Abstract
Growing evidence points to the controlled irrigation (CI) and biochar application (BA) having agricultural economic value and ecological benefits, but their synergistic effect and microbial mechanism of nitrogen conversion remain unknown in paddy fields. The effects of different BA (0, 20, 40 t/hm2) on the soil nitrogen functional transformation microbial genes (nifH, AOA-amoA, AOB-amoA) in different irrigation (CI, flooding irrigation) were clarified. After one seasonal growth of paddy, the correlation between the abundance of functional genes OUT and soil nitrogen transformation environment factors during the typical growth period was analyzed. High-throughput sequencing results illustrated that the application of CC (40 t/hm2 biochar) increased the nifH genes bacterial community abundance; the abundance of dominant microorganism increased by 79.68~86.19%. Because biochar can potentially control the rates of N cycling in soil systems by adsorbing ammonia and increasing NH4+ storage, it increased soil NH4+–N and NO3−–N content by 60.77% and 26.14%, improving microbial nitrogen fixation. Rare species Nitrosopumilus, Nitrosococcus, and Methylocystis appeared in biochar treatments group, which increased the diversity of microbial in paddy. The combined use of CI and BA affected soil inorganic nitrogen content, temperature (T), pH, Eh, etc., which affected urease, urea hydrolysis, and nitrogen functional transformation microorganism genes. Correlation analysis shows that soil NH4+–N, T, and Eh, respectively, are significant factors for the formation of nifH, AOA-amoA, and AOB-amoA soil bacterial communities, respectively. This study suggests that to maintain the biodiversity of soil and realize the sustainable development of rice cultivation, CI is of great importance in combination with BA.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Agricultural non-point source pollution is an important part of eutrophic pollution sources of various lakes in China (Ongley et al. 2010). Improper application of nitrogen fertilizer and extensive irrigation in traditional rice farming have resulted in a large quantity of nitrogen (N) loss and reduced N use efficiency of crops, thus aggravating agricultural pollution (Srinivasarao et al. 2014). China is one of the mainly rice-producing countries, accounting for nearly 20% of the rice planting area worldwide and stably supplying about 30% of the world’s rice production (Zhe et al. 2021). Therefore, scholars have carried out much research on water-fertilizer optimum management, including water-saving irrigation (WSI), controlled drainage technology, fertilizer management practices, microorganisms improvement, and soil amendments (Chen et al. 2021; Deng et al. 2019; Liu et al. 2019a, 2019b). Controlled irrigation (CI) is one of the WSI technologies. Nowadays, the research found that rice CI can mitigate the greenhouse effects, reduce nitrogen and phosphorus losses, improve crop productivity, but the alternating process of wet and dry water of rice CI can also speed up the soil organic matter decomposition (Hale et al. 2021; Yang et al. 2018). The utilization and efficiency enhancement technology of straw charcoal-based fertilizer was listed as one of the top ten leading promotion technologies in 2020 by the MARA (Ministry of Agriculture and Rural Affairs). Biochar application easily forms large aggregates in soil, which have a good adsorption effect on nitrogen ions and can reduce nitrogen volatilization (Hale et al. 2021), so biochar application plays a positive role in soil nitrogen fixation and reducing nitrogen loss. Controlled irrigation can reduce the amount of infiltration and thus decrease the loss of nitrogen ions with infiltration (Yang et al. 2013). Therefore, combining biochar with CI is an effective method for achieving sustainable utilization of water and nitrogen resources in paddy fields. At present, most studies focused on the effects of different biochar contents on soil nitrogen concentration in farmland (Cui et al. 2019; Guo et al. 2021). The mechanism of action on nitrogen transformation-related microorganisms of biochar addition in CI paddy fields is still unclear.
Soil microorganisms are quite sensitive to response to soil ecological changes and environmental stresses and play vital roles in soil structural dynamics, nutrient cycling, and some other ecological processes (Cruz-Paredes et al. 2017; Yan et al. 2022). The changes in soil microbial properties can widely monitor soil quality trends and maintain soil fertility (Guo et al. 2019). Nitrogen-fixing microorganisms undertake the role of regulating nitrogen nutrition and are one of the important functional microbial flora in the rice field ecosystem (Wang et al. 2019). All nitrogen-fixing microorganisms contain the nifH gene ferritin in encoding molybdenum-dependent nitrogenase, which is the best marker gene to study community structure (Kang et al. 2013). Ammonia oxidation is the rate-limiting process of nitrification, mainly catalyzed by ammonia-oxidizing archaea (AOA) and ammonia-oxidizing bacteria (AOB), which oxidize ammonia to nitrite (Fu et al. 2020). The functions of nitrogen-fixing microorganisms and ammonia-oxidizing microorganisms widely studied in agro-ecosystems are mainly affected by soil environmental factors, including temperature, water content, pH, soil texture, and fertilization (Gao et al. 2020; Lai et al. 2019; Yang et al. 2020). Although the abundance and uniformity of microorganisms can be improved when water is sufficient (Cai et al. 2020), AOA is more adaptable to water-deficient environments than AOB (Yang et al. 2017). Without a doubt, it has a profound effect on the microbial community structure of nitrogen transformation, soil nutrient forms, and migration patterns with different irrigation methods.
Biochar is a farmland carbon management measure with great emission reduction potential. Biochar can adsorb soil inorganic N and affects the microbial degradation process in the soil, reducing the loss of ammonia and nitrate, while potentially controlling the slow release of N to plant roots and improving N use efficiency (Haider et al. 2017; Wang et al. 2016; Yu et al. 2017). Biochar input leads to an increase in soil pH, which affects the abundance and activity of AOA and AOB (Zhang et al. 2019). For example, Song et al. (2014) found that cotton stalk biochar increased the diversity of AOB and reduced the diversity of AOA. Liu et al. (2019a, 2019b) study demonstrated that low-volume biochar (20 t/hm2) and compound fertilizer amendment increased the nifH gene abundance and altered the community structure of soil nifH. Same studies on the effects of applicated biochar on soil microbial activity have drawn inconsistent conclusions, mainly because the biochar type, amount, and use time were different (Maroušek et al. 2020; Stávková and Maroušek, 2021). Currently, most studies only focused on the impact of a single factor of irrigation pattern or biochar application on soil microorganisms (Jiang et al. 2021; Wang et al. 2021). Therefore, under the synergistic effect of biochar and CI, there are few reports on the microbial mechanism of soil nitrogen transformation from the gene level.
The aim of this paper clarified the dynamic changes of soil available N (NH4+–N and NO3−–N) in paddy soil under the joint control of BA and CI. In-depth exploration of the response mechanism and regulation process of nitrogen-fixing and ammonia-oxidizing microorganisms in paddy soil analyzes the environmental factors affecting the microbial OUT abundance of functional genes during growth. The results provide a new perspective on the mechanism of nitrogen migration and transformation, realizing a scientific basis for the sustainable utilization of soil and water resources in paddy fields.
Materials and methods
Description of the study area
The experimental site was located at the Kunshan Experimental Station (34°15′21″N, 121°05′22″E) of the State Key Laboratory of Hydrology-Water Resources and Hydraulic Engineering of Hohai University, in the east Taihu Lake region of China (Fig. 1). This area belongs to the subtropical monsoon climate zone in southeastern China. The annual average temperature, evaporation, precipitation, and sunshine hours are 15.5 °C, 1375.9 mm, 1097.1 mm, and 2104.9 h, respectively, and the frost-free period is 232 days/year. The locals were accustomed to the rice-wheat rotation. The soil type of the experimental field was dark-yellow hydromorphic paddy soil. The soil bulk density of the surface layer 0~30 cm was 1.32 g/cm3. The biochar prepared by straw pyrolysis used in this experiment was obtained from Zhejiang Biochar Engineering Technology Research Center. The biochar was spread in the pots manually and incorporated into the soil (about 20 cm) using a shovel 1 day before the transplantation of rice in 2016. The basic properties of soil and biochar were shown in Table 1.
Experimental design
The tested rice variety was Su Xiang Jing, the plant spacing and rowing were 25cm, and the emergence quantity per hole was 3~4. Rice was transplanted on June 25 and harvested on October 24 in 2019. The experiment adopted a randomized block design in 12 lysimeters (2.5×2 m). Four treatments with different combinations of irrigation and biochar were designed. There were two irrigation modes, namely controlled irrigation (CI) and flooding irrigation (FI). Under CI, the three biochar treatments were CA (0 t/hm2 biochar), CB (20 t/hm2 biochar), and CC (40 t/hm2 biochar); under FI conditions were FC (40 t/hm2 biochar) during the whole growth period of rice (re-greening stage to harvest stage).
In the CI treatment, the water depth of the paddy field was only kept at 5~25 mm in the re-greening stage; the paddy field avoided the water layer after irrigation in other stages. The irrigation quota and irrigation time of other stages were determined by taking the soil moisture of the root-soil layer as the control index. The upper limit of soil moisture control was the saturated water content, and the lower limit was 60~80% of the saturated soil moisture content (Liu et al. 2019a, 2019b). Except for the late tillering and yellow maturity stages, the water depth of the paddy field after transplanting was maintained at 30~50 mm in the FI treatment. The fertilization time and fertilization amount were carried out according to local farmers’ habits (Table 2).
Soil sample collection and measurement
The three-point mixing method was used to collect soil samples at 0–10cm during the re-greening stage (7/12), tillering stage (7/23), and harvest stage (10/25), which were marked by 1, 2, 3, respectively. A total of 36 soil samples were sequenced, including 4 treatments (CA, CB, CC, FC), 3 growth stages, and 3 replicates. The collected soil samples were taken back to the laboratory after removing plant roots and gravel. The average T during the soil sampling period was 20.4 °C. A part of the fresh soil was leached with 1mol/L KCL solution, which determines the NH4+–N, NO3−–N content; the other soil was air-dried and passed through a 20mm mesh sieve and a 100mm mesh sieve to determine the soil urease activity (Urease). In addition, it took the soil of about 2g from each sampling point, mixed, sealed, and stored in a refrigerator at −20 °C for soil microbial genome extraction and analysis. The ultraviolet spectrophotometry measured the content of NH4+–N and NO3−–N. The phenol-sodium hypochlorite colorimetric method determined the content of soil urease, simultaneously monitoring conventional physical and chemical indicators such as Eh, soil moisture (θ) of soil samples.
Soil DNA extraction and PCR amplification
The overall process of soil microbial genome extraction and analysis includes 6 parts: DNA extraction, synthetic primer barcode, PCR amplification and purification, PCR product quantification, PCR free sequencing libraries, and computer sequencing, provided by the Genepioneer Biotechnologies (Nanjing). High-throughput sequencing based on the Illumina MiSeq PE250 platform was used to study nifH, AOA, and AOB gene diversity composition spectrum sequencing. Functional gene-specific primers are shown in Table 3.
Raw FASTQ files were screened for high-quality sequences (Phred Quality Score > 20) by removing low-quality reads, unrecognized reverse primers, and any ambiguous base calls. Effective bacterial sequences were clustered into OTUs (Operational Taxonomic Units) at a 97% sequence similarity level using UPARSE software; then, the dominant sequence was chosen as the representative sequence for each OUT (Guo et al. 2019). Calculate the alpha diversity index based on the abundance of non-rare OUT in each sample, and obtain the species classification information corresponding to each OTU by comparing the OTU sequence to the proGenomes (http://progenomes.embl.de/) database.
Analytical methods
Microbiome data and drawing-related charts were processed using Microsoft 2019. R 3.1.0 software was used to calculate the high-throughput sequencing diversity index. Statistical analysis was performed using SPSS 22.0, which determines the effects of irrigation treatment and biochar on alpha-diversity indices and microbiome OTU in different periods. Heatmap analysis uses color gradients to represent the size of values in the data matrix and can be clustered according to species or sample abundance similarity. Linear regression analyses, including Pearson correlation coefficients, were used to examine the correlations between microbial diversity and soil properties under different biochar and irrigation treatments.
Results
Effect of biochar application and irrigation on soil NH4 +–N and NO3 −–N
The soil NH4+–N content of different BA treatments under different irrigation conditions showed a single peak change trend and reached the peak at the rice tillering stage (Fig. 2). The average concentrations of soil NH4+–N in CA2, CB2, and CC2 were 21.844mg/kg, 29.452mg/kg, and 47.148mg/kg, respectively. During the whole growth period of rice (from the re-greening stage to the harvest stage), under CI conditions, the average concentration of soil NH4+–N of CA, CB, CC were 8.465mg/kg, 10.755mg/kg, 16.565mg/kg, respectively. So this shows that the application of biochar had different soil NH4+–N retention effects in CI rice fields. The application of high amounts of biochar had the best adsorption effect on soil NH4+–N and can reduce the risk of NH4+–N leaching. Under the biochar management conditions, the average content of soil NH4+–N of CC2 was 20.278 mg/kg higher than that of FC2; the average content of soil NH4+–N of CC was 6.261 mg/kg higher than that of FC during the whole growth period. It is mainly related to controlling water in the rice field. On the one hand, the CI are beneficial in improving the utilization efficiency of nitrogen fertilizer and can inhibit the ammonia volatilization loss after urea hydrolysis, so the soil colloid can absorb more NH4+–N. On the other hand, FI may promote water infiltration, which causes soluble ions such as NH4+–N to migrate to deep soil layers.
Under CI conditions, the soil NO3−–N content of different BA rates reached the peak of rice (Fig. 2), CA2, CB2, CC2 were 18.792mg/kg, 23.013mg/kg, 25.403mg/kg, respectively. Compared with CA1, CB1, CC1, the contents of NO3−–N all decreased by 71.62%, 55.77%, 59.41%, respectively. During the whole growth period of paddy, the average soil NO3−–N content of each treatment group (CA, CB, CC, FC) was 14.179mg/kg, 15.182mg/kg, 15.641mg/kg, and 12.399mg/kg, respectively, so under CI conditions, the application of biochar soil NO3−–N content increased by 7.07% ~10.31%. The average level of soil NO3−–N concentration under FC was lower than under CC, with an average reduction of 3.242 mg/kg, indicating that CC can effectively reduce the loss of soil NO3−–N. The biochar improved the holding capacity of soil NH4+–N and NO3−–N under CI, and NH4+–N was more affected by biochar during the whole growth period. In terms of irrigation management, compared with traditional FI, the contents of NH4+–N and NO3−–N were higher in the surface soil of the paddy field under CI, so losing nitrogen ions with the infiltration water was few. The joint regulation of biochar and CI enhanced soil retention of soil NH4+–N and NO3−–N, thus reducing nutrient leaching.
Gene alpha-diversity analyses under biochar application and irrigation
Alpha-diversity of the nifH gene
After a series of preprocessing such as quality control filtering, splicing and chimera removal, the final effective sequence number was 80023~103020. The OUT number was 3305~4193 by using effective sequence for clustering (Table 4). The Good’s coverage for the observed nifH gene OTUs was 0.99, indicating that the species sample was nearly all covered. There were differences in the α-diversity of bacterial communities under different BA and irrigation. During the whole growth period of rice, with the increase in BA, the Chao1 index showed an increasing trend, and the content of CA, CB, CC was 3884, 4100, 4309, respectively. The Chao1 index of the CB and CC groups increased significantly by 5.57% and 10.95% compared with the CA group. The Shannon index was the highest at the tillering stage, followed by the re-greening stage and the lowest harvest stage. In the tillering stage of rice, the CB2, CA3 of chan1 and Shannon were higher than the CA2, increasing by 18.58~26.55% and 6.96~7.41%, respectively. The FC average index of Chao1 was 4398, which increased by 2.06% compared with CC. At different times, the Shannon of FI was the highest. The Shannon indexes of FC1, FC2, FC3 were 9.61, 9.71, 9.32, respectively, which increased by 0.11, 0.18, and 0.04 compared with CC1, CC2, CC3. Except for the control group, the Shannon index of each treatment group showed a trend of increasing and then decreasing. In summary, the high-carbon treatment had the highest abundance of nifH, but CI did not improve the abundance and diversity of nifH in the rice field.
Alpha-diversity of the AOA-amoA gene and AOB-amoA gene
The AOA-amoA and AOB-amoA gene sequencing results and the a-diversity index are shown in Table 5. The Good’s coverage for the observed OTUs was 0.995~1; the sequencing results can reflect the true condition of the microorganisms in the sample. NH3/NH4+ was the common substrate for AOA and AOB to participate in the ammonia oxidation reaction. Therefore, the AOA and AOB abundance and relative contribution rate were easily affected by the concentration of soil fertility in the paddy. The AOA-amoA gene OUT number increased at the re-greening stage and tillering stage, and then decreased by 73.88~133.87% at the harvest stage. The AOB-amoA gene number of OTUs of each sample was the highest during the re-greening stage and then decreased. When the soil fertility content was higher in the tillering stage, AOA dominated the ammonia oxidation reaction because of its high affinity for ammonia in the paddy soil ecosystem; in the harvest stage, when the soil fertility content was low, AOB accounted for domination (Fig. 3).
The chao1 index was higher from the AOA-amoA gene in Table 5 during the re-greening stage and tillering stage, because of the large and frequent application of base fertilizer and tillering fertilizer. During the harvest stage, the soil N content dropped to a low level; chao1 index was lower. So, fertilization was valid and conducive to increasing the abundance of AOA in paddy fields. From the perspective of community diversity, there was no significant change in the Shannon index of each treatment at the re-greening stage and tillering stage. During the re-greening stage, the medium amount of biochar (20 t/hm2) had inhibition on the abundance and diversity of AOA, and the high amount of biochar (40 t/hm2) showed the promoting effect. Under the same BA conditions, the chao1 and Shannon indexes of the CC1 were higher than the FC1, increasing by 7.57%, 4.51%, respectively. During the whole growth period, the chao1 index of CC increased by 0.36% compared with the FC. It shows that there was no significant influence in the composition of the AOA community by irrigation methods.
From the AOB-amoA gene in Table 5, the chao1 and Shannon indexes of each treatment decreased to varying degrees at the re-greening stage to tillering stage, and there was a significant rise at the harvest stage. The abundance and diversity of the AOB at the tillering stage were the smallest. Compared with the CA2, the CB2 had a promotion effect on chao1 and Shannon indexes, increasing by 1.69% and 1.16%; the CC2 had an inhibitory effect on chao1, decreasing by 10.42%. In irrigation modes, the chao1 index of CC1 was higher than that of FC1; the chao1 index of FC2 and FC3 was significantly higher than that of CC2 and CC3. The CC2 Shannon index was the highest in the tillering stage; FC1 and FC3 were the highest in other stages. The Shannon index of FC was higher than CC, an increase of 6.09%, showing that AOB was more susceptible to irrigation patterns than AOA. The chao1 index of CC was higher than CA, increasing by 4.91%, indicating that the application of high amounts of biochar increased the abundance of AOB.
Bacterial community structure under biochar application and irrigation
Comparison of dominant microbial
The relative abundance of dominant microbial species in each of the columns at the genus level showed the top 20 species abundance classification results, which contain the other microbial (Fig. 4a). The species composition of nitrogen-fixing bacteria encoded by the nifH gene in paddy soils under different water and carbon management modes was similar. The dominant microbial population in the columns were Bradyrhizobium, Unassigned, Geobacter, Anaeromyxobacter, and Geoalkalibacter, respectively. These five species accounted for nearly 80% of all the soil systems. In the CI mode, the cumulative abundance of the top 5 dominant microbial in CA1 was the lowest (76.83%); CB1, CC1 were 82.74%, and 85.67%, respectively, indicating that microbial promoted the growth of dominant microbial in the re-greening stage. The abundance of Bradyrhizobium and Geobacter increased in CB2 and CC2 compared with CB1, and the abundance of Anaeromyxobacter decreased, but the cumulative total amount of dominant microbial had little difference. In the harvest stage, the N content of paddy soil was at a relatively low level. The CA3, CB3, CC3, and FC3 nitrogen-fixing microbial communities had the highest abundance of Bradyrhizobium, 41.03%, 33.60%, 43.75%, and 43.32%, respectively. The cumulative abundance of the dominant microorganism in the harvest stage was higher than that in the other two stages. In each growth period, the abundance of dominant microorganism in CI was significantly higher than that of flooded irrigation, indicating that the inadequate irrigation mode increased soil permeability, which was beneficial to increase the microbial quantity and promote nitrogen fixation. Especially after fertilization, the cumulative abundance of the dominant bacteria in FC1 and FC2 was lower than that of CC1 and CC2, which were reduced by 5.68% and 5.33%.
Figure 4b shows the top 9 species abundance classification results for the relative abundance of AOA-amoA dominant microbial species in each column at the genus level. The species composition of the AOA-amoA gene was similar under different treatment modes in paddy soil. The dominant microbial population were Candidatus Nitrosocosmicuss and Nitrososphaera. These two species accounted for 85.65~92.34% of all the soil samples. From the time scale, there was little difference in the microbial species distribution of the samples at the re-greening and tillering stages. Except for the CC2, Nitrososphaera had the highest relative abundance, accounting for 47.39%, and the remaining treatments were Candidatus Nitrosocosmicus with the highest relative abundance, accounting for 43.17~50.83%. In the harvest stage, the distribution of AOA-amoA microbial changed significantly, Nitrososphaera had the highest relative abundance, and the cumulative abundance of dominant species of CA3, CB3, and CC3 increased slightly, to 90.42%, 92.34%, and 90.40%, respectively. Compared with CA1, CA2, and CA3, new microbial Candidatus Nitrosotenuis, Nitrosarchaeum, and Nitrosopumilus appeared in CC1, CC2, and CC3 groups, respectively. It showed that the application of biochar was beneficial to the increase of AOA-amoA microbial diversity. Under the same BA conditions, the cumulative abundance of dominant microbial in CC was higher than that of FC, which increased by 3.03%.
The relative abundance of the top 8 species of AOB-amoA dominant microbial species at the genus level is shown in Fig. 4c. The Unassigned were the dominant microbial population, and relative abundance was 56.58~87.32% in the re-greening stage and harvest stage, and it was 99.8% at the tillering stage. In the re-greening stage and harvest stage, Nitrosomonas was widely present but not detected in samples at the tillering stage. Mycolicibacterium only existed in each treatment sample at the tillering stage. In the CI mode, BA had little effect on the distribution and abundance of dominant species of AOB-amoA microbial, but it will stimulate the increase of species diversity. CC1 and CB1 samples detect 5 and 4 bacterial microbial, respectively, but CA1 only contains 3. The rare microbial of Nitrosococcus and Methylocystis appeared in the application biochar treatments, indicating that the addition of biochar increased the diversity of AOB-amoA microorganisms in rice soil. Analyzing the influence of water-carbon methods on the AOB-amoA community composition, the relative abundance of the optimal microbial community in flooded irrigation was higher than that of CI in different periods, increasing by 0.07~30.57%. Compared with AOA-amoA microbial composition, rare microbial of the AOB-amoA only appears in samples at certain specific periods. The number of microbes detected by AOB-amoA in a single sample was lower than that in AOA-amoA, indicating that the species diversity of AOA was more abundant.
Cluster analysis of microbial abundance
To visualize general differences in the composition of microbial communities among different treatments, the heatmap was used to cluster analyze the nifH, AOA-amoA, AOB-amoA genes at the family level. The resulting heatmap showed similarities in the abundance of various species or samples. The nifH communities showed more clusters than AOA-amoA and AOB-amoA communities, indicating greater complexity and diversity of nifH communities (Fig. 5). Each treatment affected nifH gene species composition in different periods (Fig. 5a). The nifH gene communities of each treatment were divided into 5 clusters: the first type was FC3, CA3, CC3; the second type was CA1; the third type was CC1 and CA2; the fourth type was FC2, CB1, CB2; the fifth type was CB3, FC1, CC2. Figure 5b shows that each sample of the AOA-amoA gene communities was divided into 4 clusters: the first type was FC3; the second type was FC2, FC1, CB1, CB2, CC2, CC1, CA1, CA2 in the re-greening and tillering stages; the third type was CB3; the fourth type was CA3 and CC3. The species composition of different clusters was similar. During the harvest stage, the abundance of FC3 was significantly higher than that of CC3, which was consistent with the conclusions in Table 5. Figure 5c showed that each sample of the AOB-amoA gene communities was divided into 2 clusters. The first category was all treatment groups at the tillering stage, in which the AOB-amoA gene was greatly affected by external fertilization. The second type was all treatment groups in the re-greening stage and harvest stage, indicating that the species composition of these two periods was similar.
Environmental factors related to soil nitrogen transformation genes
Effect of biochar application and irrigation on urease
The urease of the paddy field was higher during the rice tillering stage and then declined, remaining at the initial level (Fig. 6). Affected by tillering fertilizer, the urease of each treatment was the largest in the tillering stage, and it was positively correlated with the amount of biochar applied. The CA2, CB2, and CC2 treatments were 0.779, 0.903, 1.004 mg/g, respectively. The Urease under FI treatment was higher than that under CI, and FC2 increased by 0.159 mg/g compared to CC2. During the whole growth period of rice, the average urease of each treatment (CA, CB, CC, FC) was 0.442, 0.488, 0.548, 0.614 mg/g, respectively. The biochar treatments (CB, CC) increased soil urease by 10.31~24.00% compared with the CA, indicating that biochar was beneficial for urease. FC increased urease than CC 11.99%, indicating that the urease activity under FI was higher. In short, Fig. 2 shows that the content of increasing soil NH4+–N and NO3−–N of CC was higher than that of CB, which was 2.29 mg/kg and 0.459 mg/kg, respectively, during the whole growth period of paddy. The urease of CC was higher than that of CB in Fig. 7. In addition, the bacterial community structure under biochar application was similar. In the future, we propose to treat the soil with CC (40 t/hm2 biochar).
Correlation analysis of soil nitrogen transformation genes
The effects of biochar and CI on soil nitrogen transformation function were further investigated. The relationship among nifH, AOA-amoA, AOB-amoA, urease, NH4+–N and NO3−–N content, Eh, T, θ was analyzed through the Pearson correlation and further visualized in Fig. 7. Each treatment group had different correlations with their parameters. The abundance of AOA-amoA gene OTU was positively correlated with T, NH4+–N, and NO3−–N, and the correlation coefficients were 0.78~0.87, 0.26~062, and 0.62~0.86. In Fig. 7b, urease was significantly positively correlated with NH4+–N and NO3−–N, with correlation coefficients of 0.94** (P<0.01) and 0.92* (P<0.05). Comparing Fig. 7a, b, the correlation between the OTU abundance of nifH gene and soil NH4+–N and NO3−–N increased in the 40 t/hm2 biochar treatment group (CC), and the correlation coefficients were 0.73 and 0.71. Under FC conditions, the correlations of other parameters except for AOA-amoA and AOB-amoA genes were reduced, indicating that NH4+–N and NO3−–N were greatly affected by water-saving irrigation. In Fig. 7b, the AOA-amoA and AOB-amoA gene OTU abundances had higher correlations with θ than other treatment groups; AOA-amoA was greater than AOB-amoA because the AOA was more able to adapt to the water-deficient environment than AOB. The combined use of CI and biochar affected soil permeability, soil organic matter content, T, pH, Eh, etc., which affected urease, urea hydrolysis, and ammonia volatilization processes.
Discussion
Effects of BA and CI on nitrogen-transforming functional genes and inorganic nitrogen
The application of biochar affects the microbial degradation process, the abundance, activity, and diversity of a series of soil nitrogen cycle-related microorganisms such as nitrogen-fixing bacteria, AOA and AOB. Anderson et al. (2011) added different biochar in pots, and the results showed biochar increased the relative abundance of many known nitrogen-fixing bacteria, including Forlan’s population and Rhizobium. This experiment showed that the 40 t/hm2 biochar treatment had the highest abundance of nifH under CI; the abundance of dominant microbial (Bradyrhizobium, Unassigned, Geobacter, Anaeromyxobacter, and Geoalkalibacter) increased by 79.68–86.19%. This was consistent with the results of biochar compound fertilizer amendment that increased the nifH gene abundance and altered the community structure of soil nitrogen-fixing bacteria (Liu et al. 2019a, 2019b). Firstly, because biochar as a kind of organic C can potentially manipulate the rates of N cycle in soil systems through adsorption of ammonia and increase of NH4+ storage, thereby reducing NO3−–N leaching and improving microbial nitrogen fixation (Lan et al. 2022). Second, because the CI of repeated alternating dry and wet irrigation significantly increases the amount of dissolved oxygen and redox potential inside and outside the rhizosphere of rice (Cheng et al. 2022) enhances nitrification and promotes the transformation of nitrogen forms, thereby increasing the content of NH4+ and NO3− in paddy soil. Under CI, the soil immobilization of NH4+–N, NO3−–N can increase by 60.77% and 26.14% compared with FI in this paper.
The transformation process of soil nitrogen, especially the intensity of nitrification, determines whether the dominant form of soil inorganic nitrogen is dominated by NH4+–N or NO3−–N (Zhang et al. 2019a). In this paper, we found that the use of biochar increased the diversity of the AOA community, and Nitrosopumilus appeared in CB3 and CC3 (Fig. 4b). Because NH4+–N was between 1.190 ~2.410 at the harvest stage, the low NH4+–N environments favored the growth of AOA (Liu et al. 2017). In Fig. 2, we found that the transformation rate of NH4+–N to NO3−–N in the CC3 treatment group was the highest, which was 80.84%. This is consistent with Wu et al. (2018) enhancement of the nitrification rate by biochar amendment. Daims et al. (2015) point to completely nitrifying Nitrospira as key components of nitrogen-cycling microbial communities. The use of biochar increased the abundance of Nitrospira and promoted the diversity of the AOB community (Fig. 4c), because biochar incorporation into the soil may increase the bulk of the soil pH, elevate NH4+ and dissolved organic C, and thereby enhance nitrification activity via AOB activity (Mierzwa-Hersztek et al. 2018). An increased pH due to biochar input into soils usually had affected the abundance and activity of AOA and AOB genes (Zhang et al. 2019). In this study area, soil pH was 7.4, which was weakly alkaline. The application of a high amount of biochar willed increase the abundance of AOB, but the change of AOA was not obvious. As an alkaline material, biochar increases soil pH through its composition of alkaline substances and adsorption of exchangeable acid cations (Wu et al. 2020). Therefore, ammonia-oxidizing microorganisms also have different response mechanisms to the changes and stress in environmental conditions.
Different irrigation modes, such as internal drainage, alternate wetting and drying irrigation, soil water potential, would affect the ecological distribution of microorganisms in rice rhizosphere soil (Zhang et al. 2016). Ma et al. (2019) found that the number of OTUs of the AOA-amoA gene was larger than that of the AOB-amoA gene under different irrigation water salinity treatments. This is consistent with AOB being more susceptible to irrigation patterns than AOA under the same BA in our research because AOB dominates over AOA in drylands, while AOA dominates in waterlogged soils (Li et al. 2021). The abundances, diversity, and community structure of AOA and AOB varied largely among all the samples, which is attributable to the application of CI that affects the niche differentiation of AOA and AOB by changing the water content and oxygen concentration of the soil (Ma et al. 2019).
Influencing factors of soil nitrogen transformation genes
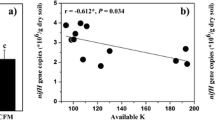
The Pearson correlation analysis showed that the properties of soil showed various significant correlations with bacterial community composition. This paper found that the correlation coefficient between AOA gene OTU abundance and T was 0.78~0.87. This is mainly because the average T during the sampling period was 20.4 °C, and the suitable growth T of ammonia-oxidizing microorganisms is 20–37 °C (Onodera et al. 2010), so the nitrification rate was positively correlated with T. Du et al. (2019) found that the application of biochar affected the correlation between AOA and pH, SOM, AN, NH4+–N, and NO3−–N. This paper found that AOA is positively correlated with NH4+–N and NO3−–N because biochar can slow down the reduction of NH4+–N by promoting organic nitrogen mineralization or reducing microbial assimilation of inorganic nitrogen. Biochar also can promote soil nitrification rate and increase the abundance of nitrifying crops in the soil (Prommer et al. 2014; Ball et al. 2010). Therefore, the abundance and diversity of AOA were more sensitive to soil environmental factors, and the AOA-amoA genes OTU abundance was highly correlated with T and θ, while AOB was low in Fig. 7. Numerous studies have demonstrated the cellular, genomic, and physiological differences between AOA and AOB (Prosser and Nicol 2012), and their divergent nitrification pathways and responses to environmental and climatic factors (Hu et al. 2015), which might lead to differential patterns between AOA and AOB in paddy soils.
The correlation coefficients between nifH gene OTU abundance and soil NH4+–N and NO3−–N in Fig. 7b were the largest, which were 0.73 and 0.71. This is because CI promoted the growth of nitrogen-fixing bacteria and reduced the leaching of nutrients (Zhao et al. 2022; Meng et al. 2014), under biochar application, thus decreasing the loss of soil NH4+–N and NO3−–N. Under CC, urease was significantly positively correlated with NH4+–N (P<0.001) and NO3−–N (P<0.05), which were 0.94** and 0.92*. The same research found that the urease activity of the treatment with biochar was increased by 0.64 to 42.17%, and the increase in urease activity was the most by the treatment with 4500 kg/hm2 of biochar (Chen et al. 2020). The effect of biochar on urease is mainly through the adsorption of reaction substrates or enzyme molecules to promote the progress of enzymatic reactions, thereby affecting the increase of soil enzyme activity. At the same time, the improvement of nitrogen nutrition promoted the reproduction of soil microorganisms, so that they secreted more urease into the soil (Petra et al. 2003). In general, BA and CI affected the correlation between soil properties and soil nitrogen transformation functional genes and caused community changes.
Conclusions
Biochar is a farmland carbon management measure with great emission reduction potential, which can reduce the impact of CI on ecology footprint. The BA improves the soil NH4+–N and NO3−–N contents of CI paddy fields, increasing by 27.05 ~ 95.68% and 7.07 ~ 10.31%, respectively. That is because BA can improve the richness and diversity of nifH at tillering stage and promote biological nitrogen fixation. High-throughput sequencing results illustrated that the application of high-volume biochar increased the Chao1 indexes of nifH and AOB-amoA by 10.95% and 4.91% under CI, but AOA-amoA did not change significantly. Because biochar can increase soil pH, AOA-amoA was accountable for ammonia oxidation in a low pH environment. The rare microbial of Nitrosopumilus, Nitrosococcus, and Methylocystis separately appeared in the BA treatments under CI, showing the diversity of AOA-amoA and AOB-amoA microorganisms increase in paddy. The combination of CI and BA increased the correlation between nifH, AOA-amoA and NH4+–N, NO3−–N. In summary, this paper creatively analyzes the nitrogen transformation mechanism in the combined BA and CI from the perspective of microbial genes, which can provide data and theoretical support for the sustainable use of farmland water and soil resources in the future.
Data Availability
The datasets used or analyzed during the current study are available from the corresponding author on reasonable request.
References
Anderson CR, Condron LM, Clough TJ, Fiers M, Stewart A, Hill RA, Sherlock RR (2011) Biochar induced soil microbial community change: implications for biogeochemical cycling of carbon, nitrogen and phosphorus. Pedobiologia 54(5-6):309–320. https://doi.org/10.1016/j.pedobi.2011.07.005
Ball PN, Mackenzie MD, Deluca TH, Montana WED (2010) Wildfire and charcoal enhance nitrification and ammonium-oxidizing bacterial abundance in dry montane forest soils. J Environ Qual 39(4):1243–1253. https://doi.org/10.2134/jeq2009.0082
Cai JM, Liu JY, Qiu HS, Lu MC, Zhou XG (2020) Effects of drip irrigation patterns and biochar addition on soil mineral nitrogen and microbial regulation of greenhouse. Environmental. Science 41(8):3836–3845. https://doi.org/10.13227/j.hjkx.201912156
Chen X, Jiang ZW, Ding J, Cai M, Yang SH (2020) Impacts of biochar application on soil nitrogen content and urease activity in water-saving irrigation paddy fields. Jiangsu. Agric Sci 19:268–274. https://doi.org/10.15889/j.issn.1002-1302.2020.19.056
Chen X, Yang SH, Jiang ZW, Ding J, Xiao S (2021) Biochar as a tool to reduce environmental impacts of nitrogen loss in water-saving irrigation paddy field. J Clean Prod 290:125811. https://doi.org/10.1016/j.jclepro.2021.125811
Cheng HM, Shu K, Zhu TY, Wang L, Liu X, Cai W, Qi ZM, Feng S (2022) Effects of alternate wetting and drying irrigation on yield, water and nitrogen use, and greenhouse gas emissions in rice paddy fields. J Clean Prod 349:131487. https://doi.org/10.1016/j.jclepro.2022.131487
Cruz-Paredes C, Wallander H, Kjoller R, Rousk J (2017) Using community trait-distributions to assign microbial responses to pH changes and Cd in forest soils treated with wood ash. Soil Biol Biochem 112:153–164. https://doi.org/10.1016/j.soilbio.2017.05.004
Cui H, Wang LX, Ou Y, Yan BX, Han L, Li YX, Jiang S (2019) Effect of the combined application of biochar and chemical fertilizer on the migration and transformation of nitrogen and phosphorus in paddy soil. Journal of Agro-Environment. Science 38(2):412–421. https://doi.org/10.11654/jaes.2018-0515
Daims H, Lebedeva EV, Pjevac P, Han P, Herbold C, Albertsen M, Jehmlich N, Palatinszky M, Vierheilig J, Bulaev A, Kirkegaard RH, Bergen MV, Rattei T, Bendinger B, Nielsen PH, Wagner M (2015) Complete nitrification by Nitrospira bacteria. Nature 528:504–509. https://doi.org/10.1038/nature16461
Deng SW, Wipf HML, Pierroz G, Raab TK, Khanna R, Coleman-Derr D (2019) A plant growth-promoting microbial soil amendment dynamically alters the strawberry boot bacterial microbiome. Sci Rep 9:17677. https://doi.org/10.1038/s41598-019-53623-2
Du Y, Wang TY, Anane PS, Li Q, Liu SX, Wang CY (2019) Effects of different types of biochar on basic properties and bacterial communities of black soil. Appl Ecol Environ Res 17(2):5305–5319. https://doi.org/10.15666/aeer/1702_53055319
Fu QL, Xi RZ, Zhu J, Hu HQ, Xing ZQ, Zuo JC (2020) The relative contribution of ammonia oxidizing bacteria and archaea to N2O emission from two paddy soils with different fertilizer N sources: a microcosm study. Geoderma 375:114486. https://doi.org/10.1016/j.geoderma.2020.114486
Gao SJ, Zhou GP, Liao YL, Lu YH, Nie J, Cao WD (2020) Contributions of ammonia-oxidising bacteria and archaea to nitrification under long-term application of green manure in alkaline paddy soil. Geoderma 374:114419. https://doi.org/10.1016/j.geoderma.2020.114419
Guo AN, Ding LJ, Tang Z, Zhao ZQ, Duan GL (2019) Microbial response to CaCO3 application in an acid soil in southern China. J Environ Sci 79:321–329. https://doi.org/10.1016/j.jes.2018.12.007
Guo LL, Yu HW, Niu WQ, Kharbach M (2021) Biochar promotes nitrogen transformation and tomato yield by regulating nitrogen-related microorganisms in tomato cultivation soil. Agronomy-Basel 11(2):381. https://doi.org/10.3390/agronomy11020381
Haider G, Steffens D, Moser G, Müller C, Kammann CI (2017) Biochar reduced nitrate leaching and improved soil moisture content without yield improvements in a four-year field study. Agric Ecosyst Environ 237:80–94. https://doi.org/10.1016/j.agee.2016.12.019
Hale L, Curtis D, Azeem M, Montgomery J, Crowley DE, McGiffen ME (2021) Influence of compost and biochar on soil biological properties under turfgrass supplied deficit irrigation. Appl Soil Ecol 168:104134. https://doi.org/10.1016/j.apsoil.2021.104134
Hu HW, Zhang LM, Yuan CL, Zheng Y, Wang JT, Chen DL, He JZ (2015) The large-scale distribution of ammonia oxidizers in paddy soils is driven by soil pH, geographic distance, and climatic factors. Front Microbiol 6:938. https://doi.org/10.3389/fmicb.2015.00938
Jiang ZW, Yang SH, Pang QQ, Xu Y, Chen X, Sun X, Qi ST, Yu WQ (2021) Biochar improved soil health and mitigated greenhouse gas emission from controlled irrigation paddy field: Insights into microbial diversity. J Clean Prod 318:128595. https://doi.org/10.1016/j.jclepro.2021.128595
Kang WL, Tai XS, Li SW, Dong K, Liu GX, Zhang W (2013) Research on the number of nitrogen-fixing microorganism and community structure of nitrogen-fixing (nifH) genes in the alkali soils of alpine steppe in the Qilian mountains. J Glaciol Geocryol 35(1):208–216. https://doi.org/10.7522/j.issn.1000-0240.2013.0025
Lai TV, Farquharson R, Denton MD (2019) High soil temperatures alter the rates of nitrification, denitrification and associated N2O emissions. J Soils Sediments 19:2176–2189. https://doi.org/10.1007/s11368-018-02238-7
Lan T, Huang YX, Song X, Deng OP, Zhou W, Luo L, Tang XY, Zeng J, Chen GD, Gao XS (2022) Biological nitrification inhibitor co-application with urease inhibitor or biochar yield different synergistic interaction effects on NH3 volatilization, N leaching, and N use efficiency in a calcareous soil under rice cropping. Environ Pollut 293:118499. https://doi.org/10.1016/j.envpol.2021.118499
Li HF, Abbas T, Cai M, Zhang QC, Wang JW, Li Y, Di HJ, Tahir M (2021) Cd bioavailability and nitrogen cycling microbes interaction affected by mixed amendments under paddy-pak choi continued planting. Environ Pollut 275:116542. https://doi.org/10.1016/j.envpol.2021.116542
Liu HF, Wu X, Wang Q, Wang S, Liu D, Liu GH (2017) Responses of soil ammonia oxidation and ammonia-oxidizing communities to land-use conversion and fertilization in an acidic red soil of southern China. Eur J Soil Biol 80:110–120. https://doi.org/10.1016/j.ejsobi.2017.05.005
Liu Q, Liu B, Zhang Y, Hu T, Lin Z, Liu G, Wang X, Ma J, Wang H, Jin H, Ambus P, Amonette JE, Xie Z (2019b) Biochar application as a tool to decrease soil nitrogen losses (NH3 volatilization, N2O emissions, and N leaching) from croplands: options and mitigation strength in a global perspective. Glob Chang Biol 25(6):2077e2093. https://doi.org/10.1111/gcb.14613
Liu XY, Liu C, Gao WH, Xue C, Guo ZH, Jiang L, Li F, Liu Y (2019a) Impact of biochar amendment on the abundance and structure of diazotrophic community in an alkaline soil. Sci Total Environ 688(20):944–951. https://doi.org/10.1016/j.scitotenv.2019.06.293
Ma LJ, Zhang HM, Hou ZA, Min W (2019) Effects of long-term saline water drip irrigation on the abundance and community structure of ammonia oxidizers. Journal of Agro-Environment. Science 38(12):2797–2807. https://doi.org/10.11654/jaes.2019-0604
Maroušek J, Rowland Z, Valášková K, Král P (2020) Techno-economic assessment of potato waste management in developing economies. Clean Techn Environ Policy 22(12):937–944. https://doi.org/10.1007/s10098-020-01835-w
Meng Y, Wang H, Song YU, Liu Z, Zhao C, Tao Y (2014) Effect of biochar on nitrogen forms and related microorganisms of rhizosphere soil of seedling maize. Chin J Eco-Agric 22(3):270–276. https://doi.org/10.3724/SP.J.1011.2014.30750
Mierzwa-Hersztek M, Klimkowicz-Pawlas A, Gondek K (2018) Influence of poultry litter and poultry litter biochar on soil microbial respiration and nitrifying bacteria activity. Waste and Biomass Valorization 9(3):379–389. https://doi.org/10.1007/s12649-017-0013-z
Ongley ED, Zhang XL, Yu T (2010) Current status of agricultural and rural non-point source pollution assessment in China. Environ Pollut 158(5):1159–1168. https://doi.org/10.1016/j.envpol.2009.10.047
Onodera Y, Nakagawa T, Takahashi R, Tokuyama T (2010) Seasonal change in vertical distribution of ammonia-oxidizing archaea and bacteria and their nitrification in temperate forest soil. Microbes Environ 25(1):28–35. https://doi.org/10.1264/jsme2.ME09179
Petra M, Ellen K, Bernd M (2003) Structure and function of the soil microbial community in a long-term fertilizer experiment. Soil Biol Biochem 35(3):453–461. https://doi.org/10.1016/S0038-0717(02)00297-3
Prommer J, Wanek W, Hofhansl F, Trojan D, Offre P, Urich T, Schleper C, Sassmann S, Kitzler B, Soja G, Hood-Nowotny RC (2014) Biochar decelerates soil organic nitrogen cycling but stimulates soil nitrification in a temperate arable field trial. PLoS One 9(1):e863881. https://doi.org/10.1371/journal.pone.0086388
Prosser JI, Nicol GW (2012) Archaeal and bacterial ammonia-oxidisers in soil: the quest for niche specialisation and differentiation. Trends Microbiol 20:523–531. https://doi.org/10.1016/j.tim.2012.08.001
Song YJ, Zhang XL, Ma B, Chang SX, Gong J (2014) Biochar addition affected the dynamics of ammonia oxidizers and nitrification in microcosms of a coastal alkaline soil. Biol Fertil Soils 50(2):321–332. https://doi.org/10.1007/s00374-013-0857-8
Srinivasarao CH, Venkateswarlu B, Lal R, Singh AK, Kundu S, Vittal KPR, Patel JJ, Patel MM (2014) Long-term manuring and fertilizer effects on depletion of soil organic carbon stocks under pearl millet-cluster bean-castor rotation in western India. Land Degrad Dev 25:173–183. https://doi.org/10.1002/ldr.1158
Stávková J, Maroušek (2021) Novel sorbent shows promising financial results on P recovery from sludge water. Chemosphere. 276:130097. https://doi.org/10.1016/j.chemosphere.2021.130097
Wang B, Zhao SS, Qin ZJ, Gao Q, Lou YJ, Liu SX (2016) Effect of biochar on adsorption-desorption characteristics of nitrate nitrogen in black soil. J Agro-Environ Sci 35(1):115–121. https://doi.org/10.11654/jaes.2016.01.016
Wang C, Chen D, Shen J, Yuan Q, Fan F, Wei W, Li Y, Wu J (2021) Biochar alters soil microbial communities and potential functions 3-4 years after amendment in a double rice cropping system. Agric Ecosyst Environ 311:107291. https://doi.org/10.1016/j.agee.2020.107291
Wang Q, Chen X, Man YU, Shen A (2019) Research progress on effects of straw returning on nitrogen cycling microbes and functional genes in paddy soil. Acta Agriculturae Zhejiangensis 31(2):333–342. https://doi.org/10.3969/j.issn.1004-1524.2019.02.20
Wu Z, Zhang QQ, Zhang X, Duan PP, Yan XY, Xiong ZQ (2020) Biochar-enriched soil mitigated N2O and NO emissions similarly as fresh biochar for wheat production. Sci Total Environ 701:134943. https://doi.org/10.1016/j.scitotenv.2019.134943
Wu Z, Zhang X, Dong YB, Xu X, Xiong ZQ (2018) Microbial explanations for field-aged biochar mitigating greenhouse gas emissions during a rice-growing season. Environ Sci Pollut Res Int 25:31307–31317. https://doi.org/10.1007/s11356-018-3112-x
Yan TT, Xue JH, Zhou ZD, Wu YB (2022) Biochar and compost amendments alter the structure of the soil fungal network in a karst mountainous area. Land Degrad Dev 33(5):685–697. https://doi.org/10.1002/ldr.4148
Yang D, Xiao X, He N, Zhu WB, Liu MD, Xie GX (2020) Effects of reducing chemical fertilizer combined with organic amendments on ammonia-oxidizing bacteria and archaea communities in a low-fertility red paddy field. Environ Sci Pollut Res 27(3):29422–29432. https://doi.org/10.1007/s11356-020-09120-5
Yang SH, Peng SZ, Xu JZ, Hou HJ, Gao XL (2013) Nitrogen loss from paddy field with different water and nitrogen managements in Taihu Lake Region of China. Commun Soil Sci Plant Anal 44(16):2393–2407. https://doi.org/10.1080/00103624.2013.803564
Yang SH, Xiao YN, Xu JZ (2018) Organic fertilizer application increases the soil respiration and net ecosystem carbon dioxide absorption of paddy fields under water-saving irrigation. Environ Sci Pollut Res 25:9958–9968. https://doi.org/10.1007/s11356-018-1285-y
Yang YD, Wang ZM, Hu YG, Zeng ZH (2017) Irrigation frequency alters the abundance and community structure of ammonia-oxidizing archaea and bacteria in a northern Chinese upland soil. Eur J Soil Biol 83:34–42. https://doi.org/10.1016/j.ejsobi.2017.09.005
Yu YL, Yang LZ, Alfred OO, Xue LH, He SY (2017) Influence of calcium carbonate and biochar addition on soil nitrogen retention in acidified vegetable Soil. Environmental. Science 38(9):3851–3859. https://doi.org/10.13227/j.hjkx.201702194
Zhang JB, Cheng Y, Cai ZC (2019a) The mechanisms of soil regulating nitrogen dynamics. Advances. Earth Sci 34(1):11–19. https://doi.org/10.11867/j.issn.1001-8166.2019.01.0011
Zhang JP, Zhou XH, Chen L, Chen ZG, Chu JY, Li YM (2016) Comparison of the abundance and community structure of ammonia oxidizing prokaryotes in rice rhizosphere under three different irrigation cultivation modes. World J Microbiol Biotechnol 32(5):85. https://doi.org/10.1007/s11274-016-2042-3
Zhang X, Duan P, Wu Z, Xiong Z (2019) Aged biochar stimulated ammonia-oxidizing archaea and bacteria-derived N2O and NO production in an acidic vegetable soil—ScienceDirect. Sci Total Environ 687:433–440. https://doi.org/10.1016/j.scitotenv.2019.06.128
Zhao ZT, Zhang YS, Sun P, Wang Q, Ruan YZ (2022) Effects of biochar and chemical fertilizer amendment on diazotrophic abundance and community structure in rhizosphere and bulk soils. Environ Sci Pollut Res. https://doi.org/10.1007/s11356-022-20086-4
Zhe NA, Wang XY, Du YA, Charles SM, Shang XS (2021) Critical period and pathways of water borne nitrogen loss from a rice paddy in northeast China. Sci Total Environ 753:142116. https://doi.org/10.1016/j.scitotenv.2020.142116
Acknowledgements
I would like to thank all research participants included in this paper for their efforts to help gather data on the relevant results. The support from Professor Yang’s funds is also acknowledged.
Funding
This work was supported by the Fundamental Research Funds for the Central Universities (B210204016), the National Natural Science Foundation of China (51879076), and the Water Conservancy Science and Technology Project of Jiangxi Province (201921ZDKT06, 202124ZDKT09).
Author information
Authors and Affiliations
Contributions
JH: conceptualization, writing—original draft, data curation. JW: investigation, writing—review and editing. SY: funding acquisition, writing—review and editing. SQ: investigation, validation. ZJ: methodology, data curation. HD: methodology, data curation. JZ: data curation, investigation.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Responsible Editor: Diane Purchase
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hu, J., Wang, J., Yang, S. et al. Soil nitrogen functional transformation microbial genes response to biochar application in different irrigation paddy field in southern China. Environ Sci Pollut Res 30, 7770–7785 (2023). https://doi.org/10.1007/s11356-022-22728-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-022-22728-z