Abstract
Agroforestry practices aim to achieve environmentally friendly land use. Fungi play a primarily role in soil organic carbon and nutrient maintenance, while the response of the soil fungi community to land use changes is little explored. Here, a high-throughput sequencing method was applied to understand the fungal community structure distinction in ginkgo agroforestry systems and adjacent croplands and nurseries. Our results showed that the agroforestry systems achieved better soil fertility and carbon contents. The agroforestry practices significantly altered the composition of soil fungal communities comparing with pure gingko plantation, adjacent cropland, and nursery. The dominant fungal phyla were always Ascomycota and Basidiomycota. The relative abundance of Ascomycota was correlated with the TN and AP, while the abundance of Basidiomycota was negatively correlated with the TN and NN. The soil organic carbon, total nitrogen, and nitrate nitrogen explained 59.80% and 63.36% of the total variance in the fungal community composition in the topsoil and subsoil, and the available phosphorus also played a key role in the topsoil. Considering soil fertility maintenance and fungal community survival and stability, the agroforestry systems achieved better results, and the ginkgo and wheat system was the best among the five planting systems we studied. In the ginkgo and wheat system, applying readily available mineral nitrogen fertilizer either alone or in combination with organic amendments will improve the soil quality and fertility.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Soil is the most important and complex ecosystem on Earth. Soil microbes are important components of the soil ecosystem and global biodiversity, contributing significantly to terrestrial ecosystem functions by mediating carbon (C) and nitrogen (N) pools and nutrient cycling (Daniel 2005; Bardgett and van der Putten 2014; Delgado-Baquerizo et al. 2016). Owing to their high abundance and the diversity and their critical ecological roles, bacteria, fungi, and archaea are receiving more and more attention (Liu et al. 2015b; Fierer 2017). Fungi are key components of soil biota, and they deliver key ecosystem services such as antagonizing pathogens, stimulating decomposition processes, and improving plant nutrient acquisition (Buee et al. 2009; Ehrmann and Ritz 2014). For example, ectomycorrhizal fungi infect plant roots and form highly branched hyphae, thereby improving the ability of plants to access nutrients and water (Dhilhon and Anderson 1993; Lehto and Zwiazek 2011). Arbuscular mycorrhizal fungi are primarily involved in transporting nutrients to the plant and availing themselves of carbon compounds to promote their microbial activities (Smith and Read 2008). There is evidence that fungi are the dominant degraders of refractory organic matter and pollutants and they are mediators of slower carbon cycling (Jeffries et al. 2003; Rinnan and Bååth 2009). Soil fungi act as potentially valuable indicators and participants in soil fertility maintenance and plant productivity improvement (van der Heijden et al. 1998; Chen et al. 2017). Moreover, fungi have certain positive and negative effects on plant communities by improving the adaptability of certain plant species through their ability to access nutrients, by deteriorating the adaptability of certain plants by causing plant disease, or by altering resource allocation patterns (Rodriguez et al. 2009).
Fungal richness and diversity are believed to be ecosystem-dependent and plant-dependent because of the soil physical and chemical properties and root exudate specificity; these factors are vital for the survival of soil microbes (Schappe et al. 2017). Recent studies have suggested that the community composition of soil fungi is significantly correlated with the plant species richness, mean annual temperature, water availability, litter quality, soil pH, soil organic carbon (SOC), nitrate, and total nitrogen (TN) in grassland ecosystems and forest soils (Chen et al. 2017; Huhe et al. 2017). These studies were primarily focused on fungal community composition patterns across large geographical distances, and most of these studies were focused on the surface soil (0–20 cm). Compared with annual herbs, perennial plants customarily produce more abundant resources through rhizodeposition, and they favor larger populations of more diverse microbes. Studies have shown that soil properties play key roles in the changes in soil fungal communities, such as fungal communities that vary with the soil depth (Prober et al. 2015) and soil nutrient contents (Lin et al. 2012; Liu et al. 2012). Research by Kim et al. (2015) indicated that nitrogen fertilizer applications significantly changed the fungal community structure. Additionally, many studies have demonstrated that plant community characteristics and local climate were also key factors (Peay et al. 2010; Li et al. 2018).
Pure agricultural systems are negatively affected by land use pressure, climate deterioration, and water resource depletion, which then threaten food production (Anderson and Zerriffi 2012). Finding better solutions to achieve sustainable development and environmentally friendly land use is of critical importance. Agroforestry is a practice that artificially integrates trees with crops and/or livestock to achieve diverse benefits, such as conserving biodiversity, enhancing ecosystem service provision, and counteracting resource degradation, making it a good candidate for addressing food security, water security, sustainable livelihoods, and climate change objectives (McNeely and Schroth 2006; Nair 2008; Koohafkan et al. 2012). In addition, the trees and the intercrops utilize the available resources such as sunlight, underground water, and soil nutrients more effectively, and intercropping reduces the incidence of diseases and insect pests (Yu et al. 2015). Changes in land use will lead to variations in the physical structure of the environment and to variations in plants and herbs (e.g., convert cropland to grassland, cropland to forest, and grassland to forest), causing variations in the plant litter species and microenvironments; these variations will affect the soil microorganism biodiversity and cause significant consequences for ecosystem functions (Loreau et al. 2001; Meier and Bowman 2008; Poeplau and Don 2013). Despite the importance of its ecological benefit, there is a lack of information on the effect of agroforestry practices on the soil fungal community composition. As a whole, tree and intercrop litter and root exudates provide important carbon resources for soil microbial growth, and changes in these plant-derived organics affect soil microbial communities. There is ample evidence to suggest that increased organic content, improved litter quality and quantity, and improved soil fertility in agroforestry systems were observed comparing with cropland which will be beneficial for the soil enzyme activity, which is closely related to fungal function (Caldwell 2005; Udawatta et al. 2009).
Ginkgo biloba L. is known as a “living fossil” and is valuable for a wide variety of uses. Ginkgo is native to China and has been introduced to many areas of the world because of its broad environmental adaptability (Major 1967). In China, the primary means of ginkgo cultivation is agroforestry. The agroforestry practices are primarily to help diversify farm incomes, on one hand provide short-term gain through the intercropping crops or landscape seedling products, and on the other harvest ginkgo trees as ornamental trees or timber products in the long run. We have little information about how fungal communities respond to different intercropping strategies, and previous studies have generally been restricted to the primary rooting zone (0–20 cm). Therefore, this study attempts to investigate the effect of agroforestry practices on the soil fungi community α-diversity down to a soil depth of 40 cm. High-throughput sequencing methods were applied to elucidate the fungal community composition and structure in five planting systems and to explore their interactions with soil physical and chemical properties. We hypothesized that (1) agroforestry practices will modify the soil fungal community composition and increase fungal diversity and that (2) the soil properties would explain a considerable proportion of the variations among the planting systems.
Materials and methods
Experimental sites and soil sampling
The study field site is in the Yellow Sea Forest Park in Dongtai County, Jiangsu Province, China (32° 51′–32° 52′ N, 120° 48′–120° 50′ E). The study area is 5 m above sea level. It has a northern subtropical monsoon climate with a mean annual temperature and an annual rainfall of 15.0 °C and 1061.2 mm, respectively (Guo et al. 2016). The field soil was classified as a coastal sandy saline-alkali soil.
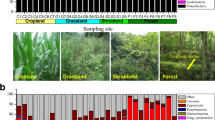
Local farmers usually interplant ginkgo with crops and seedlings; ginkgo plantations were established in 2002, and agroforestry systems were applied in 2004. Before the ginkgo agroforestry systems were applied, summer maize and winter wheat rotation systems were applied prior to 2002. We selected five popular planting systems for this study, namely, G (a pure ginkgo system), W (a wheat (Triticum aestivum L.) field), M (a pure Metasequoia (Metasequoia glyptostroboides Hu et Cheng) seedling system), GW (a ginkgo and wheat system), and GM (a ginkgo and Metasequoia system). The W, GW, and GM systems were in close proximity to each other. The G system and M system were in close proximity to each other and about 300 m from the other three systems. In order to avoid their interaction and stand edge effect, the plots in each planting system avoided the stand edge area. There were three replicate plots (10 m × 10 m) for each planting system, which were arrayed in a randomized block design. The ginkgo agroforestry systems were characterized by intra- and inter-row spacings of 2 m × 8 m, and the Metasequoia seedlings in the GM system were characterized by intra- and inter-row spacings of 0.8 m × 0.8 m. The pure gingko system and the Metasequoia seedling system were characterized by intra- and inter-row spacings of 3 m × 3 m and 0.8 m × 0.8 m, respectively. In the M and GM systems, metasequoias were planted in 2010. Corn was planted as a rotation crop after wheat. The farmers applied fertilizers twice per year (mid-May and mid-October). The amount of fertilizer was approximately 0.45 t ha−1 per application for a total of 0.9 t ha−1 year−1 (compound fertilizer with N:P2O5:K2O = 15:15:15). No pesticides, bactericides, or herbicides were applied to the systems.
Soil samples were collected on May 16, 2017. Within each plot, five soil samples were randomly collected using a 2-cm-diameter soil corer at depths of 0–20 cm and 20–40 cm, representing the topsoil and subsoil, respectively. The samples were mixed to generate a single soil sample for each layer (a total of 30 samples). The samples were immediately placed on ice in an icebox and were transported to the laboratory, where they were passed through a 2.0-mm sieve. After the sieving, one part was stored at 4 °C for soil physical and chemical property analyses, and another part was stored at − 81 °C for DNA extraction.
Basic physical and chemical properties of the soil
The soil pH was determined in a soil suspension with a soil:water ratio of 1:2.5 (w/v) using a portable pH meter. The SOC content was determined by digestion using the potassium dichromate oxidation-external heating method (Fan et al. 2017). The soil TN was measured using the micro-Kjeldahl method with digestion in H2SO4 followed by steam distillation. The soil ammonium-N (AN) and nitrate-N (NN) were extracted with 2 M KCl and determined using UV spectrophotometry (Shibata et al. 2011). The available P (AP) was extracted with 0.5 M NaHCO3 and determined using the molybdenum-blue method with an atomic absorption spectrophotometer. The available K (AK) (extracted by 1 M NH4OAc) concentrations were determined using a flame photometer (FP6450, Shanghai, China) (Lu 1999).
DNA extraction, PCR, and high-throughput sequencing of 18S rRNA genes
To extract the total soil DNA, 0.5 g of soil from each sample was extracted using the PowerSoil DNA Isolation Kit (Mo Bio, Solana Beach, CA, USA) following the manufacturer’s protocols. The final DNA concentration and purification were determined using a NanoDrop 2000 UV-vis spectrophotometer (Thermo Scientific, Wilmington, USA), and the DNA quality was detected by 1% agarose gel electrophoresis. Illumina sequencing was performed in triplicate using the primers SSU0817F (5′-TTAGCATGGAATAATRRAATAGGA-3′) and 1196R (5′-TCTGGACCTGGTGAGTTTCC-3′) in a thermocycler PCR system (GeneAmp 9700, ABI, USA); these 18S primers have been shown to be fungal-specific, and they target a region of the 18S rRNA gene that is variable between major taxa and can be aligned (Rousk et al. 2010). Each PCR reaction was performed in triplicate in a 20-μL mixture containing 4 μL of 5 × FastPfu Buffer, 2 μL of 2.5 mM dNTPs, 0.8 μL of each primer (5 μM), 0.4 μL of FastPfu Polymerase, and 10 ng of template DNA. The PCR reactions were conducted using the following program: 3 min of denaturation at 95 °C, 35 cycles of 30 s at 95 °C, 30 s for annealing at 55 °C, 45 s for elongation at 72 °C, and a final extension at 72 °C for 10 min. The resulting PCR products were extracted from a 2% agarose gel and further purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA), washed with Tris-HCl, and quantified using QuantiFluor™-ST (Promega, USA) according to the manufacturer’s protocol. The sequencing was conducted by Shanghai Majorbio Bio-Pharm Technology (Shanghai, China) using an Illumina MiSeq platform (San Diego, CA, USA). The raw reads in this study were deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database (accession number: SRP124182).
Bioinformatics and statistical analysis
Raw FASTQ files obtained by MiSeq sequencing were demultiplexed, quality-filtered by Trimmomatic, and merged by FLASH (Magoc and Salzberg 2011). The operational taxonomic units (OTUs) were clustered at a 97% similarity cutoff using Usearch (version 7.1 http://drive5.com/uparse/). The taxonomy of 18S rRNA gene sequence was analyzed with the RDP Classifier algorithm (http://rdp.cme.msu.edu/) against Silva (Release128 http://www.arb-silva.de) using a confidence threshold of 70%. The alpha diversity was estimated using mothur (version v.1.30.1). The rarefaction curve was estimated using mothur (version v.1.30.1), and it reflected the fungal diversity of different samples in different sequencing numbers and was drawn using R software. According to the principal of minimum sequence sample numbers, 28760 sequences per sample were selected to analyze the structure of the fungal community. A community column diagram was used to detect the community composition at the phylum and class levels, and Venn diagrams were used to detect unique and shared fungal OTUs between different planting systems, as calculated using R software (version 3.4.0, R Core Team 2017). The β-diversity was examined by principal coordinate analysis (PCoA) to estimate the fungal community structure using spearman_approx dissimilarities. The relationships between the soil fungal community structure and each soil property were analyzed by RDA using CANOCO 5.0 software. Pearson correlation analyses were calculated using R software 3.4.0 (vegan package, http://www.r-project.org).
SPSS (version 19.0, Chicago, IL, USA) was used to calculate the means and standard deviations from three replicates. The effects of the planting system and soil depth on the soil properties and alpha diversity were analyzed by two-way analysis of variance (ANOVA), and Duncan’s multiple range test was used to separate the means when a significant treatment effect was found (P < 0.05). The effects of the planting system on the soil properties and alpha diversity were analyzed by one-way ANOVA within the same soil layer (P < 0.05). The normality of the distribution and the homogeneity of the variance were tested before performing the ANOVA.
Results
Soil properties
The major physical and chemical characteristics of the soil from the five planting systems are summarized in Table 1. The pH of the five planting systems were ranged from 7.94 to 8.16 in the topsoil and 8.15 to 8.39 in the subsoil, respectively. The SOC (28.15 and 10.19 g kg−1) and TN (1.12 and 0.65 g kg−1) were highest in the GW system, both in the topsoil and in the subsoil. The AN (10.97 mg kg−1) in the GM system was significantly higher than that in the other systems in the topsoil; the significantly higher AN (7.80 mg kg−1) was observed in the G system. The relatively higher NN contents were obtained in the GW and W systems. The two-way ANOVA indicated that the planting system and soil depth significantly affected the pH, SOC, AN, NN, AP, and AK (P < 0.01) (Table 1). Only the soil depth significantly affected the TN (P < 0.01). The one-way ANOVAs conducted within the same soil layer indicated that the SOC, AN, NN, and AK were significantly different within the five planting systems in both the topsoil and the subsoil, while TN and AP only showed significant differences in the topsoil (P < 0.05). The TN and NN were significantly lower in the G system than those in the other systems (P < 0.05).
Fungal α-diversity
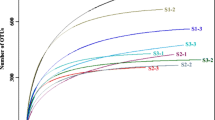
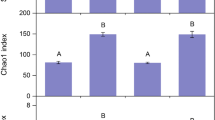
The MiSeq platform was used to reveal the α-diversity of the soil fungal communities in the different planting systems. A total of 556443 and 561702 quality-filtered and optimized 18S rRNA gene sequences were obtained from the topsoil and subsoil, respectively (Table S1). Four hundred and fifty fungal OTUs were obtained from the topsoil and subsoil. A Venn diagram analysis of the OTUs at 97% sequence similarity showed that all the systems shared 93 and 77 OTUs and that these OTUs accounted for 20.67 and 17.11% of the total OTUs observed in the topsoil and subsoil, respectively (Supplementary Fig. 1). The richness index values (ACE and Chao1; α-diversity) and the diversity (Shannon; α-diversity) index values were compared in the fungal community among the different systems within the same soil layer (Table 2). The Chao1 and ACE estimators indicated that the G system exhibited a significantly lower fungal OTU number than the M and GW systems in the topsoil (P < 0.05). The higher Shannon index of the M and GW systems resulted in greater fungal diversity than those in the G and W systems in the subsoil (P < 0.05). When considering the α-diversity indices between the topsoil and subsoil within the same planting systems, only the OTUs, Chao1, and ACE in GW system and Shannon index in GM system were significantly different (P < 0.05); significantly higher values were obtained in the topsoil. The coverage indexes were always greater than 0.999, which reflected sequencing results large enough to represent the actual situation of each soil sample (Table 2). Additionally, the rarefaction curves and the Shannon-Wiener index of the fungal communities supported the idea that the sequencing data represented the actual situation of each soil sample (Supplementary Fig. 2).
Fungal community composition
Our results showed that Ascomycota (accounting for averages of 63.88% and 52.41% of the fungal sequences in the topsoil and subsoil, respectively) was the most dominant phylum, followed by Basidiomycota (accounting for averages of 11.18% and 10.94% of the fungal sequences in the topsoil and subsoil, respectively). These trends were largely consistent among the five planting systems (Fig. 1). However, differences in the relative abundances among the five planting systems were observed in the same soil layer (P < 0.05, Supplementary Fig. 3). At the class level, Sordariomycetes was the most abundant class (40.87% of fungal sequences in the topsoil), although it accounted for a relatively smaller percentage in the subsoil (27.64%), followed by Dothideomycetes (Supplementary Fig. 4). The PCoA clearly distinguished among the fungal communities in the subsoil according to the five planting systems, while the GM and M systems did not differ from one another in the topsoil (Fig. 2a and b). The total variance explained by the first two axes (PC1 and PC2) were 43.05 and 42.09% of the fungal communities in the topsoil and subsoil in the five planting systems sampled, respectively (Fig. 2).
Relationship between the fungal community structure and soil characteristics
Differences in the fungal community structures and environmental characteristics were observed among the five planting systems. Therefore, to investigate whether the fungal community structure and soil physical and chemical characteristics are related in both the topsoil and the subsoil, the primary soil physical and chemical characteristics (including the pH, SOC, TN, AN, NN, AP, and AK) were included in the redundancy analysis (RDA). The RDA of the MiSeq data and the soil physical and chemical characteristics revealed that the TN (P < 0.01), NN (P < 0.05), and AP (P < 0.01) explained the significant changes in the fungal community composition in the topsoil, while the TN (P < 0.01) and NN (P < 0.01) explained the variation in the subsoil. The first two axes of the RDA accounted for 59.80% and 63.36% of the total variance in the fungal community composition in the topsoil and subsoil, respectively (Fig. 3). A Pearson correlation analysis was conducted, and the results showed that the relative abundance of Ascomycota was significantly correlated with the TN and AP (P < 0.01, r = 0.680, and P < 0.01, r = 0.769), while the abundance of Basidiomycota was negatively correlated with the TN and NN (P < 0.01, r = − 0.728, and P < 0.01, r = − 0.68) (Fig. 4).
Discussion
Soil physical and chemical properties have always received the most attention from foresters who wish to assess site conditions and enhance stand productivity (Schoenholtz et al. 2000). In this study, significant differences were found in all the selected soil physical and chemical characteristics, except for TN, among the five planting systems. Higher SOC, TN, and NO3–N contents were observed in the GW system and a higher NH4–N content was observed in the GM system. Agroforestry systems are both structurally and functionally more complicated than arable land and forests, resulting in better nutrient cycles and increased carbon sequestration (Nair 2007; Torralba et al. 2016). Soil preparation before sowing (litter turned over to the subsoil and litter decomposition) and the abundant resources produced by perennial plants (ginkgo) may contribute to the significantly higher SOC content in the subsoil within the GW system (Liang et al. 2012). In addition, all the selected basic soil properties were significantly depth-dependent. The significantly higher SOC and TN contents in the topsoil may be caused by the relatively higher plant litter and crop residue input on the soil surface, their gradual decomposition, and wheat rhizodeposition (Müller et al. 2016), and the lower TN content in the subsoil may be explained by higher rates of N losses (Gu et al. 2017; Mushinski et al. 2017). In accordance with previous studies, the NH4–N and NO3–N decreased with the depth in forest soils in which ammonification typically occurs (Liu et al. 2015a; Tanner et al. 2016). Considering the low solubility of phosphorus in slightly alkaline soil, the AP in the topsoil was higher than that in the subsoil (El-Baruni and Olsen 1979). Additionally, agroforestry systems can potentially improve the P cycling relative to pure plantations by producing much more stable P fractions (Wang et al. 2017a). The relatively lower AK contents in the subsoil might have been due to the high mobility of K in the soil. In our study, the basic soil properties were system-dependent and depth-dependent.
The fungal diversities in the topsoil and subsoil showed similar variation patterns and were also similar to the variation patterns in soil nutrients within the five systems. In accordance with previous studies, this result showed that the soil nutrient content plays a key role in the soil microbial survival, species composition, and metabolism (Kerfahi et al. 2016; Zhang et al. 2016). Comparing with pure planting systems, clearly, the interplanting crops or seedlings that supplied extra available substrate that was exuded by roots will be beneficial for fungal growth. Different plant species will cause different rhizosphere nutrient availability and thus lead to different fungal phylogenetic compositions (Wang et al. 2017b). In addition, Wang et al. (2017a) confirmed that agroforestry systems behaved more like natural forests than pure plantations. The plant and fungal richness were positively correlated in most regions at a fine scale, because different planting systems directly provided different quantities and qualities of organic matter and indirectly changed the soil properties, thus affecting the soil fungal communities (Tedersoo et al. 2014a; Zhou et al. 2017). These correlations will be overridden at a broader spatial scale by differences in environmental factors (Tedersoo et al. 2014b). We also found that the Shannon index showed a decreasing trend with the increasing soil depth in three of the systems. This decrease could be explained by the limited nutrient resources in the subsoil, resulting in lower nutrient source chemical heterogeneity, with a consequent decrease in the fungal alpha diversity (Voříšková et al. 2014). The C and N availability was the primary factor determining the microbial community abundance (Yoshitake et al. 2013).
Fungi are commonly involved in the cellulose decomposition process, and the saprotrophic Ascomycota and Basidiomycota are the key participants (Baldrian et al. 2012). The higher abundance of Ascomycota obtained in our study was similar to that of previous studies, which were conducted in semi-arid grasslands (Porras-Alfaro et al. 2011), temperate grasslands (Prober et al. 2015), and global dry lands (Maestre et al. 2015). However, our results differed from a recent global soil survey that showed a relatively higher abundance of Basidiomycota (55.7%) and lower abundance of Ascomycota (31.3%) in natural terrestrial ecosystems (Tedersoo et al. 2014a). As described by Chen et al. (2017), Ascomycota may be capable of tolerating stressful conditions and achieving a more efficient resource use in challenging environments (Hannula et al. 2012; Chen et al. 2017). The proportion of Ascomycota decreased with the increasing soil depth. By contrast, the proportion of Basidiomycota was lower in the topsoil than in the subsoil in most planting systems, which is consistent with previous studies (Clemmensen et al. 2013; Voříšková et al. 2014). The predominant phylum of the fungal communities was generally consistent, but the specific ratio of different fungal phyla varied across the five planting systems. When considering the class level, a majority of Sordariomycetes species are saprotrophic, and the lower proportion in the G system was most likely due to the lower litter input and plant diversity. Additionally, Sordariomycetes (order Sordariales) are also considered to be primarily straw residue decomposers (Ma et al. 2013). The accumulated evidence indicates that many of the Dothideomycetes contribute to a tolerance to extreme environmental conditions, and some species produce enzymes that help degrade rocks (Ohm et al. 2012; Qin et al. 2014; Treseder and Lennon 2015). Notably, the relative dominance of saprotrophic fungi was beneficial for the allocation of photosynthetic products between aboveground and underground portions in forests (Högberg et al. 2003). Overall, the fungal composition structures in the soils of the agroforestry systems were probably driven by tree leaf litter and intercrop residue decomposition processes.
The importance of soil properties in determining the soil microbial abundance and composition has been reported in numerous studies (Kerfahi et al. 2016; Zhang et al. 2016). This importance can be explained by the key roles fungi have played in soil carbon and nitrogen cycling (Read and Perez-Moreno 2003). In the present study, the RDA analysis showed that the major soil properties contributed over 50% of the shift in the fungal community, indicating that these factors were essential in constructing the fungal community composition, which was in accordance with our hypothesis. Significant direct relationships between fungal community structures and soil TN and NO3–N supported previous findings by Huhe et al. (2017). Additionally, in the study of Schmidt et al. (2014), the fungal survival relies more on C and N sources than bacteria and archaea. In this study, wheat straw that was returned to the field was a source of C and N, which is necessary for the survival of plants and microbes, and it increased the organic matter. For example, wheat straw is supplied in agroforestry systems as an organic substrate, although it is characterized as having a relatively low quality because it has a relatively high C:N ratio (~ 85:1) (Guo et al. 2018). Positive correlations between the C:N ratio and fungal community abundance and composition were reported by Schlatter et al. (2017). Fungal communities are thought to respond more rapidly to the existence of wheat residues when a higher N content is also available (Kerfahi et al. 2016). Additionally, we also found that AP was significantly related to the variations in the soil fungal community structure in the topsoil. This result was consistent with a previous study showing that the soil fungal diversity was closely related to different P fractions (labile P, moderate labile P, and Ca P) (Wang et al. 2017a). A Pearson correlation analysis was conducted, and it showed the positive correlation between the relative abundance of Ascomycota and TN and AP, while the abundance of Basidiomycota was negatively correlated with TN and NO3–N. Together, the soil properties have lasting effects on the soil fungal communities, and soil N and P play key roles in the construction of soil fungal communities. Similar results have been reported, showing that fungal communities are inextricably bound up with the soil C, N, and P cycling (Li et al. 2018). According to previous studies, the litter with different properties and the root exudates should be responsible for the different fungal community composition in the topsoil and subsoil, respectively (Du et al. 2017). Taken together, our study elucidated that different agroforestry practices have certain impact on the soil nutrient contents as well as the composition of fungal community both in the topsoil and in the subsoil. Agroforestry systems achieved better soil nutrients contents, and the GW system was the best planting system. The soil properties played key roles in fungal community survival, while the plant species and soil microbial community interactions should be taken into account during future studies. Besides, identification of keystone species of fungal community involved in the soil nutrient cycling will deepen our understanding and will have important directive to production practices.
Conclusions
In conclusion, we explored the changes in the soil physical and chemical properties and the distribution patterns of soil fungal communities in both the top- and subsoil in ginkgo agroforestry systems and adjacent croplands and nurseries. Our results suggest that most soil properties are system-dependent and depth-dependent. The agroforestry systems achieved better soil fertility and carbon stocks, and the GW system was the best one among these systems. Our results indicated that Ascomycota and Basidiomycota were the most dominant phyla and that Sordariomycetes and Dothideomycetes were the most dominant classes. The soil C, N, and P played key roles in the variations in soil fungal communities. In addition, significant direct relationships between the fungal community structures and soil TN and NO3–N were elucidated in the present study. Considering the soil fertility and fungal community structure and diversity, agroforestry systems achieved better results, and the GW system was the best. Additionally, applying nitrogen fertilizer either alone or in combination with organic amendments may drive the shifts in soil fungal communities. Further work is needed to characterize the seasonal variations in fungal community abundance and composition with the litter layer included.
References
Anderson EK, Zerriffi H (2012) Seeing the trees for the carbon: agroforestry for development and carbon mitigation. Clim Chang 115:741–757. https://doi.org/10.1007/s10584-012-0456-y
Baldrian P, Kolarik M, Stursova M, Kopecky J, Valaskova V, Vetrovsky T, Zifcakova L, Snajdr J, Ridl J, Vlcek C, Voriskova J (2012) Active and total microbial communities in forest soil are largely different and highly stratified during decomposition. Isme J 6:248–258. https://doi.org/10.1038/ismej.2011.95
Bardgett RD, van der Putten WH (2014) Belowground biodiversity and ecosystem functioning. Nature 515:505–511. https://doi.org/10.1038/nature13855
Buee M, Reich M, Murat C, Morin E, Nilsson RH, Uroz S, Martin F (2009) 454 Pyrosequencing analyses of forest soils reveal an unexpectedly high fungal diversity. New Phytol 184:449–456. https://doi.org/10.1111/j.1469-8137.2009.03003.x
Caldwell BA (2005) Enzyme activities as a component of soil biodiversity: a review. Pedobiologia 49:637–644. https://doi.org/10.1016/j.pedobi.2005.06.003
Chen Y-L, Xu TL, Veresoglou SD, Hu HW, Hao ZP, Hu YJ, Liu L, Deng Y, Rillig MC, Chen BD (2017) Plant diversity represents the prevalent determinant of soil fungal community structure across temperate grasslands in northern China. Soil Biol Biochem 110:12–21. https://doi.org/10.1016/j.soilbio.2017.02.015
Clemmensen KE, Bahr A, Ovaskainen O, Dahlberg A, Ekblad A, Wallander H, Stenlid J, Finlay RD, Wardle DA, Lindahl BD (2013) Roots and associated fungi drive long-term carbon sequestration in boreal forest. Science 339:1615–1618. https://doi.org/10.1126/science.1231923
Daniel R (2005) The metagenomics of soil. Nat Rev Microbiol 3:470–478. https://doi.org/10.1038/nrmicro1160
Delgado-Baquerizo M, Maestre FT, Reich PB, Jeffries TC, Gaitan JJ, Encinar D, Berdugo M, Campbell CD, Singh BK (2016) Microbial diversity drives multifunctionality in terrestrial ecosystems. Nat Commun 7:10541. https://doi.org/10.1038/ncomms10541
Dhilhon SS, Anderson RC (1993) Growth dynamics and associated mycorrhizal fungi of little bluestem grass [Schizachyrium scoparium (Michx.) Nash] on burned and unburned sand prairies. New Phytol 123:77–91. https://doi.org/10.1111/j.1469-8137.1993.tb04533.x
Du C, Geng Z, Wang Q, Zhang T, He W, Hou L, Wang Y (2017) Variations in bacterial and fungal communities through soil depth profiles in a Betula albosinensis forest. J Microbiol 55:684–693. https://doi.org/10.1007/s12275-017-6466-8
Ehrmann J, Ritz K (2014) Plant: soil interactions in temperate multi-cropping production systems. Plant Soil 376:1–29. https://doi.org/10.1007/s11104-013-1921-8
El-Baruni B, Olsen SR (1979) Effect of manure on solubility of phosphorus in calcareous soils. Soil Sci 128:219–225. https://doi.org/10.1097/00010694-197910000-00005
Fan D, Zhang Y, Qin S, Wu B (2017) Relationships between Artemisia ordosica communities and environmental factors following sand-dune stabilization in the Mu Us desert, northwest China. J For Res 28:115–124. https://doi.org/10.1007/s11676-016-0289-z
Fierer N (2017) Embracing the unknown: disentangling the complexities of the soil microbiome. Nat Rev Microbiol 15:579–590. https://doi.org/10.1038/nrmicro.2017.87
Gu Y, Wang Y, Lu S, Xiang Q, Yu X, Zhao K, Zou L, Chen Q, Tu S, Zhang X (2017) Long-term fertilization structures bacterial and archaeal communities along soil depth gradient in a paddy soil. Front Microbiol 8:1516. https://doi.org/10.3389/fmicb.2017.01516
Guo J, Wu Y, Wang B, Lu Y, Cao F, Wang G (2016) The effects of fertilization on the growth and physiological characteristics of Ginkgo biloba L. Forests 7:293. https://doi.org/10.3390/f7120293
Guo J, Wang G, Geng Q, Wu Y, Cao F (2018) Decomposition of tree leaf litter and crop residues from ginkgo agroforestry systems in Eastern China: an in situ study. J Soils Sediments 18:1424–1431. https://doi.org/10.1007/s11368-017-1870-6
Hannula SE, Boschker HT, de Boer W, van Veen JA (2012) 13C pulse-labeling assessment of the community structure of active fungi in the rhizosphere of a genetically starch-modified potato (Solanum tuberosum) cultivar and its parental isoline. New Phytol 194:784–799. https://doi.org/10.1111/j.1469-8137.2012.04089.x
Högberg MN, Bååth E, Nordgren A, Arnebrant K, Högberg P (2003) Contrasting effects of nitrogen availability on plant carbon supply to mycorrhizal fungi and saprotrophs – a hypothesis based on field observations in boreal forest. New Phytol 160:225–238. https://doi.org/10.1046/j.1469-8137.2003.00867.x
Huhe CX, Hou F, Wu Y, Cheng Y (2017) Bacterial and fungal community structures in Loess Plateau grasslands with different grazing intensities. Front Microbiol 8:606. https://doi.org/10.3389/fmicb.2017.00606
Jeffries P, Gianinazzi S, Perotto S, Turnau K, Barea J-M (2003) The contribution of arbuscular mycorrhizal fungi in sustainable maintenance of plant health and soil fertility. Biol Fertil Soils 37:1–16. https://doi.org/10.1007/s00374-002-0546-5
Kerfahi D, Tripathi BM, Dong K, Go R, Adams JM (2016) Rainforest conversion to rubber plantation may not result in lower soil diversity of bacteria, fungi, and nematodes. Microb Ecol 72:359–371. https://doi.org/10.1007/s00248-016-0790-0
Kim YC, Gao C, Zheng Y, He XH, Yang W, Chen L, Wan SQ, Guo LD (2015) Arbuscular mycorrhizal fungal community response to warming and nitrogen addition in a semiarid steppe ecosystem. Mycorrhiza 25:267–276. https://doi.org/10.1007/s00572-014-0608-1
Koohafkan P, Altieri MA, Gimenez EH (2012) Green agriculture: foundations for biodiverse, resilient and productive agricultural systems. Int J Agric Sustain 10:61–75. https://doi.org/10.1080/14735903.2011.610206
Lehto T, Zwiazek JJ (2011) Ectomycorrhizas and water relations of trees: a review. Mycorrhiza 21:71–90. https://doi.org/10.1007/s00572-010-0348-9
Li S, Shakoor A, Wubet T, Zhang N, Liang Y, Ma K (2018) Fine-scale variations of fungal community in a heterogeneous grassland in Inner Mongolia: effects of the plant community and edaphic parameters. Soil Biol Biochem 122:104–110. https://doi.org/10.1016/j.soilbio.2018.04.007
Liang C, Jesus EDC, Duncan DS, Jackson RD, Tiedje JM, Balser TC (2012) Soil microbial communities under model biofuel cropping systems in southern Wisconsin, USA: impact of crop species and soil properties. Appl Soil Ecol 54:24–31. https://doi.org/10.1016/j.apsoil.2011.11.015
Lin X, Feng Y, Zhang H, Chen R, Wang J, Zhang J, Chu H (2012) Long-term balanced fertilization decreases arbuscular mycorrhizal fungal diversity in an arable soil in North China revealed by 454 pyrosequencing. Environ Sci Technol 46:5764–5771. https://doi.org/10.1021/es3001695
Liu Y, Shi G, Mao L, Cheng G, Jiang S, Ma X, An L, Du G, Johnson NC, Feng H (2012) Direct and indirect influences of 8yr of nitrogen and phosphorus fertilization on Glomeromycota in an alpine meadow ecosystem. New Phytol 194:523–535. https://doi.org/10.1111/j.1469-8137.2012.04050.x
Liu J, Sui Y, Yu Z, Shi Y, Chu H, Jin J, Liu X, Wang G (2015a) Soil carbon content drives the biogeographical distribution of fungal communities in the black soil zone of northeast China. Soil Biol Biochem 83:29–39. https://doi.org/10.1016/j.soilbio.2015.01.009
Liu X, Chen C, Wang W, Hughes JM, Lewis T, Hou E, Shen J (2015b) Vertical distribution of soil denitrifying communities in a wet sclerophyll forest under long-term repeated burning. Microb Ecol 70:993–1003. https://doi.org/10.1007/s00248-015-0639-y
Loreau M, Naeem S, Inchausti P, Bengtsson J, Grime JP, Hector A, Hooper DU, Huston MA, Raffaelli D, Schmid B, Tilman D, Wardle DA (2001) Biodiversity and ecosystem functioning: current knowledge and future challenges. Science 294:804–808. https://doi.org/10.1126/science.1064088
Lu RK (1999) Analytical methods of soil agrochemistry. China Agricultural Science and Technology Press, Beijing
Ma A, Zhuang X, Wu J, Cui M, Lv D, Liu C, Zhuang G (2013) Ascomycota members dominate fungal communities during straw residue decomposition in arable soil. PLoS One 8:e66146. https://doi.org/10.1371/journal.pone.0066146
Maestre FT, Delgado-Baquerizo M, Jeffries TC, Eldridge DJ, Ochoa V, Gozalo B, Quero JL, García-Gómez M, Gallardo A, Ulrich W, Bowker MA, Arredondo T, Barraza-Zepeda C, Bran D, Florentino A, Gaitán J, Gutiérrez JR, Huber-Sannwald E, Jankju M, Mau RL, Miriti M, Naseri K, Ospina A, Stavi I, Wang D, Woods NN, Yuan X, Zaady E, Singh BK (2015) Increasing aridity reduces soil microbial diversity and abundance in global drylands. Proc Natl Acad Sci USA 112:15684. https://doi.org/10.1073/pnas.1516684112
Magoc T, Salzberg SL (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. https://doi.org/10.1093/bioinformatics/btr507
Major RT (1967) The ginkgo, the most ancient living tree. The resistance of Ginkgo biloba L. to pests accounts in part for the longevity of this species. Science 157:1270–1273. https://doi.org/10.1126/science.157.3794.1270
McNeely JA, Schroth G (2006) Agroforestry and biodiversity conservation – traditional practices, present dynamics, and lessons for the future. Biodivers Conserv 15:549–554. https://doi.org/10.1007/s10531-005-2087-3
Meier CL, Bowman WD (2008) Links between plant litter chemistry, species diversity, and below-ground ecosystem function. Proc Natl Acad Sci USA 105:19780. https://doi.org/10.1073/pnas.0805600105
Müller K, Kramer S, Haslwimmer H, Marhan S, Scheunemann N, Butenschön O, Scheu S, Kandeler E (2016) Carbon transfer from maize roots and litter into bacteria and fungi depends on soil depth and time. Soil Biol Biochem 93:79–89. https://doi.org/10.1016/j.soilbio.2015.10.015
Mushinski RM, Gentry TJ, Dorosky RJ, Boutton TW (2017) Forest harvest intensity and soil depth alter inorganic nitrogen pool sizes and ammonia oxidizer community composition. Soil Biol Biochem 112:216–227. https://doi.org/10.1016/j.soilbio.2017.05.015
Nair PKR (2007) The coming of age of agroforestry. J Sci Food Agric 87:1613–1619. https://doi.org/10.1002/jsfa.2897
Nair PR (2008) Agroecosystem management in the 21st century: it is time for a paradigm shift. J Trop Agric 46:1–12
Ohm RA, Feau N, Henrissat B, Schoch CL, Horwitz BA, Barry KW, Condon BJ, Copeland AC, Dhillon B, Glaser F, Hesse CN, Kosti I, LaButti K, Lindquist EA, Lucas S, Salamov AA, Bradshaw RE, Ciuffetti L, Hamelin RC, Kema GHJ, Lawrence C, Scott JA, Spatafora JW, Turgeon BG, de Wit PJGM, Zhong S, Goodwin SB, Grigoriev IV (2012) Diverse lifestyles and strategies of plant pathogenesis encoded in the genomes of eighteen Dothideomycetes fungi. PLoS Pathog 8:e1003037. https://doi.org/10.1371/journal.ppat.1003037
Peay KG, Garbelotto M, Bruns TD (2010) Evidence of dispersal limitation in soil microorganisms: isolation reduces species richness on mycorrhizal tree islands. Ecology 91:3631–3640. https://doi.org/10.1890/09-2237.1
Poeplau C, Don A (2013) Sensitivity of soil organic carbon stocks and fractions to different land-use changes across Europe. Geoderma 192:189–201. https://doi.org/10.1016/j.geoderma.2012.08.003
Porras-Alfaro A, Herrera J, Natvig DO, Lipinski K, Sinsabaugh RL (2011) Diversity and distribution of soil fungal communities in a semiarid grassland. Mycologia 103:10–21. https://doi.org/10.3852/09-297
Prober SM, Leff JW, Bates ST, Borer ET, Firn J, Harpole WS, Lind EM, Seabloom EW, Adler PB, Bakker JD, Cleland EE, DeCrappeo NM, DeLorenze E, Hagenah N, Hautier Y, Hofmockel KS, Kirkman KP, Knops JMH, la Pierre KJ, MacDougall AS, McCulley RL, Mitchell CE, Risch AC, Schuetz M, Stevens CJ, Williams RJ, Fierer N (2015) Plant diversity predicts beta but not alpha diversity of soil microbes across grasslands worldwide. Ecol Lett 18:85–95. https://doi.org/10.1111/ele.12381
Qin H, Wang H, Strong PJ, Li Y, Xu Q, Wu Q (2014) Rapid soil fungal community response to intensive management in a bamboo forest developed from rice paddies. Soil Biol Biochem 68:177–184. https://doi.org/10.1016/j.soilbio.2013.09.031
R Core Team (2017) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria
Read DJ, Perez-Moreno J (2003) Mycorrhizas and nutrient cycling in ecosystems – a journey towards relevance? New Phytol 157:475–492. https://doi.org/10.1046/j.1469-8137.2003.00704.x
Rinnan R, Bååth E (2009) Differential utilization of carbon substrates by bacteria and fungi in tundra soil. Appl Environ Microbiol 75:3611–3620. https://doi.org/10.1128/AEM.02865-08
Rodriguez RJ, White JF, Arnold AE, Redman RS (2009) Fungal endophytes: diversity and functional roles. New Phytol 182:314–330. https://doi.org/10.1111/j.1469-8137.2009.02773.x
Rousk J, Baath E, Brookes PC, Lauber CL, Lozupone C, Caporaso JG, Knight R, Fierer N (2010) Soil bacterial and fungal communities across a pH gradient in an arable soil. Isme J 4:1340–1351. https://doi.org/10.1038/ismej.2010.58
Schappe T, Albornoz FE, Turner BL, Neat A, Condit R, Jones FA (2017) The role of soil chemistry and plant neighbourhoods in structuring fungal communities in three Panamanian rainforests. J Ecol 105:569–579. https://doi.org/10.1111/1365-2745.12752
Schlatter DC, Schillinger WF, Bary AI, Sharratt B, Paulitz TC (2017) Biosolids and conservation tillage: impacts on soil fungal communities in dryland wheat-fallow cropping systems. Soil Biol Biochem 115:556–567. https://doi.org/10.1016/j.soilbio.2017.09.021
Schmidt SK, Nemergut DR, Darcy JL, Lynch R (2014) Do bacterial and fungal communities assemble differently during primary succession? Mol Ecol 23:254–258. https://doi.org/10.1111/mec.12589
Schoenholtz SH, Miegroet HV, Burger JA (2000) A review of chemical and physical properties as indicators of forest soil quality: challenges and opportunities. For Ecol Manag 138:335–356. https://doi.org/10.1016/S0378-1127(00)00423-0
Shibata H, Urakawa R, Toda H, Inagaki Y, Tateno R, Koba K, Nakanishi A, Fukuzawa K, Yamasaki A (2011) Changes in nitrogen transformation in forest soil representing the climate gradient of the Japanese archipelago. J For Res 16:374–385. https://doi.org/10.1007/s10310-011-0288-z
Smith SE, Read D (2008) Mycorrhizal symbiosis. Academic Press, London
Tanner EVJ, Sheldrake MWA, Turner BL (2016) Changes in soil carbon and nutrients following 6 years of litter removal and addition in a tropical semi-evergreen rain forest. Biogeosciences 13:6183–6190. https://doi.org/10.5194/bg-13-6183-2016
Tedersoo L, Bahram M, Dickie IA (2014a) Does host plant richness explain diversity of ectomycorrhizal fungi? Re-evaluation of Gao et al. (2013) data sets reveals sampling effects. Mol Ecol 23:992–995. https://doi.org/10.1111/mec.12660
Tedersoo L, Bahram M, Polme S, Koljalg U, Yorou NS, Wijesundera R, Ruiz LV, Vasco-Palacios AM, Thu PQ, Suija A, Smith ME, Sharp C, Saluveer E, Saitta A, Rosas M, Riit T, Ratkowsky D, Pritsch K, Poldmaa K, Piepenbring M, Phosri C, Peterson M, Parts K, Partel K, Otsing E, Nouhra E, Njouonkou AL, Nilsson RH, Morgado LN, Mayor J, May TW, Majuakim L, Lodge DJ, Lee SS, Larsson KH, Kohout P, Hosaka K, Hiiesalu I, Henkel TW, Harend H, Guo LD, Greslebin A, Grelet G, Geml J, Gates G, Dunstan W, Dunk C, Drenkhan R, Dearnaley J, de Kesel A, Dang T, Chen X, Buegger F, Brearley FQ, Bonito G, Anslan S, Abell S, Abarenkov K (2014b) Fungal biogeography. Global diversity and geography of soil fungi. Science 346:1256688. https://doi.org/10.1126/science.1256688
Torralba M, Fagerholm N, Burgess PJ, Moreno G, Plieninger T (2016) Do European agroforestry systems enhance biodiversity and ecosystem services? A meta-analysis. Agric Ecosyst Environ 230:150–161. https://doi.org/10.1016/j.agee.2016.06.002
Treseder KK, Lennon JT (2015) Fungal traits that drive ecosystem dynamics on land. Microbiol Mol Biol Rev 79:243–262. https://doi.org/10.1128/mmbr.00001-15
Udawatta RP, Kremer RJ, Garrett HE, Anderson SH (2009) Soil enzyme activities and physical properties in a watershed managed under agroforestry and row-crop systems. Agric Ecosyst Environ 131:98–104. https://doi.org/10.1016/j.agee.2008.06.001
van der Heijden MGA, Klironomos JN, Ursic M, Moutoglis P, Streitwolf-Engel R, Boller T, Wiemken A, Sanders IR (1998) Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature 396:69–72. https://doi.org/10.1038/23932
Voříšková J, Brabcová V, Cajthaml T, Baldrian P (2014) Seasonal dynamics of fungal communities in a temperate oak forest soil. New Phytol 201:269–278. https://doi.org/10.1111/nph.12481
Wang J, Ren C, Cheng H, Zou Y, Bughio MA, Li Q (2017a) Conversion of rainforest into agroforestry and monoculture plantation in China: consequences for soil phosphorus forms and microbial community. Sci Total Environ 595:769–778. https://doi.org/10.1016/j.scitotenv.2017.04.012
Wang Z, Li T, Wen X, Liu Y, Han J, Liao Y, DeBruyn JM (2017b) Fungal communities in rhizosphere soil under conservation tillage shift in response to plant growth. Front Microbiol 8:1301. https://doi.org/10.3389/fmicb.2017.01301
Yoshitake S, Fujiyoshi M, Watanabe K, Masuzawa T, Nakatsubo T, Koizumi H (2013) Successional changes in the soil microbial community along a vegetation development sequence in a subalpine volcanic desert on Mount Fuji, Japan. Plant Soil 364:261–272. https://doi.org/10.1007/s11104-012-1348-7
Yu Y, Stomph T-J, Makowski D, van der Werf W (2015) Temporal niche differentiation increases the land equivalent ratio of annual intercrops: a meta-analysis. Field Crop Res 184:133–144. https://doi.org/10.1016/j.fcr.2015.09.010
Zhang C, Liu G, Xue S, Wang G (2016) Soil bacterial community dynamics reflect changes in plant community and soil properties during the secondary succession of abandoned farmland in the Loess Plateau. Soil Biol Biochem 97:40–49. https://doi.org/10.1016/j.soilbio.2016.02.013
Zhou X, Zhang J, Gao D, Gao H, Guo M, Li L, Zhao M, Wu F (2017) Conversion from long-term cultivated wheat field to Jerusalem artichoke plantation changed soil fungal communities. Sci Rep `7:41502. https://doi.org/10.1038/srep41502
Funding
This work was supported by the Agricultural Science and Technology Independent Innovation Funds of Jiangsu Province (CX(16)1005), the National Key Research and Development Program of China (2017YFD0600700), the Priority Academy Program Development of Jiangsu Higher Education Institution (PAPD), the Postgraduate Research and Practice Innovation Program of Jiangsu Province (KYCX18_0955), and the Doctorate Fellowship Foundation of Nanjing Forestry University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Responsible editor: Zhihong Xu
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 1029 kb)
Rights and permissions
About this article
Cite this article
Guo, J., Wang, G., Wu, Y. et al. Ginkgo agroforestry practices alter the fungal community structures at different soil depths in Eastern China. Environ Sci Pollut Res 26, 21253–21263 (2019). https://doi.org/10.1007/s11356-019-05293-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-019-05293-w