1 [4], are computed by
The two-phase case of the statistical and variational multiphase level set method is considered [30, 32]. It is also called as the locally statistical level set method (LSLSM). The energy of LSLSM is derived based on Gaussian distributions of local image intensity and multiplicative noise model. The energy functions e i are defined in the first row of Eq. 4 and computed using the equivalent expression in the second and the third rows of Eq. 4.
where b, c i , and \({\sigma_{i}^{2}} \) for i = 1,2 are the bias field, the piecewise constant, and the variances, respectively. ∗ denotes the convolution operation. The function 1 K is defined as \(\int K(\mathbf {y-x}) d\mathbf {y}\) which is equal to constant 1 everywhere except near the image boundary. A truncated uniform kernel function K is defined by
with a positive constant a such that \(\int K(\mathbf {z})d\mathbf {z}=1\) and ρ is the kernel’s radius defined as the distance between the kernel center point x and other pixels y within the kernel support.
In order to reduce the overlapped intensity distribution in the image, only image intensities I(x) in a neighborhood of y are involved in the above energy functions whose size is controlled by the scale of the kernel function K. The small scale of the neighborhood enables to handle intensity inhomogeneity since the intensities involved are only in the local region at a certain scale centered at the point x [15].
The image intensities are approximated by spatially varying means and variances of Gaussian distributions. To accomplish joint segmentation and bias field correction, the means are approximated by multiplication of the bias field b and the true image signal within the window estimated by piecewise constants c i . The functions e i that represent an image partition and a bias field estimation are combined with membership function M i (ϕ) to be incorporated into the level set formulation. The total energy function of LSLSM is given by
with the first term being the regularization term to compute the arc length of the zero level set where its relative strength is controlled by the parameter ν.
By minimizing the energy function E, image segmentation and bias field estimation are jointly performed by estimating the membership functions M i (ϕ), the variances \({\sigma_{i}^{2}} \), the restored bias field b, and the constants c i . In the iterative process, the energy minimization with respect to each variable ϕ, c i , b, and σ i is obtained. These variables are derived from the equivalent expression of the energy functional in the form of convolution. The minimization problem is solved by iterating between two steps.
In step one, the variables {c i , b and σ i } are estimated. During the level set evolution, these variables are updated by minimizing the energy E with respect to one of the variables while others are fixed, respectively.
By substituting the functions e i in Eq. 4 into the energy functional E in Eq. 6, the derivative of E is taken with respect to c i for fixed ϕ, b, and σ i . Then, the resulting expressions are equated to zero, \(\frac {\partial E}{\partial c_{i}(\mathbf {x})}=0\). The optimal c i that minimizes E satisfy the following Euler-Lagrange equations:
From Eq. 7, the optimal constants c i approximating the image intensity in each region are given by
Keeping ϕ, c i and σ i fixed and minimizing the energy functional E with respect to b, and then equating the resulting expressions to zero, \(\frac {\partial E}{\partial b(\mathbf {y})}=0\), the optimal b that minimize E satisfy the following Euler-Lagrange equations:
From Eq. 9, the restored bias field b is obtained as follows
From the following Euler-Lagrange equations
The minimizer of the variances \({\sigma_{i}^{2}} \) is given by
In step two, assuming that all the variables c i , b, and σ i for i = 1,2 are known, and then the level set function ϕ is evolved, so that the energy functional E is minimized. Fixing c i , b, and σ i , the minimization of E(ϕ,c i ,b,σ i ) with respect to ϕ is obtained by solving the gradient flow equation \(\frac {\partial \phi }{\partial t}=-\frac {\partial E}{\partial \phi }\) where \(\frac {\partial E}{\partial \phi }\) is the Gâteaux derivative of the energy E. By calculus of variations, the Gâteaux derivative can be computed and the corresponding gradient flow equation is expressed as follows
To keep the level set evolution stable, for every iteration of Eq. 13, the level set function is diffused by [31]
where ϕ n is the level set function obtained from the n-th iteration of Eq. 13 and Δt represents the diffusion strength where Δ denotes the Laplacian operator.
2.3 Evaluation of segmentation accuracy
In order to evaluate the segmentation results, qualitative and quantitative segmentation assessments are performed using Cohen’s κ statistics and two validation metrics of Dice similarity coefficient (DSC) and Hausdorff distance (HD), respectively. Over the total data sets of 80 images, the manual outlines as ground truth data were compared against its isolated cartilage region obtained by the algorithm to be evaluated qualitatively and quantitatively.
2.3.1 Qualitative assessment
The overall segmentation quality of the cartilage anatomical structure in ultrasound images are validated by two experts. The segmentation quality of the cartilage area was assessed by differentiating the boundaries between the soft-tissue - cartilage interface and the cartilage - bone interface, which categorized as follows: Grade 1: excellent segmentation quality, with excellent differentiation of the boundaries between the two interfaces and no significant overlap segmentation areas. Grade 2: good segmentation quality, with good differentiation of the boundaries between the two interfaces and only small overlap segmentation areas. Grade 3: poor segmentation quality, with poor differentiation of the boundaries between the two interfaces and some overlap segmentation areas. Grade 4: bad segmentation quality, with bad differentiation of the boundaries between the two interfaces and large overlap segmentation areas.
Inter-observer agreement for the segmentation quality of the cartilage area obtained by LSLSM is expressed as the Cohen’s κ statistics [5]. The observed agreement, the chance agreement, and the κ statistics for agreement between two observers were calculated. The observed agreement is the number of occasions of complete agreement between observers divided by the total number of occasions. The chance agreement is the probability that the observers will provide the same response to an observation. The kappa coefficient is defined as the observed agreement which is above and beyond that due to chance. Different ranges for kappa values characterise the degrees of agreement. A kappa value of less than 0 implies poor agreement (agreement worse than that of chance alone), 0.00 to 0.20 slight agreement (agreement equal to that of chance alone), 0.21 to 0.40 fair agreement, 0.41 to 0.60 moderate agreement, 0.61 to 0.80 substantial agreement and 0.81 to 1.00 almost perfect agreement [14].
2.3.2 Quantitative assessment
Two different metrics adopted to examine the segmentation performance are DSC and HD metrics. DSC quantifies area similarity between two comparing segmentation regions.
where A and B are the pixel areas of the segmented and reference contours, respectively. DSC is defined as the ratio between twice of the common pixel area of A and B and the sum of the individual areas. This metric indicates the relative locations and sizes of the contour pair. The value is bounded in [0,1] and the value of 1 implies that the contour pair has the same location and size (or area).
HD, d H (X,Y ), is used to compute the boundary mismatch between two comparing segmented boundaries.
where X and Y are sets of points extracted from the segmented and reference contours of A and B, respectively. d H is defined as the maximum of the directed Hausdorff distances d(X,Y ) and d(Y,X), which are the largest rank between the point x ∈ X to the nearest point in Y and vice versa. The small value of d H implies a minimal shape difference between two comparing contours.
2.4 Cartilage thickness computation
Several computational approaches have been proposed for estimating the cartilage thickness in the 3-D MRI images [8, 20, 22, 23]. The thickness computation was performed on the 3-D cartilage surface obtained from a 3-D reconstruction of segmented sagittal knee cartilage slices or a direct 3-D MRI cartilage segmentation. Vertical distance provides z-directional distance between points on the upper and lower surfaces [10]. Proximity method computes the shortest distance from each point on a given surface to the closest point on the opposing surface [8, 16]. Another class of methods defines the thickness as the distance between two points where the normal vector in a central axis intersects the upper and lower surfaces [22]. Since the central axis are generated by an average of the two surfaces, it does not provide a true normal thickness [23]. In [28], the thickness is defined as the length of streamlines approaching the opposing boundary from a normal direction. Instead of using the streamlines, the normal distance [10] estimates the thickness from the length of surface normal vectors between the upper and lower surfaces [23]. It provides the true normal thickness from one surface to another that yielded the most accurate estimation.
In this work, the normal distance is adapted as a technique for estimating the cartilage thickness in short axis knee cartilage of the 2-D ultrasound images. It is used to compute the true normal thickness of the cartilage from one boundary to another. The thickness computation is performed by evenly spacing m points along the boundary, then taking the normal vector from the boundary points (x i ,y i ), and the perpendicular line is created from (x i ,y i ) to the intersection points of the upper or lower boundary of the cartilage. The distance between two points from the normal vector of the upper (or lower) boundary to the intersection of the line with the lower (or upper) boundary is used to compute the cartilage thickness t i for each point, respectively. The average thickness of the cartilage \(\bar {t}\) is taken from the mean of the thickness at all m boundary points, \(\bar {t}=\frac {1}{m} {\sum }_{i=1}^{m} t_{i}\).
3 Results
3.1 Comparison with other level set methods
In this section, a comparison of different level set methods in their attempt of segmenting a real knee cartilage ultrasound image is provided. A brief summary of the other two level set methods without and with multiplicative component estimation is given as follows
The first method is the local Gaussian distribution fitting (LGDF) model [25]. Similar to LSLSM, LGDF also assumes a Gaussian distribution with spatially locally varying mean and variance. However, LGDF can only be used for segmentation since it does not estimate multiplicative component as in LSLSM which can be used for joint segmentation and bias estimation. The energy functions e i of LGDF are written as
where
and
The second method is considered as the locally weighted K-means variational level set (WKVLS) [15]. WKVLS ignores the variance component that is taken into account in LSLSM which helps to distinguish different tissues more accurately. Similar to LSLSM, WKVLS is essentially designed to simultaneously estimate the multiplicative bias field while segmenting the images. The energy functions e i of WKVLS is given by
where
and
In this experiment, LSLSM was compared to the other two methods without and with multiplicative component estimation, i.e., LGDF and WKVLS, respectively. All the methods were implemented in MATLAB R2014a in an Intel (R) Xeon (R), 2.00 GHz, 32 GB RAM with the following parameter settings. Small kernel’s radius ρ = 5 was chosen to provide more accurate boundary location. For images with intensity range in [0,255], the parameter ν was set to 0.001 × 2552. It was chosen to be small when segmenting objects of any size. The time steps for level set evolution Δt 1 and for regularization Δt 2 were set as Δt 1 = 0.01 for LGDF, Δt 1 = 0.1 and Δt 2 = 0.1 for WKVLS, and Δt 1 = 0.01 and Δt 2 = 0.01 for LSLSM. Image size is 420 × 150 pixels.
Figure 2 illustrates segmentation results of the three related level set methods that were applied for the cartilage boundary segmentation. The initial contour and the final contours are coloured in red and green, respectively. The initial contour is in circle shape with 10 pixels radius and placed just around the center of the images. These three segmentation algorithms were generally able to distinguish different tissues in the presence of noise and intensity inhomogeneity. This is due to the use of local image intensity defined in a local neighborhood that suppresses the overlapping intensity distribution. With the multiplicative component estimation, WKVLS and LSLSM were able to reduce the non-uniform textures and then locate the boundaries between different tissues correctly as seen in Fig. 2c, d. Without the multiplicative component estimation, LGDF produced unstable segmentation result where misclassified contours inside the object and some unnecessary contours around the object appear in the final contours in Fig. 2b. Both models yield satisfying segmentation results, while LSLSM that considers the variance component obtained a better segmentation result than WKVLS.
DSC and HD were computed from the manual outline and the isolated cartilage region as depicted in Fig. 3. The cartilage region was isolated from its surrounding tissues in the final contours. Connected-component labeling was used to remove other tissue regions in the binary image. This is to ensure that the validation metrics of the DSC and HD are computed based on the cartilage region and not affected by segmentation of the surrounding tissues. DSC and HD values for the segmentation results of LGDF, WKVLS, and LSLSM in Fig. 2b, c, d are summarized in the first, second, and third rows of the matrices \(\left [\begin {array}{l} 0.9027\\0.9148\\0.9423 \end {array} \right ]\) and \(\left [\begin {array}{l}6.8557\\7\\6.3246 \end {array} \right ]\), respectively. DSC value obtained by LSLSM is higher than the ones obtained by WKVLS and LGDF while HD value obtained by LSLSM is smaller than the ones obtained by WKVLS and LGDF. In addition, the total computational time of LGDF, WKVLS, and LSLSM required for 500 iterations are 54.82, 13.77, and 12.97 seconds, respectively.
3.2 Knee cartilage ultrasound image segmentation
In this subsection, the three different level set methods were applied to segment a set of 80 real knee cartilage ultrasound images. The data sets comprise of four repeated scans of the cartilage imaged from both left and right knee joints of the ten subjects. Figure 4 depicts a subset of 10 segmentation outcomes of LSLSM for left and right knee cartilages from a subset of five subjects, respectively. Segmentation results of LGDF, WKVLS, and LSLSM were evaluated qualitatively and quantitatively over the total data sets of 80 images. While the qualitative segmentation assessment is performed using Cohen’s κ statistics, the quantitative segmentation assessment is performed using validation metrics of DSC and HD. The manual outlines as ground truth data were compared against its isolated cartilage region obtained by the algorithm to be evaluated qualitatively and quantitatively. An expert segmented the cartilage manually from each ultrasound image scan. The isolated cartilage region as in Fig. 3 was extracted from its surrounding tissues in the final contours using connected-component labeling.
Left and right columns represent segmentation results obtained by LSLSM for left and right knee cartilages of a subset of five subjects, respectively. The initial contours are represented by the red circles with 10 pixels radius and placed just around the center of the image. The green lines represent the final segmentation contours
The number of observed agreements is 67 images (83.75% of the observations), as grade 1 (excellent) in 39 images (48.75%), as grade 2 (good) in 21 images (26.25%), as grade 3 (poor) in 5 images (6.25%), as grade 4 (bad) in 2 images (2.5%). The number of agreement due to chance is 32.05 images. An overall segmentation quality of the cartilage areas for all 10 subjects rated by two experts indicates a substantial agreement with κ = 0.73.
Figure 5 illustrates segmentation performance of LGDF, WKVLS, and LSLSM validated using DSC and HD values over the total data sets of 80 images. Figure 5a shows the computed DSC values for 80 images fall in the range between 0.84 and 0.94, 0.29 and 0.95, and 0.82 and 0.95 for LGDF, WKVLS, and LSLSM, respectively. The higher value of DSC indicates that the two comparing contours have a good agreement in size and location which correspond to more accurate segmentation results. The computed HD values in Fig. 5b are ranging from 4.47 to 8.83, 5.39 to 19.10, and 4.69 to 8.25 pixels for LGDF, WKVLS, and LSLSM, respectively. The smaller HD values correspond to the least difference in shape between two comparing contours.
The means, standard deviations, and p values of DSC and HD values for all methods over 80 data sets are summarized in Table 1. It indicates that LSLSM obtained higher average value of DSC for all available data sets than LGDF and WKVLS. Meanwhile, the average value of HD obtained by LSLSM is smaller than obtained by LGDF and WKVLS. It implies that LSLSM provides a good area similarity and a minimally different contour shape which illustrates the satisfactory segmentation outcomes for all available data sets. In addition, p values in Table 1 indicate that LGDF and LSLSM have means significantly different from WKVLS (p < 0.0001). Meanwhile, it is found that the mean of LGDF is not statistically significant from LSLSM where p-values for DSC and HD metrics are 0.69 and 0.27, respectively.
Segmentation errors were mainly due to the overlapped intensity distribution between different tissues. The boundary between different tissues is not sufficiently distinct, particularly around the interfaces of soft tissue-cartilage and cartilage-bone. The variances of the Gaussian distributions that are taken into account in LGDF and LSLSM helps to distinguish the two interfaces more satisfactorily. Although WKVLS considers the multiplicative component, it does not take into account the variance component, thus tends to misclassify the two interfaces. In addition, the degree of inhomogeneity is varied between the scanned images. These may be the cause of the less satisfactory segmentation performance indicated by DSC values below 0.8 and HD values above 7 pixels in the graphs.
3.3 Cartilage thickness measurement
In this subsection, the cartilage thickness is computed based on the segmented images using the normal distance. The cartilage images were acquired four times each from both left and right knee joints of the ten subjects as described in Section 2.1. The cartilage images were firstly segmented using LSLSM described in Section 2.2. Connected-component labelling was used to remove surrounding tissue regions from the final contour results. An example of an isolated cartilage region extracted from the segmentation outcome is illustrated in Fig. 3. Using this isolated cartilage ensures the thickness measurements are performed in the cartilage region only. The cartilage thickness was calculated over 80 image data sets using the normal distance as described in Section 2.4. The true thickness of the cartilage is estimated by the length of boundary normal vectors between the upper and lower boundaries of the cartilage [22, 23] as depicted in Fig. 6b, c, d, and e. The measurements were made at every pixel on its upper and lower boundaries.
The obtained measurements of the cartilage thickness ranged from 1.35 to 2.72 mm, 1.36 to 2.45 mm, 1.33 to 2.17 mm, 1.68 to 2.39 mm, and 1.35 to 2.42 mm for manual thickness measurement, the normal distance in the cartilage area segmented by the manual outline, LGDF, WKVLS, and LSLSM, respectively. It reflects the robustness of the segmentation algorithms to various cartilage thickness. The statistics such as mean, standard deviation, and the intraclass correlation coefficient (ICC) in Table 2 were computed to determine the accuracy and reproducibility of the cartilage thickness computation using the normal distance. ICC values were determined from the thickness measurements of the four repeated scans for all methods. Higher value of ICC indicates a good reproducibility between the measurement sets.
From Table 2, it can be seen that the thickness measurement obtained manually is found more reproducible than the ones computed by the normal distance. Moreover, the thickness computation based on the manual segmentation results are found to be more reproducible than the ones based on the segmentation results obtained by the algorithms. The thickness computation obtained by LSLSM is found more reproducible than LGDF and WKVLS. This is attributed to segmentation accuracy obtained by LSLSM that is higher than LGDF and WKVLS. The variations of the intensity inhomogeneity across all data sets make the segmentation algorithms difficult to accurately capture each cartilage boundary and causes the less satisfactory segmentation results. Meanwhile, the thickness measurement and the segmentation results that are obtained manually are less affected by the variations in the intensity inhomogeneity for all data sets.
The cartilage thickness computed by the normal distance on the cartilage area segmented by the manual outline, LGDF, WKVLS, and LSLSM was compared to the results obtained by the manual measurement using Bland-Altman plot. The manual measurement is provided by drawing the perpendicular line between the hyper-echoic lines at the soft tissue-cartilage interface and at the cartilage-bone interface [13, 19]. In each knee joint, three separate measurements were performed at three locations, i.e., the trochlear notch, two-thirds lateral (two-thirds of the distance from the trochlear notch to the convexity of the lateral trochlea), and two-thirds medial (two-thirds of the distance from the trochlear notch to the convexity of the medial trochlea) as illustrated in Fig. 6a. The average value is taken from the manual measurement at the three locations.
Bland-Altman plots depicted in Fig. 7a, b, c, and d are used to illustrate an agreement of the cartilage thickness obtained by two measurement methods. The mean differences for all pairs of the thickness measurements were 0.02 ± 0.17, 0.19 ± 0.20, − 0.04 ± 0.22, and 0.05 ± 0.18 mm for the manual outline, LGDF, WKVLS, and LSLSM, respectively. Small mean difference indicates no significant bias for both comparing methods. In addition, p values for the manual outline, LGDF, WKVLS, and LSLSM are 0.70, < 0.0001, 0.20, and 0.19, respectively. There was no significant difference in the thickness measurements obtained manually and by the normal distance in the cartilage area segmented by the manual outline, WKVLS, and LSLSM. Meanwhile, the measurement result by the normal distance in the cartilage area segmented by LGDF is significantly different from the manual measurement result. It can also be observed that nearly all differences between measurements by the two methods lie within the 95% limit of agreement, i.e., 0.34 to − 0.31, 0.59 to − 0.20, 0.39 to − 0.48, and 0.39 to − 0.29 mm for the manual outline, LGDF, WKVLS, and LSLSM, respectively. Meanwhile, there were only several differences between measurements by both compared methods that fall outside the limits of agreement.
4 Discussion
The cartilage boundary segmentation and thickness computation in short axis knee cartilage of the 2-D ultrasound images pose a considerable challenge and clinical value. This paper studies cartilage boundary segmentation using LSLSM with the energy functional and the variables expressed in the equivalent expression of the convolution operation. The cartilage thickness is automatically computed by averaging the normal distances along the segmented cartilage area. Comparison experiment on several level set methods in segmenting the knee cartilage ultrasound image shows that LSLSM yielded a more satisfactory result than other methods. Cohen’s κ coefficient indicates a substantial agreement of the cartilage segmentation quality rated by two experts. DSC and HD metrics computed from a set of the knee cartilage ultrasound images show a very good and consistent segmentation outcome. ICC value computed from the thickness measurements of the four repeated scans indicates a good reproducibility of the thickness measurements. Bland-Altman plot demonstrates a good agreement between the measurements by the normal distance and the manual measurement.
The cartilage boundary segmentation possible with LSLSM has allowed the segmentation results to be used for making computation of the cartilage thickness in the 2-D ultrasound images. The obtained results show the accuracy and reproducibility of the segmentation and thickness estimation methods. The methods described in this work are useful to characterize the normal cartilage and can also be applied to delineate the degenerative diseases such as osteoarthritis. It can be used to assess cartilage degeneration typically seen as the cartilage thinning. Changes in the cartilage thickness can be quantified over time by comparing the true thickness at a certain time interval, i.e., six months. The robustness in segmenting and computing various cartilage thickness demonstrated in this work indicates a potential application of the methods for the assessment of the cartilage degeneration, which is left for future work.
The incorporation of the joint shape-intensity prior constraint in [24] to LSLSM could increase the robustness to capture shape and thickness variations in the cartilage. The inclusion of different grades of the cartilage degeneration to investigate the performance of the segmentation and thickness computational techniques to various degeneration progression in the cartilage is also of interest for future work. Future improvement also includes reducing or eliminating two user interactions involved in this work, i.e., the contour initialization and the extraction of the cartilage region from surrounding tissues.
Since the short-axis view of the knee cartilage in the 2-D ultrasound image is seen as a projected image plane from the real 3-D MRI volume of the knee joint, the distance between the cartilage interfaces in the 2-D ultrasound images may not be seen to be perpendicular between upper and lower 3-D cartilage surfaces. Therefore, spatial registration of the 2-D ultrasound image plane to the 3-D MRI volume to provide a precise relation of the thickness computation on both imaging modalities could also guide future work.
5 Conclusion
The knee cartilage boundary segmentation using LSLSM and thickness computation using the normal distance in short axis knee cartilage of the 2-D ultrasound images have been presented. The energy functional derived from Gaussian distributions of local image intensity and multiplicative noise model allowed LSLSM to cope with speckle noise and intensity bias thus capture the monotonous hypoechoic band between the two interfaces of the soft tissue-cartilage and the cartilage-bone that represents the cartilage region. The cartilage thickness is then computed by averaging the normal distances along the segmented cartilage area. The knee cartilage boundary segmentation possible using LSLSM has allowed the obtained segmentation results to be used for computing the cartilage thickness in the 2-D ultrasound images. The robustness in segmenting and computing cartilage of various thickness demonstrated in this work indicates a potential application of the methods for the assessment of the cartilage degeneration where the changes in the cartilage thickness can be quantified over time by comparing the true thickness at a certain time interval.
References
Abraham AM, Goff I, Pearce MS, Francis RM, Birrell F (2011) Reliability and validity of ultrasound imaging of features of knee osteoarthritis in the community. BMC Muscuskelet Disord 70(12):1471–2474
Aisen AM, McCune WJ, MacGuire A, Carson PL, Silver TM, Jafri SZ, Martel W (1984) Sonographic evaluation of the cartilage of the knee. Radiology 153(3):781–784
Buckland-Wright JC (1994) Quantitative radiography of osteoarthritis. Ann Rheum Dis 53:268–275
Chan TF, Vese LA (2001) Active contours without edges. IEEE Trans Image Process 10(2):266–277
Cohen J (1960) A coefficient of agreement for nominal scales. Educ Psychol Meas 20(1):37–46
Dodin P, Pelletier JP, Martel-Pelletier J, Abram F (2010) Automatic human knee cartilage segmentation from 3-D magnetic resonance images. IEEE Trans Biomed Eng 57(11):2699–2711
Folkesson J, Dam EB, Olsen OF, Pettersen PC, Christiansen C (2007) Segmenting articular cartilage automatically using a voxel classification approach. IEEE Trans Biomed Eng 26(1):106–115
Fripp J, Crozier S, Warfield SK, Ourselin S (2010) Automatic segmentation and quantitative analysis of the articular cartilages from magnetic resonance images of the knee. IEEE Trans Med Imag 29(1):55–63
Graichen H, Eisenhart-Rothe R, Vogl T, Englmeier KH, Eckstein F (2004) Quantitative assessment of cartilage status in osteoarthritis by quantitative magnetic resonance imaging. Arthritis Rheum 50:811–816
Heuer F, Sommer M, III JBR, Bottlang M (2001) Estimation of cartilage thickness from joint surface scans: comparative analysis of computational methods. In: Proceedings 2001 ASME bioengineering Conference, vol 50, pp 569–570
Iagnocco A, Coari G, Zoppini A (1992) Sonographic evaluation of femoral condylar cartilage in osteoarthritis and rheumatoid arthritis. Scand J Rheumatol 21(4):201–203
Jackson D, Simon T, Aberman H (2001) Symptomatic articular cartilage degeneration: the impact in the new millennium. Clin Orthop Relat Res 391:14–15
Kazam JK, Nazarian LN, Miller TT, Sofka CM, Parker L, Adler RS (2011) Sonographic evaluation of femoral trochlear cartilage in patients with knee pain. J Ultrasound Med 30(6):797–802
Landis JR, Koch GG (1977) The measurement of observer agreement for categorical data. Biometrics 33 (1):159–174
Li C, Huang R, Ding Z, Gatenby C, Metaxas DN, Gore JC (2011) A level set method for image segmentation in the presence of intensity inhomogeneities with application to MRI. IEEE Trans Image Process 20 (7):2007–2016
Maurer CR, Qi R, Raghavan V (2003) A linear time algorithm for computing exact Euclidean distance transforms of binary images in arbitrary dimensions. IEEE Trans Pattern Anal Mach Intell 25(2):265–270
Moller I, Bong D, Naredo E, Filippucci E, Carrasco I, Moragues C, Iagnocco A (2008) Ultrasound in the study and monitoring of osteoarthritis. Osteoarthr Cartil 16:S4–S7
Mukherjee S, Acton ST (2015) Region based segmentation in presence of intensity inhomogeneity using legendre polynomials. IEEE Signal Process Lett 22(3):298–302
Naredo E, Acebes C, Moller I, Canillas F, de Agustin JJ, de Miguel E, Filippucci E, Iagnocco A, Moragues C, Tuneu R, Uson J, Garrido J, Delgado-Baeza E, Saenz-Navarro I (2009) Ultrasound validity in the measurement of knee cartilage thickness. Ann Rheum Dis 68:1322–1327
Pakin SK, Tamez-Pena JG, Totterman S, Parker KJ (2002) Segmentation, surface extraction, and thickness computation of articular cartilage. In: Proceedings SPIE Medical Imaging 2002: Image Processing, San Diego, USA, vol 4686, pp 155–166
Roemer FW, Crema MD, Trattnig S, Guermazi A (2011) Advances in imaging of osteoarthritis and cartilage. Radiology 260(2):332–354
Solloway S, Hutchison C, Waterton J, Taylor C (1993) The use of active shape models for making thickness measurements of articular cartilage from MR images. Magn Reson Med 13(5):943–952
Tang J, Millington S, Acton ST, Crandall J, Hurwitz S (2006) Surface extraction and thickness measurement of the articular cartilage from MR images using directional gradient vector flow snakes. IEEE Trans Biomed Eng 53(5):896–907
Wang J, Cheng Y, Guo C, Wang Y, Tamura S (2016) Shape-intensity prior level set combining probabilistic atlas and probability map constrains for automatic liver segmentation from abdominal CT images. Int J Comput Assist Radiol Surg 11(5):817–826
Wang L, He L, Mishra A, Li C (2009) Active contours driven by local Gaussian distribution fitting energy. Signal Process 89(12):2435–2447
Wang XF, Min H, Zou L, Zhang YG (2015) A novel level set method for image segmentation by incorporating local statistical analysis and global similarity measurement. Pattern Recogn 48(1):189–204
Xiao G, Brady M, Noble JA, Zhang Y (2002) Segmentation of ultrasound B-mode images with intensity inhomogeneity correction. IEEE Trans Med Imag 21(1):48–57
Yezzi A, Prince J (2003) An eulerian PDE approach for computing tissue thickness. IEEE Trans Med Imag 22(10):1332–1339
Yin Y, Zhang X, Williams R, Wu X, Anderson DD, Sonka M (2010) LOGISMOS–Layered optimal graph image segmentation of multiple objects and surfaces: cartilage segmentation in the knee joint. IEEE Trans Med Imag 29(12):2023–2037
Zhang K, Zhang L, Zhang S (2010) A variational multiphase level set approach to simultaneous segmentation and bias correction. In: Proceedings IEEE International Conference Image Processing, Hong Kong, pp 4105–4108
Zhang K, Zhang L, Song H, Zhang D (2013) Reinitialization-free level set evolution via reaction diffusion. IEEE Trans Image Process 22(1):258–271
Zhang K, Zhang L, Lam KM, Zhang D (2016) A level set approach to image segmentation with intensity inhomogeneity. IEEE Trans Cybern 46(2):546–557
Acknowledgements
This work was supported in part by the University of Malaya Research Grant (RP020A-13AET), in part by the International Graduate Research Assistantship Scheme, and in part by the Postgraduate Research Grant (PG003-2014B).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Faisal, A., Ng, SC., Goh, SL. et al. Knee cartilage segmentation and thickness computation from ultrasound images. Med Biol Eng Comput 56, 657–669 (2018). https://doi.org/10.1007/s11517-017-1710-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-017-1710-2