Abstract
Biochar has been widely accepted as a soil amendment to improve nitrogen (N) use efficiency, but the effect of biochar on N transformation metabolic pathways is unclear. A field experiment was conducted to evaluate the effect of biochar on N transformation in drip-irrigated cotton field. Four treatments were set as (1) no N fertilization (CK), (2) N fertilizer application at 300 kg ha−1 (N300), (3) N fertilizer application plus cotton straw (N300+ST), and (4) N fertilizer application plus cotton straw–derived biochar (N300+BC). Result showed that soil total N in N300+ST and N300+BC was 16.3% and 24.9% higher than that in N300, respectively. Compared with N300+ST, the nitrate N (NO3−-N) in N300+BC was significantly increased. Acidolyzable N and non-acidolyzable N in N300+ST and N300+BC were higher than those in CK and N300, while N300+BC performed better than N300+ST. Furthermore, the N fertilizer use efficiency of cotton in N300+ST and N300+BC was 15.1% and 23.2% higher than that in N300, respectively. Both N fertilizer incorporations with straw and biochar significantly altered the microbial community structures and N metabolic pathways. Genes related to denitrification and nitrate reduction in N300+ST were higher than those in N300, and N300+BC significantly increased nitrification and glutamate synthesis genes. Therefore, N fertilizer application plus cotton straw–derived biochar changed the microbial community composition, increased nitrification and glutamate synthesis enzyme genes which were beneficial to the accumulation of soil N content, and improved soil N retention capacity thus to increase N fertilizer use efficiency.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Nitrogen is an important nutrient element for crop growth and yield improvement. Soil N includes inorganic N and organic N. Inorganic N is the main source of nutrient uptake by crops, which directly affects the growth and morphogenesis of crops (Raven et al. 2004). Organic N is the main form of soil N, accounting for more than 90% of total N. Most of the organic N is unavailable to plants, but it serves both as an important mineralization substrate and a source of N to a variety of plants in ecosystems (Kielland et al. 2007; Xu et al. 2003). Soil N cycle is a collection of important biogeochemical pathways, including N mineralization, immobilization, and various redox reactions (Gruber and Galloway 2008).
Biochar has a large surface area, high hydrophilicity, and high cation exchange capacity, which is conducive to soil water conservation and prevention of nutrient loss (Lehmann et al. 2011; Ajayi et al. 2016; Liu et al. 2016). Many studies have shown that biochar addition increased soil nitrogen and promoted N uptake and N fertilizer use efficiency (NUE) of crops (Deluca et al. 2006; Huang et al. 2014; Piash et al. 2019). Therefore, biochar play an important role in soil N cycle (Bi et al. 2018).
Soil N transformation is mainly driven by soil microbial community. Previous study found that biochar induced shifts in the composition and functional genes of soil microbial community (Bai et al. 2015). Biochar is conducive to the survival of microbes, thus promoting the transformation of soil N (Liao et al. 2016). The effect of biochar on microbial-mediated N-cycling processes (including nitrification, denitrification, and N fixation) has been previously investigated in several systems (Ducey et al. 2013; Anderson et al. 2014; Bai et al. 2015; Liu et al. 2017). Biochar addition improves the abundance and activity of nitrifying bacteria (such as Nitrospirae) and stimulates potential nitrification rates (Ball et al. 2010; Prommer et al. 2014; Sorrenti et al. 2017). It has been also reported that biochar addition affects the denitrifying bacteria community structure (Zhang et al. 2017). High pyrolysis temperature biochars significantly affected the abundance of narG and nosZ genes (Zhong et al. 2018). Wang et al. (2018a) found that the highest abundance of the nifH gene was observed in the biochar-amended soils. Previous studies on the N transform from the perspective of genes mainly focused on specific N-transforming gene families (Tu et al. 2017). However, the direction and magnitude of the effect of biochar on multiple N-transform process gene abundance are still poorly understood.
The evaluation of microbial community structure and metabolic characteristics of N-transforming microorganisms is necessary to elucidate the mechanism of soil N transformation caused by biochar addition. In order to know the effect of biochar on soil N transformation mechanism in drip-irrigated cotton field, we investigated the effects of cotton straw– and straw-derived biochar on (1) soil N nutrient and cotton N uptake, (2) composition of microbial community, and (3) N metabolic pathways using high-throughput metagenomic sequencing technology. We hypothesized that cotton straw–derived biochar changed the composition of soil microbial community, increased the microbial assimilation of N, and improved soil N retention thus to increase N uptake of cotton.
Materials and methods
Experiment site and biochar preparation
The field experiment was conducted from 2014 to 2018, at an agricultural experimental station of Shihezi University (44° 18′ N, 86° 02′ E), Shihezi, Xinjiang province of China. The region is classified as a temperate arid zone with a continental climate. The soil is a gray desert soil (calcaric fluvisol in the FAO/UNESCO System), and the texture is loam. The topsoil (0–20 cm) properties at the beginning of the study were as follows: pH 8.58, organic matter (OM) 12.2 g kg−1, total nitrogen (TN) 0.98 g kg−1, ammonium nitrogen (NH4+-N) 1.7 mg kg−1, nitrate nitrogen (NO3−-N) 6.9 mg kg−1, available phosphorus (AP) 16.4 mg kg−1, and available potassium (AK) 364 mg kg−1. The biochar was prepared by pyrolysis of cotton straw in a muffle oven at 450 °C during 6 h. The cotton (Gossypium hirsutum L.) cultivar used in this study was Xinluzao 61.
Experiment design
According to the management of N fertilization, field experiment was set up as 4 treatments: (1) no N fertilization (CK), (2) N fertilizer application at 300 kg ha−1 (N300), (3) N fertilizer application plus cotton straw (N300+ST), and (4) N fertilizer application plus cotton straw biochar (N300+BC). In this experiment, the amount of straw application was 6 t ha−1, and biochar was 3.7 t ha−1 (the dose of organic carbon addition of biochar was equivalent to straw returning to the field). The treatments were arranged in a randomized complete block design with three replications. A total of 12 plots were established in 2014, and the size was 10 m × 6 m. Our study was conducted in 2018.
Every year after the cotton harvest, the straw was all pulled out. After drying and crushing, cotton straw and biochar were spread evenly on the ground and plowed into the 0–20-cm soil depth before sowing. In each plot, there were three drip irrigation lines and six rows of cotton plants, and the row spacing was 66 cm + 10 cm + 66 cm + 10 cm + 66 cm + 10 cm. The cotton plants were sown at 10-cm intervals within each row to obtain a population of 263,000 plants ha−1. This planting pattern is commonly used for drip-irrigated cotton in the region. Cotton was sown in late April and harvested in mid-October each year. A flow meter was installed in each plot to control the irrigation amount. The plots were all drip-irrigated with 45 mm of water at sowing to improve germination and seedling establishment. During the growing season of cotton, 9 times of irrigation were conducted, and the amount of irrigation was 450 mm. Nitrogen fertilizer (urea) was applied through the drip irrigation system. A total of 360-kg N ha−1 was applied with six applications from early June to early August. P and K fertilizers were used as the base fertilizer with P2O5 105 kg ha−1 and K2O 75 kg ha−1 before sowing, respectively.
Sample collection
Topsoil samples (0–20-cm depth) were collected on July 25, 2018. The soil samples were packed with ice packs and immediately brought back to the laboratory. A portion of each sample was used to determine the N content of soil. The remainder of the soil was stored at − 80 °C through a 2-mm sieve for high-throughput sequencing analysis.
At the boll-opening stage (August 20, 2018), the cotton plants were cut at the soil surface and washed with deionized water, which were divided into three parts: stems, leaves, and bolls. After being killed out at 105 °C for 30 min, plant samples were dried at 75 °C to constant weight. The samples were smashed and screened through a 1-mm sieve to determine the N concentration of plants.
Soil and plant N analyses
The soil TN was measured by using the semi-micro-Kjeldahl technique (Bremner and Mulvaney 1982). The contents of NH4+-N and NO3−-N were extracted from the soil by shaking samples in 2 mol L−1 KCl solution (soil:liquid ratio 1:10) for 1 h and then filtered. The NO3−-N and NH4+-N concentrations in the extracts were measured using an auto-discrete analyzer system (Smartchem 140, AMS Company, Italy). Soil organic N was determined by using an improved Bremner method with sealed-tube hydrolysis. Soil hydrolysate was prepared by using the 6 mol L−1 HCl sealed-tube hydrolysis method, and total acidolytic N was determined by using the acid-hydrolysate digestion–Kjeldahl method. Non-acidolytic N was calculated by difference subtraction, non-acidolytic N = total N − total acidolytic N (Stevenson 1982).
Plant N concentrations of each sample were measured using an auto-Kjeldahl system (B-339, Buchi Labortechnik AG, Switzerland). The NUE of cotton was calculated as follows:
Soil DNA extraction and metagenome sequencing
Total microbial genomic DNA samples were extracted using the DNeasy PowerSoil Kit (Qiagen, Inc., Netherlands), following the manufacturer’s instructions, and stored at − 20 °C prior to further assessment. The quantity and quality of extracted DNAs were measured using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and agarose gel electrophoresis, respectively. The extracted microbial DNA was processed to construct metagenome shotgun sequencing libraries with insert sizes of 400 bp by using the Illumina TruSeq Nano DNA LT Library Preparation Kit. Each library was sequenced by using the Illumina HiSeq X-ten platform (Illumina, USA) with the PE150 strategy at Personal Biotechnology Co., Ltd. (Shanghai, China).
Raw sequencing reads were processed to obtain quality-filtered reads for further analysis. First, sequencing adapters were removed from sequencing reads using Cutadapt (v1.2.1) (Martin 2011). Secondly, low-quality reads were trimmed by using a sliding window algorithm. Thirdly, reads were aligned to the host genome using BWA (http://bio-bwa.sourceforge.net/) (Li and Durbin 2009) to remove host contamination. Once quality-filtered reads were obtained, they were de novo assembled to construct the metagenome for each sample by using IDBA-UD (iterative De Bruijn graph assembler for sequencing data with highly uneven depth) (Peng et al. 2012). All coding regions (CDS) of metagenomic scaffolds longer than 300 bp were predicted by using MetaGeneMark (http://exon.gatech.edu/GeneMark/metagenome) (Zhu et al. 2010). CDS sequences of all samples were clustered by CD-HIT (Fu et al. 2012) at 90% protein sequence identity, to obtain non-redundant gene catalog. Gene abundance in each sample was estimated by using soap.coverage (http://soap.genomics.org.cn/) based on the number of aligned reads. The best hit to the NCBI microbial taxonomy database of the non-redundant genes was obtained by aligning them against the NCBI-nr database by BLASTN (e value < 0.001). The functional profiles of the non-redundant genes were obtained by annotating against the eggNOG (Evolutionary Genealogy of Genes: Non-supervised Orthologous Groups, Version 4.0) protein databases, by using the DIAMOND (Buchfink et al. 2015) alignment algorithm. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotation was conducted using BLAST search against the KEGG database. The metagenomic raw reads were submitted to the NCBI Sequence Read Archive under the accession number PRJNA634262.
Statistical analysis
The changes in soil N, plant N uptake, microbial community composition, and key gene abundance were tested for difference among sites with one-way ANOVA using the SPSS 21.0 statistical software. Tukey’s method was used for multiple comparisons of the significance among different treatments (P < 0.05). To illustrate the clustering of different samples and to further determine the microbial community composition and N transform function key genes, heatmap was performed using the vegan package in R.
Results
Plant N uptake and NUE
Compared with CK, all of the N fertilization treatments (N300, N300+ST, N300+BC) significantly increased the N uptake of plants (Table 1). The total N uptakes of cotton in the N300+ST and N300+BC treatments were 15.1% and 23.2% higher than that of N300, respectively. Compared with N300, the N uptakes of stems, leaves, and bolls in N300+BC significantly increased, while only N uptakes in stems and bolls were significantly promoted in N300+ST.
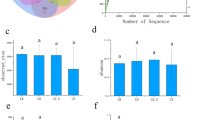
The effects of cotton straw or biochar addition on NUE of cotton are summarized in Fig. 1. NUE in the N300+ST treatment was 15.1% greater than that in the N300 treatment, and the NUE in the N300+BC treatment was increased by 23.2%. However, there was no significant difference between N300+ST and N300+BC treatments.
N fertilizer use efficiency (NUE) of cotton as affected by different treatments. Error bars represent standard error (SD; n = 3). Bars with different letters are significantly different at P < 0.05. CK, no amendment; N300, N fertilizer application at 300 kg ha–1; N300+ST, N fertilizer application plus cotton straw; N300+BC, N fertilizer application plus cotton straw biochar
Soil N content
There was no significant difference for soil TN between the N300 and CK treatments. The N300+ST and N300+BC treatments significantly improved TN, which represented 16.3% and 24.9% higher than the N300 treatment, respectively (Fig. 2).
Soil total N (TN) as affected by different treatments. Error bars represent standard error (SD; n = 3). Bars with different letters are significantly different at P < 0.05. CK, no amendment; N300, N fertilizer application at 300 kg ha–1; N300+ST, N fertilizer application plus cotton straw; N300+BC, N fertilizer application plus cotton straw biochar
Compared with CK and N300, both straw and biochar incorporation significantly increased the soil inorganic N (NH4+-N and NO3−-N) (Fig. 3). There was no significant difference for NH4+-N concentration between N300+ST and N300+BC treatments, while the NO3−-N concentration in the N300+BC treatment was 20.7% greater than that in the N300+ST treatment.
Concentrations of soil NO3–-N (a) and NH4+-N (b) as affected by different treatments. Error bars represent standard error (SD; n = 3). Bars with different letters are significantly different at P < 0.05. CK, no amendment; N300, N fertilizer application at 300 kg ha–1; N300+ST, N fertilizer application plus cotton straw; N300+BC, N fertilizer application plus cotton straw biochar
The N300+BC treatment had the highest concentrations of acidolyzable N (AN) and non-acidolyzable N (NAN) in soil, and then the N300+ST treatment, while there were no significant differences between N300 and CK treatments (Fig. 4). Compared with N300, the AN in the N300+ST and N300+BC treatments was increased by 16.5% and 22.7%, and the NAN was increased by 16.1% and 27.8%, respectively.
Concentration of soil acidolyzable N (a) and non-acidolyzable N (b) as affected by different treatments. Error bars represent standard error (SD; n = 3). Bars with different letters are significantly different at P < 0.05. CK, no amendment; N300, N fertilizer application at 300 kg ha–1; N300+ST, N fertilizer application plus cotton straw; N300+BC, N fertilizer application plus cotton straw biochar
Overview of metagenomic sequencing
We obtained 69.9 GB of clean reads from the metagenomic sequencing, and a total of 72–82 million raw sequences per sample after quality control. We gained 73,979, 70,249, 91,533, and 69,599 contigs (> 1 kb) in CK, N300, N300+ST, and N300+BC, respectively (Table 2). To determine whether the current sequencing depth is sufficient to reflect the microbial species composition of the sample, rarefaction analysis was performed (Fig. 5). The curve in each group was near saturation, which suggested that the sequencing data were great enough with very few new species undetected.
Soil microbial community composition
BLASTN comparison was performed for the Scaftigs sequences of each sample in the NCBI-nr database; we could obtain the taxonomy annotation and abundance of the species. Microorganisms in each treatment were mainly bacteria, accounting for 96.73–97.26% (Table 3), then were archaea and eukaryotes, accounting for 0.67–1.19% and 0.26–0.31%, respectively. The relative abundance of bacteria in N300 was less than that in CK, while both archaea and eukaryotes were significantly increased. Compared with N300, N300+ST significantly increased the relative abundance of bacteria, and the relative abundance of archaea decreased. In contrast, both archaea and eukaryotes were reduced in N300+BC.
The effects of biochar addition on soil microbial community composition in drip-irrigated cotton field were investigated at the phylum and order levels. Proteobacteria, Actinobacteria, Gemmatimonadetes, Acidobacteria, Planctomycetes, Bacteroidetes, Verrucomicrobia, Firmicutes, Chloroflexi, and Nitrospirae were the top 10 species of soil bacterial communities at the phylum level, accounting for 86.9% of the total soil microorganisms (Fig. 6). The relative abundances of Chloroflexi and Nitrospirae in the N300 treatment were significantly higher than those in the CK treatment, and the abundance of Planctomycetes was less than that in CK. Compared with the CK and N300 treatments, N300+ST significantly increased the relative abundance of Proteobacteria, Bacteroidetes, and Verrucomicrobia. In the N300+BC treatment, the abundance of Nitrospirae was greater than that in CK, but there was no significant difference between N300+BC and N300 treatments.
We did further compare the changes of soil microbial community abundance at the order level (Fig. 7). The first 21 orders of relative abundance in each sample were obtained by sequence comparison. Overall, straw addition treatment (N300+ST) had the largest difference from other treatments (CK, N300, N300+BC). Compared with CK and N300, the relative abundances of Burkholderiales, Xanthomonadales, Rhizobiales, Micrococcales, Sphingomonadales, Pseudomonadales, Myxococcales, Micromonosporales, Propionibacteriales, and Rhodocyclales were increased significantly in N300+ST. In contrast, N300+BC significantly increased the relative abundances of Rhodocyclales, Streptomycetales, and Nitrosomonadales.
Functional shifts in metabolic output and nitrogen metabolic pathways
To reveal the functional profiles of microorganisms under biochar amendment, total reads were annotated by means of categories in the eggNOG databases. As shown in Table 4, functional classification based on eggNOG annotations of microorganisms in different treatments revealed that about 40% of the total reads were associated with metabolism. In the metabolism category, amino acid transport and metabolism was the most abundant category, followed by energy production and conversion, and amino and carbohydrate transport and metabolism. Compared with CK and N300, the N300+ST treatment significantly increased the relative abundance of carbohydrate transport and metabolism, and decreased amino acid transport and metabolism. However, N300+BC increased the relative abundance of amino acid transport and metabolism.
Nitrogen cycling is of great significance in soils. The functional profiles of soil microbe–associated enzymes after biochar application had not been entirely explored. In the current study, we used metagenomic analysis to investigate key enzyme genes involved in five N transformation processes, nitrification, denitrification, N fixation, glutamate synthesis, and carbamoyl phosphate synthesis (Fig. 8).
Relative abundance of functional genes in the N metabolic pathways is shown in Fig. 9. Compared with CK, the treatment of N300 significantly increased the relative abundance of nitrification (amoABC, nxrB), denitrification (nirKS, norC, and narH), N fixation (nifDH), and glutamate synthesis (GLT1) among enzyme genes related to N metabolism, but reduced the glutamate synthesis (gudB, GLUD1-2, GLU) and nitrate reduction (nrfH). From the perspective of enzyme, ammonia monooxygenase (EC: 1.4.99.39), nitrite oxidoreductase (EC: 1.7.99), nitrite reductase (NO-forming) (EC: 1.7.2.1), and nitrogenase (EC: 1.18.6.1) in N300 were significantly higher than those in CK (Fig. 10). The N300+ST treatment had the largest differences with other treatments (Fig. 9). Compared with N300, the N300+ST treatment significantly increased the relative abundance of nitrate reduction genes (napAB, nirBD, nasAB, and narB), nitric oxide reductase genes (norB), and some glutamate synthesis genes (GDH2, gltBD). Meanwhile, N300+ST significantly increased the abundance of soil nitrite reductase (EC: 1.7.1.15, EC: 1.7.7.1, and EC: 1.7.2.2), nitrate reductase (EC: 1.7.99.4), nitric oxide reductase (EC: 1.7.2.5), glutamate synthase (EC: 1.4.1.13, EC: 1.4.1.14, and EC: 1.4.7.1), and carbamoyl phosphate synthase (EC: 6.3.4.16) (Fig. 10). In contrast, the N300+BC treatment significantly increased the relative abundance of assimilatory nitrate reduction (nasB), nitrification (hao), and glutamate synthesis (gdhA, GLUD1-2, gudB, and GLU) genes (Fig. 9). Thus, hydroxylamine dehydrogenase (EC: 1.7.2.6), glutamate dehydrogenase (EC: 1.4.1.2, EC: 1.4.1.3, and EC: 1.4.1.4), glutamine synthetase (EC: 6.3.1.2), and glutamate synthase (EC: 1.4.1.13, EC: 1.4.1.14, and EC: 1.4.7.1) in N300+BC were significantly higher than those in N300 (Fig. 10).
Discussion
Many studies have reported that biochar incorporation with N fertilizer significantly increased crop N uptake and NUE under both pot (Zhu et al. 2014) and field conditions (Li et al. 2016; Huang et al. 2018). Our results also showed that cotton straw–derived biochar increased N uptake and NUE of cotton, but there was no significant difference between biochar and straw treatments. This result showed that N fertilizer application combined with straw or biochar promoted N nutrient uptake and N fertilizer use efficiency in cotton.
In the present study, both cotton straw and biochar application with N fertilizer increased the soil TN and inorganic N (NH4+-N and NO3–-N). This finding is consistent with previous studies showing that straw or biochar application improved soil total N (Wei et al. 2015; Biederman and Harpole, 2013; Ma et al. 2016). Compared with N300+ST, biochar treatment (N300+BC) significantly increased TN and NO3−-N contents by 7.6% and 20.7%, respectively. The results indicated that biochar could enhance the N retention of soil, which was in agreement with previous reports (Yang et al. 2018; Joseph et al. 2020; Bai et al. 2020). Soil organic N content affects the N supply capacity of soil. AN is a part of soil organic N with strong activity, while NAN is a more stable organic N. Previous studies have already reported that straw or biochar application increased soil organic N content (Malhi et al. 2011; Prommer et al. 2014). Compared with CK and N300, N300+ST and N300+BC significantly increased the AN and NAN contents in soil, and the effects were more pronounced by biochar amendment (Fig. 4). A possible mechanism of biochar-increased soil N retention is the stimulation of microbial immobilization of N and increased nitrates recycling due to higher availability of carbon (Verheijen et al. 2009). Zhang et al. (2014) found that biochar increased soil microbial biomass N (MBN) significantly after 4 years of consecutive application. However, Prommer et al. (2014) reported that biochar application decreased soil organic N transformation rates and led to the build-up of soil organic N. Therefore, it is necessary to further determine whether biochar can increase soil organic N by promoting microbial N immobilization.
Soil microbial communities play a vital role in soil nutrient cycling. Biochar could stimulate the abundance of a variety of important soil microorganisms associated with soil N cycling (Liu et al. 2018). In this study, bacteria were the predominated microbial community in each treatment soils, followed by archaea and eukaryotes via metagenomic analyses (Table 3). The most dominant phyla were Proteobacteria, Actinobacteria, Gemmatimonadetes, Acidobacteria, Planctomycetes, Bacteroidetes, Verrucomicrobia, Firmicutes, Chloroflexi, and Nitrospirae. Those results were consistent with former reports (Castaneda and Barbosa 2017; Yang et al. 2018; Zhang and Lv 2020). As described in our study, for the relative abundances of major bacteria at both the phylum and order levels, straw addition treatment (N300+ST) had the largest difference from other treatments (CK, N300, N300+BC). Straw addition significantly increased the relative abundance of Proteobacteria, Bacteroidetes, and Verrucomicrobia, while there was no significant difference in microbial composition between N300+BC and N300 treatments at the phylum level (Fig. 6), At the order level, the N300+ST treatment increased the relative abundance of Burkholderiales, Xanthomonadales, Rhizobiales, Micrococcales, Sphingomonadales, Pseudomonadales, Myxococcales, Micromonosporales, and Propionibacteriales, which play important role in denitrification and cellulose degradation (Tu et al. 2017). Some studies also found that straw incorporation increased the abundances of Rhodocyclales, Pseudomonadales, Xanthomonadales, and Burkholderiales (Wang et al. 2018b; Su et al. 2019). Compared with CK and N300, N300+BC increased the relative abundance of Rhodocyclales, Streptomycetales, and Nitrosomonadales, which related to nitrification and turnover of organic matter at later stages of litter decomposition (Šnajdr et al. 2011). Similar effects have been reported by Jaiswal et al. (2018) based on a pre-conditioned growth medium with the biochar-amended pot experiment. Those results indicated that both straw and biochar application altered the structure of microbial communities, while the effects of straw on the relative abundances of major bacterial phyla and orders were higher than those of biochar.
To reveal the functional profiles of straw and biochar addition, the soil metagenomic reads were annotated by means of categories in the eggNOG and KEGG databases. We found that most of the eggNOG annotations were involved in microbial metabolism, and the relative abundance of amino acid transport and metabolism was the highest among the metabolism category (Table 4). Compared with CK and N300, the relative abundance of amino acid transport and metabolism genes was significantly increased in N300+BC, but decreased in N300+ST. This result suggested that biochar addition promoted the amino acid transport and metabolism of the corresponding bacteria.
In this study, metagenomic analysis was used to investigate key enzyme genes involved in nitrification, denitrification, nitrogen fixation, and glutamate synthesis. Compared with CK, N300 significantly increased the relative abundance of nitrification (amoABC, nxrB) and denitrification genes (nirKS, norC, and narH) (Fig. 9). Previous studies have already reported that N fertilization stimulated nitrifying bacteria (amoA) and the abundance of denitrification genes (nirK, nirS, and nosZS) (Kelly et al. 2013; Yang et al. 2018). In addition, our results also demonstrated that N300 significantly reduced the relative abundances of glutamate synthesis (gudB, GLUD1-2, GLU) and nitrate reduction (nrfH), compared with CK. Therefore, single N application promoted soil microbial nitrification and denitrification, and decreased glutamate synthesis, which might led to the risk of N losses.
We found that the relative abundance of functional genes related to the N metabolic pathways in the N300+ST treatment was significantly different from other treatments (CK, N300, N300+BC). N300+ST significantly increased the relative abundance of napAB, nirBD, nasAB, narB, and norB, which represented that the copy numbers of soil nitrite reductase (EC: 1.7.1.15, EC: 1.7.7.1, EC: 1.7.2.2), nitrate reductase (EC: 1.7.99.4), and nitric oxide reductase (EC: 1.7.2.5) were promoted (Fig. 10). It was in agreement with previous studies showing that N plus straw increased the abundance of nirB, nirD, and norB (Fracetto et al. 2017; Zheng et al. 2019), but Fracetto et al. (2017) found N plus straw also increased the abundance of nirK, nirS, and nosZ. In the present study, the relative abundance of key enzyme genes related to nitrification and denitrification in the N300+BC treatment was sim
Relative abundances of key enzymes in the N metabolic pathways as affected by different treatments. Error bars represent standard error (SD; n = 3). Bars with different letters are significantly different at P < 0.05. CK, no amendment; N300, N fertilizer application at 300 kg ha−1; N300+ST, N fertilizer application plus cotton straw; N300+BC, N fertilizer application plus cotton straw biochar
For most microorganisms, assimilation of ammonia is accomplished through the synthesis of glutamine and glutamate (Schreier 1993). The glutamine synthetase–glutamate synthase pathway (GS-GOGAT) and glutamate dehydrogenase (GDH) play distinct roles for incorporating inorganic N into organic molecules (Schwartz et al. 1991). Those key enzymes recycle glutamate; amino groups are eventually transferred to other amino acids and utilized for protein synthesis and also for synthesis of other key biomolecules (Liu et al. 2019). In our study, N300+ST significantly increased the relative abundance of GDH2 and gltBD genes related to glutamate synthesis, which represented that the copy numbers of glutamate synthase (EC: 1.4.1.13, EC: 1.4.1.14, EC: 1.4.7.1) were significantly higher than those in the N300 treatment. However, the copy numbers of glutamate dehydrogenase (EC: 1.4.1.2, EC: 1.4.1.3, EC: 1.4.1.4) and glutamine synthetase (EC: 6.3.1.2) in N300+ST were significantly lower than those in N300. In contrast, N300+BC significantly increased the relative abundance of gdhA, GLUD1-2, gudB, and GLU genes. The relative abundances of glutamate dehydrogenase (EC: 1.4.1.2, EC: 1.4.1.3, EC: 1.4.1.4), glutamine synthetase (EC: 6.3.1.2), and glutamate synthase (EC: 1.4.1.13, EC: 1.4.1.14, and EC: 1.4.7.1) in N300+BC were higher than those in the N300 treatment. Farhangi-Abriz and Torabian (2018) also reported that the application of biochar increased the activity of glutamine synthetase. Those results indicated that cotton straw–derived biochar addition increased microbial assimilation of N via promoting synthesis of glutamine and glutamate.
Conclusions
The results of five consecutive years of experiment support our hypothesis that cotton straw–derived biochar changed the composition of soil microbial community, increased the microbial assimilation of N, and improved soil N retention thus to increase N uptake of cotton. N fertilizer single application promoted nitrification and denitrification genes, but decreased the relative abundance of glutamate synthesis compared with CK. N fertilizer application combined with straw increased the carbohydrate transport and metabolism, and decreased amino acid transport and metabolism. The relative abundances of glutamate dehydrogenase and glutamine synthetase were lower than that in N300. In contrast, biochar incorporation with N fertilizer promoted the amino acid transport and metabolism. The relative abundances of glutamate dehydrogenase, glutamine synthetase, and glutamate synthase were increased. The present study proposed that straw-derived biochar may increase soil N content and improve soil N retention and availability, with better effects than straw directly returning.
References
Ajayi AE, Holthusen D, Horn R (2016) Changes in microstructural behaviour and hydraulic functions of biochar amended soils. Soil Tillage Res 155:166–175. https://doi.org/10.1016/j.still.2015.08.007
Anderson CR, Hamonts K, Clough TJ, Condron LM (2014) Biochar does not affect soil N-transformations or microbial community structure under ruminant urine patches but does alter relative proportions of nitrogen cycling bacteria. Agric Ecosyst Environ 191:63–72. https://doi.org/10.1016/j.agee.2014.02.021
Bai NL, Zhang HL, Zhou S, Sun HF, Zhao YH, Zheng XQ, Li SX, Zhang JQ, Lv WG (2020) Long-term effects of straw return and straw-derived biochar amendment on bacterial communities in soil aggregates. Sci Rep 10:7891. https://doi.org/10.1038/s41598-020-64857-w
Bai SH, Reverchon F, Xu CY, Xu ZH, Blumfield TJ, Haitao Z, Van ZL, Wallace HM (2015) Wood biochar increases nitrogen retention in field settings mainly through abiotic processes. Soil Biol Biochem 90:232–240. https://doi.org/10.1016/j.soilbio.2015.08.007
Ball PN, MacKenzie MD, DeLuca TH, Holben MWE (2010) Wildfire and charcoal enhance nitrification and ammonium-oxidizing bacterial abundance in dry montane forest soils. J Environ Qual 39(4):1243–1253. https://doi.org/10.2134/jeq2009.0082
Bi QF, Chen QH, Yang XR, Li H, Zheng BX, Zhou WW, Liu XX, Dai PB, Li KJ, Lin XY (2018) Effects of combined application of nitrogen fertilizer and biochar on the nitrification and ammonia oxidizers in an intensive vegetable soil. AMB Express 7:198. https://doi.org/10.1186/s13568-017-0498-7
Biederman L, Harpole WS (2013) Biochar and its effects on plant productivity and nutrient cycling: a meta-analysis. GCB Bioenergy 5:202–214. https://doi.org/10.1111/gcbb.12037
Bremner JM, Mulvaney C (1982) Nitrogen total. In: Page AL (ed) Methods of soil analysis, Part, vol 2. Chemical and microbiological properties. American Society of Agronomy, Soil Science Society of America, Madison, Wisconsin, pp 595–624
Buchfink B, Xie C, Huson DH (2015) Fast and sensitive protein alignment using DIAMOND. Nat Methods 12(1):59–60. https://doi.org/10.1038/nmeth.3176
Castaneda LE, Barbosa O (2017) Metagenomic analysis exploring taxonomic and functional diversity of soil microbial communities in Chilean vineyards and surrounding native forests. Peer Journal 5:e3098. https://doi.org/10.7717/peerj.3098
DeLuca TH, MacKenzie MD, Gundale MJ, Holben WE (2006) Wildfire-produced charcoal directly influences nitrogen cycling in Ponderosa Pine Forests. Soil Sci Soc Am J 70(2):448–453. https://doi.org/10.2136/sssaj2005.0096
Ducey TF, Ippolito JA, Cantrell KB, Novak JM, Lentz RD (2013) Addition of activated switchgrass biochar to an aridic subsoil increases microbial nitrogen cycling gene abundances. Appl Soil Ecol 65:65–72. https://doi.org/10.1016/j.apsoil.2013.01.006
Farhangi-Abriz S, Torabian S (2018) Biochar improved nodulation and nitrogen metabolism of soybean under salt stress. Symbiosis 74: 215–223. https://doi.org/10.1007/s13199-017-0509-0
Fracetto FJC, Fracetto GGM, Bertini SCB, Cerri CC, Feigl BJ, Neto MS (2017) Effect of agricultural management on N2O emissions in the Brazilian sugarcane yield. Soil Biol Biochem 109:209–213. https://doi.org/10.1016/j.soilbio.2017.02.004
Fu LM, Niu BF, Zhu ZW, Wu ST, Li WZ (2012) CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28(23):3150–3152. https://doi.org/10.1093/bioinformatics/bts565
Gruber N, Galloway JN (2008) An Earth-system perspective of the global nitrogen cycle. Nature 451:293–296. https://doi.org/10.1038/nature06592
Huang M, Yang L, Qin HD, Jiang LG, Zou YB (2014) Fertilizer nitrogen uptake by rice increased by biochar application. Biol Fertil Soils 50(6):997–1000. https://doi.org/10.1007/s00374-014-0908-9
Huang M, Long F, Chen JN, Jiang LG, Zou YB (2018) Continuous applications of biochar to rice: effects on nitrogen uptake and utilization. Sci Rep 8(1):11461. https://doi.org/10.1038/s41598-018-29877-7
Jaiswal AK, Elad Y, Cytryn E, Graber ER, Frenkel O (2018) Activating biochar by manipulating the bacterial and fungal microbiome through pre-conditioning. The New phytologist 219:363–377. https://doi.org/10.1111/nph.15042
Joseph UE, Toluwase AO, Kehinde EO, Omasan EE, Yetunde AT, George OO, Zhao C, Wang H (2020) Effect of biochar on soil structure and storage of soil organic carbon and nitrogen in the aggregate fractions of an albic soil. Arch Agron Soil Sci 66(1):1–12. https://doi.org/10.1080/03650340.2019.1587412
Kelly H, Tim JC, Alison S, Peter WC, Alan ER, Steven AW, Maureen O, Leo MC (2013) Effect of nitrogen and waterlogging on denitrifier gene abundance, community structure and activity in the rhizosphere of wheat. FEMS Microbiol Ecol 83(3):568–584. https://doi.org/10.1111/1574-6941.12015
Kielland K, McFarland JW, Ruess RW, Olson K (2007) Rapid cycling of organic nitrogen in taiga forest ecosystems. Ecosystems 10(3):360–368. https://doi.org/10.1007/s10021-007-9037-8
Lehmann J, Rillig MC, Thies J, Masiello CA, Hockaday WC, Crowley D (2011) Biochar effects on soil biota—a review. Soil Biol Biochem 43(9):1812–1836. https://doi.org/10.1016/j.soilbio.2011.04.022
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760. https://doi.org/10.1093/bioinformatics/btp324
Li Q, Liao N, Zhang N, Zhou G, Zhang W, Wei X, Ye J, Hou Z (2016) Effects of cotton (Gossypium hirsutum L.) straw and its biochar application on NH3 volatilization and N use efficiency in a drip-irrigated cotton field. Soil Sci Plant Nutr 62(5-6):534–544. https://doi.org/10.1080/00380768.2016.1219969
Liao N, Li Q, Zhang W, Zhou GW, Ma LJ, Min W, Ye J, Hou ZN (2016) Effects of biochar on soil microbial community composition and activity in drip-irrigated desert soil. Eur J Soil Biol 72:27–34. https://doi.org/10.1016/j.ejsobi.2015.12.008
Liu M, Tang D, Shi Y, Ma L, Li Y, Zhang Q, Ruan J (2019) Short-term inhibition of glutamine synthetase leads to reprogramming of amino acid and lipid metabolism in roots and leaves of tea plant (Camellia sinensis L.). BMC Plant Biol 19:425. https://doi.org/10.1186/s12870-019-2027-0
Liu Q, Liu BJ, Zhang YH, Lin ZB, Zhu TB, Sun R, Wang XJ, Ma J, Bei QC, Liu G, Lin XW, Xie ZB (2017) Can biochar alleviate soil compaction stress on wheat growth and mitigate soil N2O emissions? Soil Biol Biochem 104:8–17. https://doi.org/10.1016/j.soilbio.2016.10.006
Liu Q, Zhang Y, Liu B, Amonette JE, Lin Z, Liu G, Ambus P, Xie Z (2018) How does biochar influence soil N cycle? A meta-analysis. Plant Soil 426:211–225. https://doi.org/10.1007/s11104-018-3619-4
Liu YX, Lu HH, Yang SM, Wang YF (2016) Impacts of biochar addition on rice yield and soil properties in a cold waterlogged paddy for two crop seasons. Field Crop Res 191:161–167. https://doi.org/10.1016/j.fcr.2016.03.003
Ma NN, Zhang LL, Zhang YL, Yang LJ, Yu CX, Yin GH, Doane TA, Wu ZJ, Zhu P, Ma X (2016) Z (2016) Biochar improves soil aggregate stability and water availability in a mollisol after three years of field application. PLoS One 11(5):e0154091. https://doi.org/10.1371/journal.pone.0154091
Malhi S, Nyborg M, Solberg E, McConkey B, Dyck M, Puurveen D (2011) Long-term straw management and N fertilizer rate effects on quantity and quality of organic C and N and some chemical properties in two contrasting soils in Western Canada. Biol Fertil Soils 47(7):785–800. https://doi.org/10.1007/s00374-011-0587-8
Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet Journal 17(1):10–12. https://doi.org/10.14806/ej.17.1.200
Peng Y, Leung HC, Yiu SM, Chin FYL (2012) IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28(11):1420–1428. https://doi.org/10.1093/bioinformatics/bts174
Piash MI, Hossain MF, Parveen DZ (2019) Effect of biochar and fertilizer application on the growth and nutrient accumulation of rice and vegetable in two contrast soils. Acta Sci Agric 3(2):74–83
Prommer J, Wanek W, Hofhansl F, Trojan D, Offre P, Urich T, Schleper C, Sassmann S, Kitzler B, Soja G, Hood-Nowotny RC (2014) Biochar decelerates soil organic nitrogen cycling but stimulates soil nitrification in a temperate arable field trial. PLoS One 9(1):e86388. https://doi.org/10.1371/journal.pone.0086388
Raven JA, Handley LL, Andrews M (2004) Global aspects of C/N interactions determining plant-environment interactions. J Exp Bot 55:11–25
Schreier H (1993) Biosynthesis of glutamine and glutamate and the assimilation of ammonia. In: Sonenshein A, Hoch J, Losick R (eds) Bacillus subtilis and other gram-positive bacteria. ASM Press, Washington, DC, pp 281–298. https://doi.org/10.1128/9781555818388.ch20
Schwartz T, Misri BK, Fock HP (1991) The involvement of glutamate dehydrogenase and glutamine synthetase/glutamate synthase in ammonia assimilation by the basidiomycete fungus Stropharia semiglobata. J Gen Microbiol 137(9):2253–2258. https://doi.org/10.1099/00221287-137-9-2253
Šnajdr J, Cajthaml T, Valášková V, Merhautová V, Petránková M, Spetz P, Leppänen K, Baldrian P (2011) Transformation of Quercus petraea litter: successive changes in litter chemistry are reflected in differential enzyme activity and changes in the microbial community composition. FEMS Microbiol Ecol 75(2):291–303. https://doi.org/10.1111/j.1574-6941.2010.00999.x
Sorrenti G, Buriani G, Gaggìa F, Baffoni L, Spinelli F, Gioia DD, Toselli M (2017) Soil CO2 emission partitioning, bacterial community profile and gene expression of Nitrosomonas spp. and Nitrobacter spp. of a sandy soil amended with biochar and compost. Appl Soil Ecol 112:79–89. https://doi.org/10.1016/j.apsoil.2017.01.003
Stevenson FJ (1982) Nitrogen - organic forms. In: Page AL (ed) Methods of soil analysis, Part, vol 2. Chemical and microbiological properties. American Society of Agronomy, Madison, pp 625–641
Su Y, Lv JL, Yu M, Ma ZH, Xi H, Kou CL, He ZC, Shen AL (2019) Long-term decomposed straw return positively affects the soil microbial community. J Appl Microbiol 128:138–150. https://doi.org/10.1111/jam.14435
Tu QC, He ZL, Wu LY, Xue K, Zhou JZ, Xie G, Chain P, Reich PB, Hobbie SE (2017) Metagenomic reconstruction of nitrogen cycling pathways in a CO2-enriched grassland ecosystem. Soil Biol Biochem 106:99–108. https://doi.org/10.1016/j.soilbio.2016.12.017
Verheijen F G A, Jeffery S, Bastos A C, van der Velde M, Diafas I (2009) Biochar application to soils–a critical scientific review of effects on soil properties, processes and functions. EUR 24099 EN, Office for the Official Publications of the European Communities, Luxembourg, 149pp.
Wang C, Zheng MM, Hu AY, Zhu CQ, Shen RF (2018a) Diazotroph abundance and community composition in an acidic soil in response to aluminum-tolerant and aluminum-sensitive maize (Zea mays L.) cultivars under two nitrogen fertilizer forms. Plant Soil 424(1-2):463–478. https://doi.org/10.1007/s11104-017-3550-0
Wang YQ, Bai R, Di HJ, Mo LY, Han B, Zhang LM, He JZ (2018b) Differentiated mechanisms of biochar mitigating straw-induced greenhouse gas emissions in two contrasting paddy soils. Front Microbiol 9:2566. https://doi.org/10.3389/fmicb.2018.02566
Wei T, Zhang P, Wang K (2015) Effects of wheat straw incorporation on the availability of soil nutrients and enzyme activities in semiarid areas. PLoS One 10(4):e0120994. https://doi.org/10.1371/journal.pone.0120994
Xiao Z, Rasmann S, Yue L, Lian F, Zou H, Wang Z (2019) The effect of biochar amendment on N-cycling genes in soils: a meta-analysis. Sci Total Environ 696:133984. https://doi.org/10.1016/j.scitotenv.2019.133984
Xu YC, Shen QR, Ran W (2003) Content and distribution of forms of organic N in soil and particle size fractions after long-term fertilization. Chemosphere 50(6):739–745. https://doi.org/10.1016/s0045-6535(02)00214-x
Yang OY, Tiemann LK, Evans SE, Friesen ML (2018) Effect of nitrogen fertilization on the abundance of nitrogen cycling genes in agricultural soils: a meta-analysis of field studies. Soil Biol Biochem 127:71–78. https://doi.org/10.1016/j.soilbio.2018.08.024
Zhang K, Chen L, Li Y, Brookes PC, Xu J, Luo Y (2017) The effects of combinations of biochar, lime, and organic fertilizer on nitrification and nitrifiers. Biol Fertil Soils 53(1):77–87. https://doi.org/10.1007/s00374-016-1154-0
Zhang Q, Dijkstra FA, Liu X, Wang Y, Huang J, Lu N (2014) Effects of biochar on soil microbial biomass after four years of consecutive application in the North China plain. PLoS One 9(7):e102062. https://doi.org/10.1371/journal.pone.0102062
Zhang L, Lv J (2020) Metagenomic analysis of microbial community and function reveals the response of soil respiration to the conversion of cropland to plantations in the Loess Plateau of China. Glob Ecol Conserv 23:e01067. https://doi.org/10.1016/j.gecco.2020.e01067
Zheng W, Zhao Z, Lv F, Wang R, Gong QL, Zhai BN, Wang ZH, Zhao ZY, Li ZY (2019) Metagenomic exploration of the interactions between N and P cycling and SOM turnover in an apple orchard with a cover crop fertilized for 9 years. Biol Fertil Soils 55(9):1–17. https://doi.org/10.1007/s00374-019-01356-9
Zhong ML, Cheng RC, Mehran RR (2018) High pyrolysis temperature biochars reduce nitrogen availability and nitrous oxide emissions from an acid soil. GCB Bioenergy 10:930–945. https://doi.org/10.1111/gcbb.12529
Zhu QH, Peng XH, Huang TQ, Xie ZB, Holden NM (2014) Effect of biochar addition on maize growth and nitrogen use efficiency in acidic red soils. Pedosphere 24(6):699–708. https://doi.org/10.1016/S1002-0160(14)60057-6
Zhu WH, Lomsadze A, Borodovsky M (2010) Ab initio gene identification in metagenomics sequences. Nucleic Acids Res 38(12):15. https://doi.org/10.1007/978-1-4899-7478-5_440
Acknowledgments
We would like to thank anonymous reviewers for helpful comments on previous versions of this manuscript.
Funding
This work was jointly funded by the National Key Research and Development Project (2017YFD0200100, 2018YFD0800800).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Zhihong Xu
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wang, J., Long, Z., Min, W. et al. Metagenomic analysis reveals the effects of cotton straw–derived biochar on soil nitrogen transformation in drip-irrigated cotton field. Environ Sci Pollut Res 27, 43929–43941 (2020). https://doi.org/10.1007/s11356-020-10267-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-020-10267-4