Abstract
Brominated flame retardants (BFRs) are a group of widely used compounds that, due to their limited biodegradability, exhibit excessive persistence in the environment. The persistence and high toxicity of these compounds to the natural biota causes great environmental concern. We investigated the biodegradation of the BFR dibromoneopentyl glycol (DBNPG) under continuous culture conditions using a miniature membrane bioreactor (mMBR) to assess its feasibility as a bioremediation approach. This system demonstrated long-term, stable biodegradation of DBNPG (>90 days), with an average removal rate of about 50 %. Pyrosequencing of the 16S rRNA gene of the microorganisms involved in this process revealed the dominance of reads affiliated with the genus Brevundimonas of the Alphaproteobacteria class during the different mMBR operational stages. The bacterial community was also dominated by reads affiliated with the Sinorhizobium and Sphingopyxis genera within the Alphaproteobacteria class and the Sediminibacterium genus of the Sphingobacteria class. Real-time PCR used to analyze possible changes in the population dynamics of these four dominant groups revealed their consistent presence throughout the long-term mMBR biodegradation activity. Two genera, Brevundimonas and Sphingopyxis, were found to increase in abundance during the acclimation period and then remained relatively stable, forming the main parts of the consortium over the prolonged active stage.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Brominated flame retardants (BFRs) comprise a large and diverse group of chemicals that are integrated into a wide variety of products, including plastic polymers, electronic and electrical equipment, textiles, furniture, and vehicles. They are primarily used to protect materials against ignition and to prevent fire-related damage (Birnbaum and Staskal 2004). Their recalcitrant nature and high toxicity to the natural biota are attributed to the chemical properties of their bromide substituent(s). These halogen moieties generally reduce the bioavailability of the molecule while increasing its lipid solubility, ultimately resulting in the persistence and accumulation of the BFRs in the environment and in animal tissues (Häggblom and Bossert 2003). Consequently, their widespread production and extensive use over the past few decades, combined with the inappropriate treatment of industrial wastewater and waste containing BFRs, has generated great environmental concern (Segev et al. 2013).
Microbial communities are known to inhabit contaminated sites and are capable of metabolizing these recalcitrant anthropogens via complex processes that mostly rely on multispecies interactive networks (Maphosa et al. 2010a). Organohalide-respiring bacteria, for example, have been found to thrive in consortia rather than in a pure culture, suggesting a possible role of syntrophic interactions that could act as drivers for dehalogenation (Maphosa et al. 2010a). Thus, a microbial consortium (rather than a pure culture) should be advantageous for treatment strategies, providing the metabolic stability and robustness required for these applications (Tyagi et al. 2011).
Several methods, such as advanced oxidation processes and adsorption to activated carbon, were found to be effective in eliminating halo-organic compounds from contaminated sites and wastewaters (Taş et al. 2009). Nevertheless, when applicable, biological treatment technologies are preferable alternatives for economic and ecological reasons (Jain et al. 2005). Among these technologies, membrane bioreactors (MBRs) have recently emerged as a promising alternative for water applications, including wastewater treatment processes (Radjenović et al. 2009; Wan et al. 2011). Because of their complete retention of the biomass, MBRs ensure high biomass concentrations within compact reactor volumes. The high solid retention time attainable with these systems promote both the development of slow growing microbial communities and the adaptation of these communities to the recalcitrant organic substances contained in wastewaters. Therefore, compared to conventional activated sludge treatment processes that are restricted to fast-growing and floc-forming microorganisms, MBR technology may facilitate more effective bioremediation (Petrovic et al. 2009; Lipp et al. 2009; Radjenović et al. 2009).
MBR systems have been widely tested for the removal of a variety of trace organic contaminants, such as antibiotics and other pharmaceuticals, endocrine disrupting chemicals, pesticides and flame retardants, including some halogenated compounds (Lipp et al. 2009, 2012; Hai et al. 2011a, b). This technology has also been applied in the treatment of higher concentrations of several chlorinated organics (Visvanathan et al. 2005; Urgun-Demirtas et al. 2006; Carucci et al. 2007, 2010; Bolzonella et al. 2010; Zhao et al. 2011; Munro et al. 2013). Furthermore, MBRs were recently used to treat the BFR tetrabromobisphenol A (Potvin et al. 2012), but the ability of the technology to eliminate other BFRs has not been tested.
Dibromoneopentyl glycol (DBNPG) is a persistent aliphatic BFR that is mainly used as an additive during the manufacture of plastic polymers and it is a suspected human carcinogen (Ezra et al. 2010; Segev et al. 2013). Moreover, DBNPG was included in the US Environmental Protection Agency’s 1990 list of high-production-volume chemicals, indicating that it was manufactured in or imported into the USA in amounts of ≥1 million lb/year (EPA 2006). The high resistance to biodegradation conferred by its halogen moieties along with its relatively high solubility in water (20 g L−1) probably constitutes the main reason for the wide distribution of DBNPG in aqueous environments (Ezra et al. 2010).
In this study, we assessed the dynamics of a bacterial community in a miniature membrane bioreactor (mMBR) system treating DBNPG and evaluated the relationship between reactor performance and community behavior. Pyrosequencing of the 16S ribosomal RNA (rRNA) gene and real-time PCR (qPCR) analysis were used to characterize the bacterial community at different stages of mMBR operation.
Materials and methods
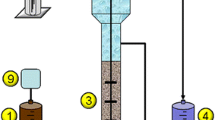
mMBR system and operation
The mMBR system was operated for a period of 126 days as previously described in detail by Segev et al. (2013). The mMBR was inoculated with microbial biomass (1:10 v/v) that demonstrated complete biodegradation of DBNPG under batch culture conditions in earlier research (Segev et al. 2009). The feed to the reactor was a sterile mineral salt medium as previously described (Segev et al. 2013) with the addition of 1 g L−1 yeast extract (YE; Sigma Aldrich) and 250 mg L−1 DBNPG. The effluent from the mMBR was kept at 4 °C until further analysis. To minimize microbial colonization of the membrane, the mMBR was operated under constant mixing conditions. The concentration of 250 mg L−1 of DBNPG was chosen based on preliminary batch culture experiments, which showed complete biotransformation of DBNPG under these conditions. Biomass samples from the mMBR were collected at different time points in the operation of the mMBR (one sample of 5 mL for each time point) through the sampling port (sealed with a silicon septum) using a sterile syringe and needle and stored at −80 °C for subsequent analysis.
Analytical methods
The biodegradation capacity of the mMBR was evaluated by measuring the DBNPG and bromide ion concentrations in the effluent using high-performance liquid chromatography (HPLC) and ion chromatography, respectively.
The DBNPG concentration was determined using an Agilent 1100 HPLC system with a diode array detector (DAD) UV-200 nm and a LiChroCART 250-4 HPLC Cartridge LiChrospher 100 RP-18 (5 μm) column (DBNPG recovery of 99 % was achieved). Prior to HPLC analysis, 0.5 mL of each sample was filtered using an Amicon Ultra—0.5 mL 3 K centrifugal filter device (3000 Da cutoff, Merck Millipore, Darmstadt, Germany) for protein elimination.
The bromide ion concentration was measured by an ion chromatograph (Dionex DX 500 system with an LC20 Chromatography Enclosure, Dionex, Thermo Scientific) equipped with an analytical column (4 × 250 mm IonPac AS9-SC, Dionex, Thermo Scientific), a guard column (4 × 50 mm IonPac AG9-HC, Dionex, Thermo Scientific), an anion electrolytic suppressor (4 mm ASRS 300, Dionex, Thermo Scientific), and a conductivity detector.
Measured DBNPG and bromide ion concentrations were corroborated by total organic carbon (TOC; Tekmar-Dohrmann Apollo 9000 TOC-analyzer, Teledyne Tekmar, Mason, OH) and adsorbable organic halide (AOX) analyses of the effluent. AOX present in effluent samples of the mMBR was analyzed on an AOX analyzer (multiX 2500, Analytik Jena, Jena, Germany) using a two-column adsorption method (APU2, Analytik Jena, Jena, Germany). Samples were prepared according to the manufacturer’s instructions.
DNA extraction and pyrosequencing
Total genomic DNA was extracted from mMBR biomass samples (one from each biomass sample) using a MoBio Power soil DNA isolation kit (MoBio Laboratories, Solana Beach, CA) according to the manufacturer’s protocol and stored at −20 °C. The DNA concentration was determined by a NanoDrop ND 1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). Eleven DNA samples from different time points were selected for 454 pyrosequencing (Roche 454 Life Sciences, Branford, CT) at the Molecular Research (MR) DNA Laboratories (Shallowater, TX). The company’s 27Fmod (AGRGTTTGATCMTGGCTCAG) primer was used to obtain bacterial reads of the hypervariable regions V1-V2 of bacterial 16S rRNA genes.
Sequence analysis
In total, 74,107 raw reads were treated using the mothur platform (v. 1.31.0) following the 454 standard operating procedure (SOP) (Schloss et al. 2011). Barcodes, primers, and adapters were trimmed. Low-quality sequences were then removed by eliminating reads shorter than 150 bps, containing ambiguous nucleotides or long homopolymers, or with average quality scores lower than 35. The resulting sequences were aligned and checked for PCR chimeras using the “chimera.uchime” command in mothur, and reads identified as chimeras were filtered out. Taxonomic classification of unique sequences was performed using the Ribosomal Database Project (RDP) classifier with a cutoff of 50 % (Wang et al. 2007). Sequences classified as chloroplast, mitochondria or “unknown” were also removed, resulting in a total of 43,615 effective bacterial sequences. To conduct the downstream analyses for all samples at the same sequencing depth, each sample size was normalized by randomly selecting 2092 sequences—the minimum number of bacterial sequences among the 11 biomass samples.
Multiple sequence alignment and cluster analysis were conducted to group the sequences into operational taxonomic units (OTUs) based on a dissimilarity threshold of 3 %. These OTUs were used to generate rarefaction curves and to calculate the richness and diversity indexes: Chao1 and inverse Simpson, and Good’s coverage estimator in mothur. Representative sequences from each OTU were phylogenetically assigned taxonomic classifications obtained from the RDP’s classifier. Raw sequencing data were deposited in the MG-RAST (metagenomics.anl.gov) archive under accession numbers 4543824.3–4543834.3.
qPCR primer design
New sets of primers were designed to track specific groups of bacteria in the consortium, generating amplicons within the 110–125 bp range as listed in Table S3. The primer sets were designed upon multiple alignments of 16S rRNA gene sequences retrieved from a clone library of the active consortium (Segev et al. 2013) using ClustalW with the MEGA 5.2 package (Tamura et al. 2011) to detect conserved regions on the 16S rRNA genes of all the targeted genera: Brevundimonas, Sediminibacterium, Sinorhizobium, and Sphingopyxis. Each primer pair was further checked using AmplifX software (v. 1.5.4, http://crn2m.univ-mrs.fr/pub/amplifx-dist). The “probe match” tool on the RDP website (Wang et al. 2007) and primer-BLAST tool (Ye et al. 2012) were used to assign the primer sequences to different taxonomy levels. Based on the obtained results (Table S4), all four primer sets showed high specificity to the targeted genera.
qPCR standards
qPCR standards were prepared by cloning PCR-amplified 16S rRNA gene fragments of the targeted bacterial groups into the pGEM-T Easy Vector System I (Promega, Madison, WI), as specified by the manufacturer. Plasmids were transformed into BioSuper CaCl2-competent HD5α Escherichia coli cells (Bio-Lab, Israel) according to the manufacturer’s protocol. Plasmids were then purified using the Wizard Plus SV Miniprep DNA purification system (Promega, Madison, WI) and verified by PCR amplification (Biometra TGradient thermocycler, Biometra, Göttingen, Germany) using the primers listed in Table S3. Plasmids harboring the respective target gene amplicons were used to obtain standard calibration curves in the subsequent qPCR analysis (six serial dilution points). PCR efficiencies, linear response, and dynamic ranges of all standard curves were within acceptable limits (Table S4).
qPCR assay
qPCR was performed with a StepOnePlus Real-Time PCR System (A&B Applied Biosystems, Life technologies, Carlsbad, CA) for quantification of 16S rRNA genes of the four bacterial groups (Brevundimonas, Sediminibacterium, Sinorhizobium, and Sphingopyxis) and of the total bacterial community. Assays proceeded through three stages. Stage 1: 1 cycle of 15 min at 95 °C; stage 2: 40 cycles of 15 s at 95 °C, 20 s at 55 °C, and 30 s at 72 °C; and stage 3: a PCR product dissociation protocol, in which the temperature was ramped from 60 to 95 °C, to verify that each used primer pair produced only a single specific product. The PCR reaction consisted of 10 μL of Absolute QPCR SYBR Green ROX Mix (Thermo Fisher Scientific, Surrey, UK), 150 nM each of forward and reverse primers, and 5.0 μL of each DNA template in a total reaction volume of 20 μL. Samples were analyzed in triplicates, and no-template controls were included. Respective target gene copy numbers were calculated using standard curves as copies per milliliter of biomass sample, as previously described (Ritalahti et al. 2006).
Statistical analysis
To examine interactions between mMBR operation time and the population structures of four key genera, classification and regression tree (CART), followed by the Tukey-Kramer HSD test, were performed (JMP Pro 11.1.1, 2013, SAS Institute Inc.). The CART analysis, a binary recursive partitioning technique (Pasipanodya et al. 2013; Gass et al. 2014), entails sequentially splitting the data into two groups of maximum homogeneity, after which the same process is repeated to create two daughter nodes. It generates a variable importance score of how much each subsequent predictor identified as a daughter node improves the primary decision node (highest-ranked node). This process continues, such that each class is homogeneous in the outcome examined, until no further partitions are possible. As such, CART constructs maximum trees that are important in identifying data structures. CART analysis was performed for both pyrosequencing and qPCR datasets (i.e., once using 16S rRNA gene copy number as the potential predictor and once using days as the predictor). Only attempts that succeeded in generating regression trees are shown (Figs. S2, S3).
The relative abundances of dominant microbial genera obtained using pyrosequencing and qPCR techniques were analyzed using a two-sided paired t test. Statistical significance was determined at a value of p < 0.05 (JMP Pro 11.1.1, 2013, SAS Institute Inc.).
Results
Process performance and DBNPG removal
The mMBR system was successfully operated for 126 days. Following an acclimation period of approximately 30 days, once the DBNPG in the reactor was stable, removal (Fig. 1) was allowed to proceed for an additional 96 days. Removal rates ranged from 32.6 to 57.3 % during the biodegradation period (Fig. 1 and Table S1). The removal rate of adsorbable organic halide (AOX) was highly similar to that estimated for DBNPG (Fig. 1 and Table S1), indicating that no brominated intermediates were produced under these operational conditions. In further support of the existence of a dehalogenation process, the reductions in both DBNPG and AOX concentrations were accompanied by an increase in bromide concentration in the effluent (average bromide release of about 54 %, Table S1). Based on prior control experiments (i.e., sterile control) conducted under batch culture conditions using the same growth media (Segev et al. 2009), abiotic factors (e.g., temperature, pH) have no effect on DBNPG concentration and do not contribute to the release of bromide ions.
By day 12, a total organic carbon (TOC) removal rate of about 78 % was demonstrated (Fig. 1). A high rate of stable TOC removal (of up to 88 %) was maintained over the complete mMBR operation period (Table S1). The initial high TOC concentration in the reactor (507 mg L−1) was mainly due to the addition of auxiliary YE, which contains various organic carbon sources, to the growth media. These auxiliary nutrients are thought to stimulate bacterial growth and, as such, to support and enhance biodegradation (Segev et al. 2009; Xiao et al. 2010).
Overall mMBR microbial community composition and diversity
The mMBR consortium was analyzed for its community structure and dynamics during the different reactor operation stages (i.e., acclimation and biodegradation) using the high throughput 454 pyrosequencing method. Pyrosequencing produced a total of 43,615 effective sequences with an average length of 365 bp. The numbers of effective sequences from the different time points varied in the range of 2092 to 12,146. All effective reads were clustered into 293 OTUs using 97 % sequence identity as a cutoff. The total numbers of OTUs estimated by the non-parametric richness estimator Chao1 (Table S2) were in the range of 39–99 in all samples, indicating that the mMBR community consisted of a relatively low number of different OTUs, thus reflecting a relatively simple and specific bacterial community structure. Coverage-based analysis using Good’s coverage estimator demonstrated that high coverage of 98–99 % was already achieved for all samples (Table S2).
At the class level, Alphaproteobacteria was the most abundant class overall during the different operation stages (i.e., inoculum on day 0, acclimation, and active biodegradation from day 31 onwards), accounting for up to 90.6 % of total effective bacterial sequences (Fig. S1). After the Alphaproteobacteria, Bacilli (0.91–67.2 % of total effective sequences, averaging 16.3 %), Acidobacteria–Gp4 (0–19.8 %, averaging 6.0 %), and Sphingobacteria (0.05–23.7 %, averaging 4.0 %) were the predominant classes.
The initial acclimation period (day 3), when no degradation activity was observed, was associated with a significant, yet temporal, shift in community composition compared to that of the inoculum (Fig. S1). At this time point, the Bacilli (67.2 %), Gammaproteobacteria (23.8 %), and Betaproteobacteria (5.9 %) classes significantly increased and dominated the population. An additional shift in population structure was observed on day 63, as “unclassified bacteria” (affiliated with the Spirochaetes class) were highly enriched and became the dominant group, accounting for 48.3 % of total effective bacterial reads. This latter change in community structure was not accompanied by a reduction in reactor performance. Bacteria belonging to the phylum Spirochaetes live chemoheterotrophically in anaerobic environments and are frequently detected in anaerobic digestion systems, but their ecophysiological function is virtually unknown (Lee et al. 2013). This temporary increase of Spirochaetes numbers could, therefore, be the result of a transient change in the aeration conditions in the mMBR.
Overall, phylogenetic stability of the consortium was observed at the active biodegradation stage, i.e., there were no major changes in the DBNPG-degrading microbial population. Furthermore, the structure of the biodegrading community in the mMBR (day 31 onwards) fairly resembled that of the active inoculum (day 0), suggesting that community composition was not influenced by the culture system (i.e., batch vs. continuous).
Microbial population dynamics during different stages of mMBR operation
The genus Brevundimonas within Alphaproteobacteria was highly abundant, representing up to 85 % of all bacterial effective reads (Fig. 2). Other genera that belong to the α-subdivision were Sinorhizobium and Sphingopyxis, the latter of which was present in all samples (except for day 3), although at relatively low abundances (4.5 % in the inoculum; 0.5–2.7 % from day 12 forward). The relative abundances of Brevundimonas and Sphingopyxis gradually increased between days 3 and 31, when active biodegradation was first observed (from 2.4 to 85.1 % and from undetectable levels to 2.6 %, respectively). The three genera of the Alphaproteobacteria class, together with the genera Brevibacillus, Sediminibacterium, and Acidobacteria–Gp4 (of the Bacilli, Sphingobacteria, and Acidobacteria classes, respectively), accounted for up to 99.2 % of total effective reads (Fig. 2) and were commonly present in at least ten of the samples.
The Sediminibacterium, Sinorhizobium, and Brevibacillus genera showed high variability in their abundances in the first three samples, which corresponded to the inoculum and acclimation periods. Reads affiliated with Sediminibacterium and Sinorhizobium were found at low levels (1.5 and 5.5 %, respectively) in the inoculum, were not detected on day 3, and increased in abundance on day 12, accounting for 23.7 and 12.8 % of total effective reads, respectively. In the active biodegrading consortium, on the other hand, they accounted for a much lower proportion (ranging from 0.14 up to 6.5 % for Sediminibacterium and from 0.09 up to 4.2 % for Sinorhizobium). Furthermore, sequences corresponding to the Sediminibacterium genus were found at very low levels on days 31, 42, 97, and 104, despite apparent DBNPG removal from the system. The Brevibacillus genus peaked on day 3 (67.2 %) despite its initially negligible presence in the inoculum (i.e., represented by only a few sequences accounting for <1 %). This increase was followed by a significant decrease (day 12 onwards), although some variations were still observed (varying from 5.1 to 28.2 %).
The inoculum was also dominated by sequences related to the genus Gp4 within the Acidobacteria (19.8 %). Nonetheless, reads affiliated with this group were missing from or were negligible in (<1 %) the three subsequent samples, including those collected at the onset of biodegradation (day 31).
qPCR quantification of 16S rRNA genes in the mMBR
To quantify changes in the population size of specific bacterial groups, newly designed sets of primers were used in qPCR analysis (Table S3). As shown in Fig. 3, the presence of four genera was confirmed in all samples. Overall, both methods—qPCR and pyrosequencing—generated comparable patterns (Table S5). Three out of the four groups showed very good correlation between both methods (R 2 between 0.857 and 0.976), and one also demonstrated highly similar relative abundance values (i.e., Sediminibacterium).
Changes in 16S rRNA gene copy numbers of the four key bacterial groups as analyzed using qPCR. A specific primer set was designed for each group. Universal 16S rRNA primers were used to estimate total bacterial 16S rRNA gene copy numbers in the mMBR biomass. Error bars represent S.D. for three qPCR reactions
In accordance with the pyrosequencing results, the outcome of the qPCR (Fig. 3) analysis also showed that the Brevundimonas genus was the most dominant (except for day 12) at all time points. The abundance of the Brevundimonas population in the mMBR was found to increase by 6.5-fold over the acclimation period (from 2.40 × 107 in the inoculum to 1.56 × 108 on day 31). Brevundimonas numbers were relatively stable throughout the following biodegradation period (around 108 copies/mL). The 16S rRNA gene copy number of Sphingopyxis dropped significantly on day 3 (1.2 × 105 copies/mL) compared to the inoculum (3.2 × 106 copies/mL). However, throughout the acclimation period they gradually increased until they were at numbers higher than those found in the initial inoculum (around 2 × 107 copies/mL on day 31). During the active biodegradation period, the Sphingopyxis population fluctuated, sometimes changing by up to one order of magnitude but without dropping below 3 × 106 copies/mL. In keeping with the observed trend, CART yielded two splits for the Sphingopyxis population, with a root node (i.e., the most important potential predictor or decision node) on day 31, when biotransformation was first reported, and a daughter node on day 78 (Fig. S2). Counts of Sinorhizobium 16S rRNA gene copy number increased on the first 12 days (8.16 × 106 copies/mL in the inoculum up to 1.96 × 108 copies/mL on day 12). Moreover, on day 12, the Sinorhizobium 16S rRNA gene copy number was higher than that observed for Brevundimonas. Similar numbers (around 108 copies/mL) were also detected on days 63 and 90. Sediminibacterium numbers varied significantly over time in the reactor and were more than two orders of magnitude lower on day 3 than at the adjacent time points (inoculum and day 7). Two additional oscillations were observed, with minimum points on days 39 and 104 and peaks on days 12–20 and 56–62. CART identified three terminal nodes in the final tree of Sediminibacterium (Fig. S3), with a terminal node designated T3 on days 12, 20, and 50–63. Insofar as they resemble the bacterial population dynamics of phage-hosts systems, these oscillations could be the result of phage predation (Shapiro and Kushmaro 2011).
Total bacterial 16S rRNA gene copy numbers were generally around 7.5 × 108 copies/mL (ranging from 8.72 × 107 copies/mL in the inoculum to 2.23 × 109 copies/mL on day 3). In support of the prevalence of some new bacterial groups on day 3 (e.g., unclassified Enterobacteriaceae and Comamonas), demonstrated in the pyrosequencing results, total bacterial 16S rRNA gene copy numbers markedly increased. This increase occurred concurrently with a major reduction of both Sediminibacterium and Sphingopyxis, while the population sizes of Brevundimonas and Sinorhizobium were not significantly altered.
Discussion
MBR technology is increasingly being used worldwide for water and wastewater treatment (Meng et al. 2009; Ma et al. 2013). Owing to the generally very good biomass retention capacity of MBRs, these systems can be used with slow growing, non-flocculating microbial consortia, such as those exploited in the study by Segev et al. (2013). For practical applications, it is important to have durable and stable treatment technology. In the present study, the long-term biotransformation of DBNPG was demonstrated using an mMBR system. To our knowledge, this is the first study showing a stable and effective removal process of this compound, with an average removal rate of about 50 %.
Previous studies (Segev et al. 2009) found that complete biotransformation of DBNPG and the BFR tribromoneopentyl alcohol (TBNPA) can occur under batch culture conditions by a bacterial consortium. Recently, an mMBR system was used to evaluate the biodegradability of DBNPG and TBNPA under continuous culture conditions (Segev et al. 2013). The mMBR system was operated for a short operational period (14 days) and reactor activity collapsed after 7 days. Although this short-term, transient elimination of both BFRs demonstrated the potential of the process, it also exposed some of the difficulties associated with the operation of these systems. In the present study, reactor performance was greatly improved in terms of stability and reactor operation time (i.e., 14 vs. >120 days, respectively), with no major decline in activity observed during the active stage of mMBR operation (>90 days).
To date, the information about natural biotic and abiotic degradation pathways for DBNPG is very limited (Segev et al. 2009; Ezra et al. 2010). Apart from bromide ions, no other intermediates or degradation products have been reported in the literature for the biotransformation of DBNPG. On the other hand, its spontaneous decomposition under alkaline conditions involves intramolecular nucleophilic substitution, which results in the release of bromide ions through a sequential formation of 3-bromomethyl-3-hydroxymethyloxetane and 2,6-dioxaspiro[3.3]heptane (Ezra et al. 2010; Kozell et al. 2015).
Because it was necessary to add auxiliary YE (Segev et al. 2009), co-metabolic biotransformation due to dehalogenation by the microbial consortium is likely the mechanism of DBNPG removal. Under aerobic conditions, oxidative biotransformation of various contaminants has been observed concurrently with the transformation of auxiliary substrates (e.g., YE). These processes, which involve the activity of oxygenase enzymes that are used by the microorganisms to initiate the oxidation of primary substrates, can fortuitously transform halogenated hydrocarbons. Bacterial isolates that belong to the Brevundimonas and Sphingopyxis genera have been previously demonstrated to be capable of metabolizing different pollutants (phenanthrene and the chlorinated compound triclosan, respectively) in the presence of auxiliary substrates supporting their growth (Xiao et al. 2010; Lee et al. 2012).
The combination of pyrosequencing and qPCR analyses enabled us to follow the dynamics of target phylotypes affiliated with four bacterial genera: Sediminibacterium from the Sphingobacteria class, and Brevundimonas, Sinorhizobium, and Sphingopyxis from the Alphaproteobacteria class. This allowed us to evaluate the relationship between mMBR performance and the behavior of these key groups, whose persistent presence in the mMBR community was confirmed by qPCR (Fig. 3). The relative abundance in the active inoculum of Brevibacillus was low (<1.0 %), while Gp4 demonstrated negligible abundance over the acclimation period and at the onset of biodegradation. Both groups were thus not included in the qPCR analysis. The elevated prevalence of Brevibacillus on day 3 is likely due to the high availability of alternative carbon sources over the first 7 days, as reflected by the high TOC levels on those days (Fig. 1), which may have facilitated the rapid proliferation of Brevibacillus.
In addition, the consortium was characterized by low diversity (as revealed by alpha-diversity analysis, Table S2). This corresponds to a specialized community adapted to DBNPG transformation, that apparently utilized the YE as a carbon and energy source. Although active DBNPG degrading microorganisms could not be distinguished from YE degraders, we believe that owing to the low diversity of the acclimated community in the mMBR, DBNPG transforming microorganisms should be found at relatively high abundance in the active stage, and those levels should persist in the mMBR system throughout this phase. It should be noted that we and others (Segev et al. 2009) have attempted to isolate debrominating bacteria on different agar plates, but the bacterial isolates that were obtained did not show biodegradation activity (Segev et al. 2009).
Several genera within the sphingomonads group, of which the Sphingopyxis genus is a member, share a number of phenotypic traits. These include their pronounced ability to degrade various recalcitrant natural and xenobiotic compounds (Aranda et al. 2003; Stolz 2009, 2014; Vaz-Moreira et al. 2011; Lee et al. 2012), such as chlorinated dibenzofurans and dibenzodioxins, polyethylene glycols, polyvinyl alcohols, and different herbicides and pesticides (Stolz 2014). Sphingomonads can occupy a variety of anthropogenically polluted environments, including wastewater treatment plants (Kämpfer et al. 2002). The metabolic versatility of these organisms suggests that they have the ability to adapt to and efficiently degrade new compounds in the environment (Tani et al. 2007; Stolz 2009). This ability is presumably mediated initially by the expression of different oxygenases (e.g., 2,3-dioxygenases) that can oxidize a wide range of organic compounds, although the substrate specificities of the various dioxygenases are still largely unknown (Stolz 2009; Lee et al. 2012). Therefore, the onset of DBNPG transformation on day 31 could be due to an increase in the bacterial expression of such genes. Previous reports have indicated that the Brevundimonas spp. are associated with the biodegradation of pollutants (Deshpande et al. 2004; Xiao et al. 2010), including a chlorinated insecticide (Shetti and Kaliwal 2012). Interestingly, the ability of Brevundimonas sp. strain X08 to biodegrade the polycyclic aromatic hydrocarbon phenanthrene was improved in the presence of 0.5 g/L YE (Xiao et al. 2010). This observation supports our findings indicating that the DBNPG transforming consortium required YE supplementation to achieve biodegradation.
A microbial population shift concomitant with the improved performance of the mMBR system was demonstrated for both the Brevundimonas and Sphingopyxis genera. In addition to their persistent presence in the mMBR system throughout its operation, both genera increased in relative abundance over the acclimation stage (Fig. 3). This relationship between population dynamics and performance parameters suggests not only that these bacterial groups may be significant players in the DBNPG biodegradation process, but also that they may be required for process stability. In light of the relatively low abundance of Sphingopyxis in the mMBR compared to other genera, it should be noted that dehalogenating populations do not always dominate their communities. For example, the organohalide-respiring bacteria Dehalococcoides, which plays a crucial role in the conversion of tetrachloroethene and trichloroethene to ethane, were found in low numbers even when bioreactor operating conditions were designed for their optimal growth and proliferation (Maphosa et al. 2010b). We also cannot rule out the possibility that the high prevalence of Brevundimonas in the system may correspond to an effective utilization of alternative carbon sources in the media rather than to the biodegradation of DBNPG.
Apart from Brevundimonas and Sphingopyxis, Sinorhizobium spp. were also previously shown to biodegrade environmental contaminants (Keum et al. 2006, 2008), including polychlorinated biphenyls (Tu et al. 2011), and they are known to possess ring-cleavage enzymes such as catechol 1,2-dioxygenase and protocatechuate 3,4- or 4,5-dioxygenase (Keum et al. 2006). The qPCR results (Fig. 3) showed that Sinorhizobium were present at high numbers throughout the entire mMBR operation period, and therefore, they could have taken part in DBNPG degradation. Knowledge of the function of members of the Sediminibacterium is limited. However, these bacteria were detected in a variety of natural and engineered habitats, including activated sludge treating wastewater from a petroleum refinery (Ayarza et al. 2014).
The results presented here, based on deep 16S rRNA sequencing, generally fit a previous structure analysis of the DBNPG transforming community in the mMBR system using a 16S rRNA clone library (ca. 50 sequences) (Segev et al. 2013). However, there was a clear discrepancy between our study and that of Segev et al.’s regarding the relative abundances of these four genera. While long-term operation of the mMBR allowed for stabilization in terms of community composition and activity, the short-term operation of the mMBR reported by Segev et al. (2013) was apparently insufficient to achieve community acclimation and stabilization. Long-term operation of the mMBR enabled us not only to investigate the dynamics of the transforming community, but also to characterize the adapted microbial community (i.e., analysis of 11 time-points vs. 3 in Segev et al. (2013)).
Conclusions
This study demonstrated the long-term, stable biodegradation of DBNPG with an average removal rate of about 50 %. The long-term operation of the mMBR allowed us to track changes in its bacterial population dynamics and characterize the core bacterial community associated with process stability. Consistent patterns emerged from the data obtained both by 454 pyrosequencing and by qPCR. Members of the Alphaproteobacteria class dominated the consortia throughout the study period. From this class, members of two genera, Brevundimonas and Sphingopyxis, were found to increase during the acclimation period, after which they remained relatively stable over the prolonged active stage, indicating that they may be involved in the biotransformation of DBNPG.
References
Aranda C, Godoy F, Becerra J, Barra R, Martínez M (2003) Aerobic secondary utilization of a non-growth and inhibitory substrate 2,4,6-trichlorophenol by Sphingopyxis chilensis S37 and sphingopyxis-like strain S32. Biodegradation 14:265–274
Ayarza JM, Figuerola EL, Erijman L (2014) Draft genome sequences of type strain Sediminibacterium salmoneum NJ-44 and Sediminibacterium sp. strain C3, a novel strain isolated from activated sludge. Genome Announc 2:e01073–13
Birnbaum LS, Staskal DF (2004) Brominated flame retardants: cause for concern? Environ Health Perspect 112:9–17
Bolzonella D, Fatone F, Pavan P, Cecchi F (2010) Poly-chlorinated dibenzo-p-dioxins, dibenzo-furans and dioxin-like poly-chlorinated biphenyls occurrence and removal in conventional and membrane activated sludge processes. Bioresour Technol 101:9445–9454
Carucci A, Manconi I, Manigas L (2007) Use of membrane bioreactors for the bioremediation of chlorinated compounds polluted groundwater. Water Sci Technol 55:209–216
Carucci A, Milia S, Cappai G, Muntoni A (2010) A direct comparison amongst different technologies (aerobic granular sludge, SBR and MBR) for the treatment of wastewater contaminated by 4-chlorophenol. J Hazard Mater 177:1119–1125
Deshpande NM, Sarnaik SS, Paranjpe SA, Kanekar PP (2004) Optimization of dimethoate degradation by Brevundimonas sp. MCM B-427 using factorial design: studies on interactive effects of environmental factors. World J Microb Biotechnol 20:455–462
EPA (2006) U.S. Environmental Protection Agency 1990 HPV Challenge Program Chemical List
Ezra S, Bilkis I, Feinstein S, Adar E, Ganor J (2010) Chemical degradation of 2,2-bis(bromomethyl)propan-1,3-diol (DBNPG) in alkaline conditions. Chemosphere 79:476–481
Gass K, Addiss DG, Freeman MC (2014) Exploring the relationship between access to water, sanitation and hygiene and soil-transmitted helminth infection: a demonstration of two recursive partitioning tools. PLoS Negl Trop Dis 8:e2945
Häggblom MM, Bossert ID (2003) Halogenated organic compounds—a global perspective. In: Häggblom MM, Bossert ID (eds) Dehalogenation—microbial processes and environmental applications. Kluwer Academic Publishers, Boston, pp 3–29
Hai FI, Tadkaew N, McDonald JA, Khan SJ, Nghiem LD (2011a) Is halogen content the most important factor in the removal of halogenated trace organics by MBR treatment? Bioresour Technol 102:6299–6303
Hai FI, Tessmer K, Nguyen LN, Kang J, Price WE, Nghiem LD (2011b) Removal of micropollutants by membrane bioreactor under temperature variation. J Membr Sci 383:144–151
Jain RK, Kapur M, Labana S, Lal B, Sarma PM, Bhattacharya D, Thakur IS (2005) Microbial diversity: application of microorganisms for the biodegradation of xenobiotics. Curr Sci India 89:101–112
Kämpfer P, Witzenberger R, Denner EB, Busse HJ, Neef A (2002) Sphingopyxis witflariensis sp. nov., isolated from activated sludge. Int J Syst Evol Microbiol 52:2029–2034
Keum YS, Seo JS, Hu Y, Li QX (2006) Degradation pathways of phenanthrene by Sinorhizobium sp. C4. Appl Microbiol Biotechnol 71:935–941
Keum YS, Seo JS, Li QX, Kim JH (2008) Comparative metabolomic analysis of Sinorhizobium sp. C4 during the degradation of phenanthrene. Appl Microbiol Biotechnol 80:863–872
Kozell A, Yecheskel Y, Balaban N, Dror I, Halicz L, Ronen Z, Gelman F (2015) Application of dual carbon–bromine isotope analysis for investigating abiotic transformations of Tribromoneopentyl alcohol (TBNPA). Environ Sci Technol 49:4433–4440
Lee DG, Zhao F, Rezenom YH, Russell DH, Chu KH (2012) Biodegradation of triclosan by a wastewater microorganism. Water Res 46:4226–4234
Lee SH, Park JH, Kang HJ, Lee YH, Lee TJ, Park HD (2013) Distribution and abundance of Spirochaetes in full-scale anaerobic digesters. Bioresour Technol 145:25–32
Lipp P, Kreißel K, Meuler S, Bischof F, Tiehm A (2009) Influencing parameters for the operation of an MBR with respect to the removal of persistent organic pollutants. Desalin Water Treat 6:102–107
Lipp P, Gross H-J, Tiehm A (2012) Improved elimination of organic micropollutants by a process combination of membrane bioreactor (MBR) and powdered activated carbon (PAC). Desalin Water Treat 42:65–72
Ma J, Wang Z, Yang Y, Mei X, Wu Z (2013) Correlating microbial community structure and composition with aeration intensity in submerged membrane bioreactors by 454 high-throughput pyrosequencing. Water Res 47:859–869
Maphosa F, de Vos WM, Smidt H (2010a) Exploiting the ecogenomics toolbox for environmental diagnostics of organohalide-respiring bacteria. Trends Biotechnol 28:308–316
Maphosa F, Smidt H, de Vos WM, Röling WF (2010b) Microbial community- and metabolite dynamics of an anoxic dechlorinating bioreactor. Environ Sci Technol 44:4884–4890
Meng F, Chae SR, Drews A, Kraume M, Shin HS, Yang F (2009) Recent advances in membrane bioreactors (MBRs): membrane fouling and membrane material. Water Res 43:1489–1512
Munro JE, Liew EF, Coleman NV (2013) Adaptation of a membrane bioreactor to 1,2-dichloroethane revealed by 16S rDNA pyrosequencing and dhlA qPCR. Environ Sci Technol 47:13668–13676
Pasipanodya JG, McIlleron H, Burger A, Wash PA, Smith P, Gumbo T (2013) Serum drug concentrations predictive of pulmonary tuberculosis outcomes. JID 208:1464–1473
Petrovic M, de Alda MJL, Diaz-Cruz S, Postigo C, Radjenović J, Gros M, Barcelo D (2009) Fate and removal of pharmaceuticals and illicit drugs in conventional and membrane bioreactor wastewater treatment plants and by riverbank filtration. Phil Trans R Soc A 367:3979–4003
Potvin CM, Long Z, Zhou H (2012) Removal of tetrabromobisphenol A by conventional activated sludge, submerged membrane and membrane aerated biofilm reactors. Chemosphere 89:1183–1188
Radjenović J, Petrović M, Barcelo D (2009) Fate and distribution of pharmaceuticals in wastewater and sewage sludge of the conventional activated sludge (CAS) and advanced membrane bioreactor (MBR) treatment. Water Res 43:831–841
Ritalahti KM, Amos BK, Sung Y, Wu Q, Koenigsberg SS, Löffler FE (2006) Quantitative PCR targeting 16S rRNA and reductive dehalogenase genes simultaneously monitors multiple Dehalococcoides strains. Appl Environ Microbiol 72:2765–2774
Schloss PD, Gevers D, Westcott SL (2011) Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One 6:e27310
Segev O, Meusel W, Friedenberger M, Brenner A, Kushmaro A (2009) Aerobic biodegradation of the brominated flame retardants, dibromoneopentyl glycol and tribromoneopentyl alcohol. Biodegradation 20:621–627
Segev O, Shapiro OH, Brenner A, Kushmaro A (2013) Application of a unique miniature MBR for screening the biodegradation of brominated flame retardants. Desalin Water Treat 51:5909–5917
Shapiro OH, Kushmaro A (2011) Bacteriophage ecology in environmental biotechnology processes. Curr Opin Biotechnol 22:449–455
Shetti AA, Kaliwal BB (2012) Biodegradation of imidacloprid by soil isolate Brevundimonas sp. MJ15. Int J Curr Res 4:100–106
Stolz A (2009) Molecular characteristics of xenobiotic-degrading sphingomonads. Appl Microbiol Biotechnol 81:793–811
Stolz A (2014) Degradative plasmids from sphingomonads. FEMS Microbiol Lett 350:9–19
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tani A, Charoenpanich J, Mori T, Takeichi M, Kimbara K, Kawai F (2007) Structure and conservation of a polyethylene glycol-degradative operon in sphingomonads. Microbiology 153:338–346
Taş N, van Eekert MHA, de Vos WM, Smidt H (2009) The little bacteria that can—diversity, genomics and ecophysiology of ‘Dehalococcoides’ spp. in contaminated environments. Microb Biotechnol 3:389–402
Tu C, Teng Y, Luo Y, Li X, Sun X, Li Z, Liu W, Christie P (2011) Potential for biodegradation of polychlorinated biphenyls (PCBs) by Sinorhizobium meliloti. J Hazard Mater 186:1438–1444
Tyagi M, da Fonseca MM, de Carvalho CC (2011) Bioaugmentation and biostimulation strategies to improve the effectiveness of bioremediation processes. Biodegradation 22:231–241
Urgun-Demirtas M, Stark BC, Pagilla KR (2006) Comparison of 2-chlorobenzoic acid biodegradation in a membrane bioreactor by B. cepacia and B. cepacia bearing the bacterial hemoglobin gene. Water Res 40:3123–3130
Vaz-Moreira I, Nunes OC, Manaia CM (2011) Diversity and antibiotic resistance patterns of Sphingomonadaceae isolates from drinking water. Appl Environ Microbiol 77:5697–5706
Visvanathan C, Thu LN, Jegatheesan V, Anotai J (2005) Biodegradation of pentachlorophenol in a membrane bioreactor. Desalination 183:455–464
Wan CY, De Wever H, Diels L, Thoeye C, Liang JB, Huang LN (2011) Biodiversity and population dynamics of microorganisms in a full-scale membrane bioreactor for municipal wastewater treatment. Water Res 45:1129–1138
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267
Xiao J, Guo L, Wang S, Lu Y (2010) Comparative impact of cadmium on two phenanthrene-degrading bacteria isolated from cadmium and phenanthrene co-contaminated soil in China. J Hazard Mater 174:818–823
Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden T (2012) Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinforma 13:134
Zhao Y, Liu ZJ, Liu FX, Li ZY (2011) Cometabolic degradation of trichloroethylene in a hollow fiber membrane reactor with toluene as a substrate. J Membr Sci 372:322–330
Acknowledgments
The authors gratefully acknowledge the financial support of the German Ministry of Education and Research (BMBF; grant no. 02WA1052), the Israeli Ministry of Science, Culture, and Sport (BMBF-MOST grant no. WT-901) and the Ramat Hovav Council, Israel. The authors thank Sheli Radoshitzky and Esti Kramarsky-Winter for their helpful comments on the manuscript, and Yigal Kotler and Tal Biton for their assistance with the statistical analysis.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Gerald Thouand
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 133 kb)
Rights and permissions
About this article
Cite this article
Zangi-Kotler, M., Ben-Dov, E., Tiehm, A. et al. Microbial community structure and dynamics in a membrane bioreactor supplemented with the flame retardant dibromoneopentyl glycol. Environ Sci Pollut Res 22, 17615–17624 (2015). https://doi.org/10.1007/s11356-015-4975-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-015-4975-8