Abstract
In this study, the mating system, contemporary pollen flow, and landscape pollen connectivity of the wild olive tree (Olea europaea subsp. cuspidata) were analyzed in a fragmented landscape of less than 4-km diameter located in north-western Ethiopia. Four remnant populations of different sizes were investigated. Eight highly polymorphic microsatellite markers were used to genotype 534 adults and 704 embryos. We used contrasting sampling schemes and different methodological approaches to analyze the pollen flow. We observed a lower rate of inbreeding and correlated mating in the fragmented vs. the non-fragmented subpopulation. Using parentage analysis, we detected a bidirectional pollen movement across subpopulations. Pollen flow was found to be directed towards small subpopulations based on parentage and anisotropic analysis. Pollen immigration amounted to more than 50%. Although most pollination occurred within a distance of less than 200 m, longer distance pollen movements of more than 3 km were also detected. Pollen dispersal in the large and dense subpopulation was reduced, and a smaller number of effective pollen sources were detected compared to a smaller fragmented subpopulation. We obtained consistent estimates for the number of effective pollen donors (approximately 6 per mother tree) using three different methods. The average pollen dispersal distance at the landscape level amounted to 276 m while at the local level, 174 m was estimated. Bigger trees were better pollen contributors than smaller trees. We showed here for the first time that pollen dispersal in wild olive follows a leptokurtic distribution.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Effective long-distance pollen movement enhances inter-population gene flow and reduces selfing or biparental inbreeding (Young et al. 1996; Wang et al. 2011; Kremer et al. 2012). Although studies in many tree species have shown that most pollination occurs within a few hundred meters, pollen movement over more than 100-km distance has been observed (Kremer et al. 2012). Due to anthropogenic impact on the genetic structure and adaptability of forest trees, an accurate estimate of contemporary pollen flow has become very relevant (Burczyk et al. 2004). Currently, two complementary methods in terms of sampling scale and research questions can be used to infer contemporary pollen flow, i.e., indirectly using genetic structure analysis of the pollen pool or using parentage analysis (Smouse and Sork 2004; Burczyk and Koralewski 2005). How much immigrating pollen arrives in a population and how far pollen travels are major questions in gene flow studies (Burczyk et al. 2004). The spatial scale of effective pollen dispersal is a function of different biological and physical processes that affect pollen quantity, movement, and viability before and during movement (Kremer et al. 2012). The rate of pollen flow into a population of a tree species depends on the mode of pollination, isolation from the nearest conspecific tree, landscape features, the size of the studied population, and its fragmentation level (Burczyk et al. 2004; Smouse and Sork 2004; Sork and Smouse 2006). Numerous studies showed a high pollen immigration and fat-tailed dispersal pattern in wind-pollinated tree species (Austerlitz et al. 2004; Burczyk et al. 2004).
Long-distance pollen movement can effectively connect isolated populations. Often, natural forests exist as small remnant patches mainly due to anthropogenic influence. Therefore, besides the rate and distance of pollen movement, understanding the dynamics of pollen flow (genetic connectivity) in a landscape has become an important research question (Sork et al. 1999; Burczyk et al. 2004; Sork and Smouse 2006; Kamm et al. 2009). The success of pollen flow across the landscape depends on the size of the fragmented (sub-)populations, the distances among them, the flowering synchrony, and the wind direction (Goodell et al. 1997; Fuchs et al. 2003; Saro et al. 2014). On top of this, theoretical results indicate that intraspecific spatial aggregation also affects effective dispersal (Robledo-Arnuncio and Austerlitz 2006). Empirical data under non-fragmented and fragmented scenarios indicated the presence of equally large-scale pollen flow (White et al. 2002; Byrne et al. 2007, 2008; Hanson et al. 2008; Ismail et al. 2012). However, the effect of gene flow in connectivity studies may not be detectable due to insufficient sampling. It is very important to consider the distance between patches with meaningful pollen exchange when analyzing genetic connectivity of fragmented (sub-)populations (Anderson et al. 2010; Mona et al. 2014). Fragmentation affects a number of mating system parameters including outcrossing rate. Although a meta-analysis of mating system studies indicated the presence of high selfing and biparental inbreeding in small and fragmented populations in general (Eckert et al. 2010), there are findings that in fragmented populations outcrossing was not reduced (Young et al. 1996; White et al. 2002; Mona et al. 2014) or even enhanced (Wang et al. 2011) compared to less fragmented populations. Another mating system parameter which can be altered by population fragmentation is the degree of correlated mating events, which is the probability that sibling pairs are fully outcrossed (Ritland 2002). In a number of studies, increased correlated mating in a fragmented population was found mainly associated with a low number of reproductive trees (e.g., Lee 2000).
In this study, we analyzed Olea europaea subsp. cuspidata, which is a native tree species widely found in the dry Afromontane sub-region of the Ethiopian highlands (Bekele-Tesemma et al. 1993). It has high economic value and is very useful for rehabilitation of a fragmented landscape in this area because of long persistence of seeds and easy regeneration (Bekele 2005). The species has been exploited for at least 3000 years, mainly for farm tool preparation and cutting for fuel; large parts of these forests were cleared for agricultural land (Darbyshire et al. 2003). At present, large to small remnant populations of wild olive are only found around older monasteries and churches and at some inaccessible mountain sites; their existence is highly scattered in the open landscape (Zegeye et al. 2011). The species is poorly investigated in comparison to its Mediterranean or Saharan counterparts and possible genetic consequences of fragmentation are unknown. Therefore, the current study was carried out in a fragmented landscape of approximately 4-km circular diameter consisting of four (two big and two very small) forest fragments using highly polymorphic microsatellite markers. The objectives were to describe the mating patterns in the fragments, to estimate distance and amount of pollen immigration, and to analyze the connectivity through pollen flow. This work is the first to investigate contemporary gene flow in this subspecies. We report the dispersal kernel in a natural population of the olive tree in order to improve our understanding of how remnant forest patches are genetically interconnected and how population density affects pollen dispersal.
Materials and methods
Study species
Wild olive tree (O. europaea L. subsp. cuspidata (Wall. ex DC.)) is naturally occurring in Eastern and Southern Africa, southwestern Asia, and some parts of China (Green 2002). In Ethiopia, it is distributed from middle to higher altitudes in the dry Afromontane sub-region and reaches an average height of up to 15 m; exceptionally, it may reach up to 25 m (Bekele-Tesemma et al. 1993). The species is a diploid (2n = 46), wind-pollinated tree with hermaphroditic or male flowers (Besnard et al. 2008; Breton and Bervillé 2013). The flowers are whitish and the fleshy fruits are oval and of purple color (Bekele-Tesemma et al. 1993). Generally, flowers of the subspecies are morphologically similar to the cultivated forms, the proportion of staminate to hermaphroditic flowers can reach approximately 10%, and both female and male gametophytes develop synchronously (Caceres et al. 2016). Flowering time in its native range is during October and December (Cuneo and Leishman 2006). From cultivated forms and Mediterranean wild counterparts, we know that pollen production is enormous in this species. Outcrossing is further enhanced through male sterility and self-incompatibility (Besnard et al. 2000; Cuevas and Polito 2004; Breton et al. 2014). Birds are the most important seed dispersers, but also rodents, the African civet, and monkeys feed on the fruits and disperse the seeds (Cuneo and Leishman 2006; Abiyu et al. 2012, 2015).
Study area
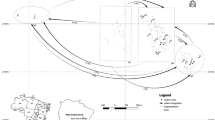
The study was done in remnant forests located in northwestern Ethiopia (Fig. 1). This landscape is intensively used for crop cultivation and pasture land and forests in this landscape are highly fragmented. All forests are found at higher altitudes compared to the agricultural land. The study site consisted of four forests, which we call subpopulations in the following. They are located within a diameter of approximately 4 km. An intensively studied plot (ISP; 2.5 ha) was sampled in the Tara Gedam monastery forest (TG) (133 ha). This unfragmented forest is Olea dominated, very dense, and accompanied by other woody species (Zegeye et al. 2011). The ISP comprised approximately 100 adult Olea individuals per hectare.
Washaendrias (WE) (23 ha) is located around 3 km west of TG and is fragmented as only 28 Olea individuals per hectare were found. The remaining two subpopulations are very small Olea-dominated patches located less than 2 km south of TG and each is less than 4 ha in size with in total approximately 60 (Silassie, SL) and 22 (Kidanemihiret, KM) Olea individuals, respectively. The two bigger stands (TG and WE) are the only large populations of wild olive within 50-km distance; however, two very small forests of not more than 50 Olea individuals are located around 10 km northeast of TG, but these trees were not sampled.
Sample collection, DNA extraction, and markers
A total of 534 individuals were sampled in the whole metapopulation. This included 299 individuals from the exhaustively sampled ISP and 138 trees randomly sampled surrounding this plot at TG. All individuals at KM (22) and random samples from WE (48) and SL (27) were also collected. Leaf samples of these trees were taken and dried in silica gel. Each potentially mature individual sample was mapped by its GPS coordinates, and measurements of tree height and diameter at breast height (DBH) were recorded.
In addition, a total of 704 fruit samples were collected from 33 seed trees. Twenty of these trees were located inside of the ISP and three outside of the ISP within TG, from which we sampled 370 and 110 fruits, respectively. Ten fruit samples were collected from a tree in the farmland very closely located south of TG and eight fruits were sampled from a tree located at the mountain escarpment located east of TG. In total, 171 fruits were sampled from six trees at WE. Nine (KM) and 24 (SL) fruit samples were collected from two trees in the remaining subpopulations (Fig. 1).
The hard seed coat (endocarp) was carefully removed from the seeds. DNA of the embryo and endosperm were extracted together by using the Qiagen DNeasy 96 Plant Kit (Qiagen, Germany). DNA from the leaf samples was extracted with the same method. Eight nuclear microsatellite markers described in Kassa et al. (2017) which are highly polymorphic with low frequency of null alleles were used for genotyping. Genotyping was outsourced to a commercial provider (ecogenics, Balgach, Switzerland) using Applied Biosystems (Foster City, CA, USA) chemistry and allele-calling tools with manual editing.
Data analysis
Genetic diversity
Genetic diversity was analyzed for adults, embryos, and subsamples of pollen donors identified using parentage analysis using FSTAT v2.9.3.2 (Goudet 2001). Parameters included allelic richness (A R ) after rarefaction (Petit et al. 1998), observed (H o ) and expected heterozygosity (H e ), and inbreeding coefficient (F is ). For comparison, genetic diversity parameters were computed between all adults with embryos and identified pollen donors. Computation was done by using 10,000 permutations of randomized datasets. In addition, the number of private alleles in adults and embryos was computed using GenAlex v6.5 (Peakall and Smouse 2006, 2012).
Inbreeding and correlated mating
Mating system parameters were computed for the whole metapopulation as well as separately for two of the subpopulations TG (unfragmented) and WE (fragmented). Computation was done based on the assumption of a mixed mating model using MLTR implementing the Expectation-Maximization (EM) algorithm (Ritland 2002). Parameters estimated were multi-locus outcrossing rate (t m), biparental inbreeding (t m -t s) as the difference between the averaged single-locus estimate (t s) and (t m), because t s is more biased downwards than t m when mating between relatives occurred, and correlation of paternity (r p). The effective number of pollen donors per mother tree (N ep) was obtained as (1/r p). One thousand bootstraps were computed to calculate the standard errors (SE). The latter were used to determine the probability whether t m was significantly smaller than 1 (mean ± 1.96 [SE] < 1) and that the other parameters (t m -t s and r p ) were greater than 0 (mean ± 1.96 [SE] > 0). A Z-test was used to evaluate the significance of differences between the mean mating system parameter estimates of the fragmented (WE) and non-fragmented (TG) subpopulations, assuming normality for t m, t m-t s, and r p , with normal approximation of the binomial for selfing rate (s) obtained from parentage analysis.
Paternity
Pollen donors were determined for 704 embryos considering all adult trees sampled as potential pollen sources. Paternity was assigned using the corrected likelihood equation method (Kalinowski et al. 2007) implemented in Cervus 3.0.7 (Marshall et al. 1998). Data were simulated using the reference frequency of genotypes to obtain the likelihood (LOD score) of true parents. The simulation was run by setting the number of mating events to 10,000. The proportion of candidate pollen sources sampled was assumed to be 50% of the potential pollen donors based on similar expected and observed parentage rates. The proportion of loci typed amounted to 0.981 in our dataset, and the mistyping proportion of loci was assumed to be 0.01. An inbreeding value of 7%, based on a previous analysis (Kassa et al. 2017), was used for simulation. The confidence of parentage during simulations was set at a relaxed (80%) and strict (95%) confidence interval. Pollen dispersal distances were calculated from the geographic (Euclidean) distance of the known mother to the first candidate pollen source determined. The frequency distribution of the distances between the mother trees and the respective observed pollen sources and the frequency distribution of the distances between mother trees with all potential pollen sources were compared by the Kolmogorov-Smirnov test (Sokal and Rohlf 1995). The probability of cryptic gene flow into the ISP was estimated as 1 − P (ex) n, where P (ex) is the combined exclusion probability and n is the total number of adults sampled (Dow and Ashley 1996).
Immigration and dispersal kernel
The level of pollen flow into the ISP and parameters of the dispersal kernel were inferred using the neighborhood mating model (Burczyk et al. 2002) implemented in NM+ (Chybicki and Burczyk 2010). The approach divides parentage probabilities into three groups: (1) selfing events within the circumscribed neighborhood, (2) outcrossing events within the circumscribed neighborhood, and (3) outcrossing events with pollen sources from outside the neighborhood. The ISP was taken as neighborhood including all adults as potential pollen donors (Chybicki and Burczyk 2010). To minimize a possible sampling bias of the background allelic frequencies, these frequencies are generally better estimated from the genotypic frequencies of the embryos of the ISP as shown by Burczyk and Chybicki (2004).
Initial estimation was done by assuming random dispersal (d p = inf) using an exponential function (b = 1). Subsequently, using an exponential function, the best initial values with the highest log likelihood were determined for distance d p by leaving it as a non-estimable parameter. With this chosen distance value, we fit the data with a simple exponential function (b = 1). Then, by selecting different initial values consecutively for each of the parameters (shape, kappa, and angle) and finally together, we fit the data to exponential power, Weibull, geometric, and 2Dt functions. Estimation of selection gradients for covariates was done for two growth parameters (DBH and tree height) using the standardized values under the assumption of a linear model (directional selection) using a simple exponential function.
Furthermore, KINDIST was used to estimate shape (b), scale (a), and average dispersal distance of the pollen dispersal distribution based on the expected decay with distance of correlated paternity among mother sibship pairs. To avoid inflation of the dispersal distance estimate (Burczyk and Koralewski 2005), sibs resulted from selfing of the parentage output were not included in this analysis. A threshold distance of 1750 m was set based on inspection of the correlated paternity plot to define unrelated pollen pools (Robledo-Arnuncio et al. 2007). An exponential power distribution was used to determine the dispersal function.
Pollen pool differentiation
Pollen pool differentiation (Φ ft ) among the pollen cloud was determined for the whole metapopulation and subpopulations TG and WE using the TWOGENER method (Smouse et al. 2001) as implemented in GenAlex v6.5 (Peakall and Smouse 2006, 2012). To compare the differentiation with an equal number of families and a comparable area size, six random families were randomly taken from the ISP. The statistical significance was tested after permutation of 999 pollen gametes among families. To reduce the effect of adult genetic structure (F is ) in subpopulations and the whole metapopulation, respectively, the population-specific Φ ft estimate was calculated as Φ ft /(1 + F is ) according to Austerlitz and Smouse (2001), where F is is the inbreeding coefficient of the adults in each subpopulation or the whole metapopulation obtained from FSTAT v 2.9.3.2 (Goudet 2001).
Results
Genetic diversity across generations
Estimates of genetic diversities are given in Table 1. In adults, a significantly higher number of alleles were observed than in the embryos. However, there was no significant difference in allelic richness between all adults and pollen donors identified in the parentage analysis. Adults shared 33 private alleles while for embryos only 16 alleles were found. Observed and unbiased expected heterozygosity as well as the inbreeding coefficient did not differ between adults and embryos, and also not between all adults and the identified pollen donors.
Inbreeding and correlated mating
For the whole metapopulation, an outcrossing rate of 0.967 was estimated. In contradiction to our expectation, we observed a significantly smaller rate of outcrossing in the unfragmented subpopulation TG compared to WE (Table 2). Biparental inbreeding and correlated paternity were significantly higher in TG compared to WE. The effective number of pollen donors per mother tree was significantly different as shown by the correlation of paternity (Table 2).
Paternity analysis
Frequencies in each distance class of observed matings and of pairs between mother trees and their potential pollen sources are shown in Fig. 2. The combined exclusion probability for the unknown candidate parent was very high (0.999). The probability of cryptic gene flow was low 0.007. At a relaxed confidence level, paternity was assigned for 424 embryos (60%). Assignment rates were higher at TG (70%), in which sampling was more exhaustive than at WE (37%) where only few trees were sampled. A total of 214 pollen donors were identified at the landscape level with on average 6.48 pollen donors per mother tree. Out of all identified mating events, 52 (12%) resulted from self-pollination. Selfing rate was higher at TG (15%) than at WE (2%). In the ISP of all female gametes, 11% were self-fertilized, 54% were outcrossed by identified pollen donors located inside the ISP, and 35% were outcrossed by pollen donors outside the ISP.
When weighted with the number of genotypes identified, 3% of total mating events in WE were attributed to pollen donors from TG, while only 1% of the mating events in TG were attributed to pollen donors originating from WE. From the total mating events at TG, seven were attributed to pollen donors from small forest fragments [KM (three), SL (four)]. One mother tree in WE was pollinated by a pollen donor from SL, and one mother tree in SL was pollinated by a WE pollen source. At KM, out of nine seeds from a single mother tree, six were fertilized by pollen coming from TG. However, out of 24 seeds from a single mother tree at SL (which is relatively far > 2 km from TG), only three were fertilized by trees from TG. From all embryos of the isolated tree located south of TG, 25% were sired by pollen donors from TG, while for all embryos of the isolated tree east of TG, no pollen donors were identified.
Mating events were significantly (P < 0.001) distance dependent. The maximum distance of observed pollen movement was 3262 m, while on average 276 m of pollen movement was estimated and more than 75% of the pollen dispersal was observed within less than 200-m distance (Fig. 2).
Immigration and dispersal kernel
At the ISP level, we obtained high (50%) estimates of pollen immigration and 14% of self-fertilization using the neighborhood model. The outcrossing rate with mates originating from the ISP was 36%. The best fit of the data was obtained by the Weibull function (log likelihood = − 8427.53). The best fitting model suggested a fat-tailed (b < 1) distribution (b = 0.7447). The average dispersal distance was estimated to be 174 m and the direction at angle of pollen dispersal (θ p ) was 326°. The results of the other dispersal kernels are shown in Table 3. Linear selection gradient coefficients for DBH (0.77) and tree height (0.62) indicated that with increasing size, also the mating success will enhance. Average growth of the sampled trees in each remnant subpopulation can be found in Supplementary Table S1. Trees were relatively small in KM.
At the landscape level, a significant (P < 0.001) decline of correlated paternity of sibship pairs over distance was observed (Spearman correlation coefficients r = − 0.181) using KINDIST. The average pollen dispersal obtained when assuming an exponential power distribution was 417 m. The shape (b = 0.175) parameter estimates indicated a leptokurtic distribution of pollen dispersal.
Pollen pool differentiation
We found a moderate global differentiation among the pollen clouds of the sampled females (Φ ft = 0.090). The estimate was threefold higher at TG (Φ ft = 0.0936) than at WE (Φ ft = 0.0335). The estimate was translated into an effective number of pollen donors per mother tree at WE (14.9), TG (5.3), and the whole metapopulation (5.6).
Discussion
Compared to the findings from other studies using microsatellites in Olea subspecies, our estimates of genetic diversity were similar (Besnard et al. 2013) or higher (Baali-Cherif and Besnard 2005; Besnard et al. 2009; García-Verdugo et al. 2010). However, the different numbers of markers used and different populations studied as well as different sampling must be taken into account. Our relative high estimates must also be interpreted in view of historic bottlenecks caused by intensive deforestation which probably affected both SL and KM (Kassa et al. 2017). Although a loss of genetic diversity, especially measured as allelic richness, is expected in fragmented populations (Mona et al. 2014), high pollen movement such as observed in this study might have counteracted diversity loss. This was also found in other species (Saro et al. 2014).
Heterozygosity estimates were similar among generations. However, a significantly smaller number of alleles were found in embryos compared to adults, indicating that as expected not all adults contributed to the embryonic pool. Allelic richness in the subsamples of pollen donors was only slightly lower than that of all adults, which implies that the lower allelic richness in the embryos was mainly due to the analysis of only 33 maternal trees. A significant loss of allelic richness in embryos compared to the adults was also reported in Laperrine’s olive (Besnard et al. 2009). Our observation of an excess of heterozygotes in the embryos (F is = − 0.036) in comparison to the adults (F is = 0.033) is only tentative as the difference between these cohorts was not significant (P < 0.08). Nevertheless, the latter finding is probably the result of predominant outbreeding due to random mating from unrelated pollen sources and relatively isolated, distant maternal trees.
Breton et al. (2014) showed that a self-incompatibility system exists in olive cultivars, and the authors suggested a sporophytic incompatibility system with several differently dominant S-alleles which prevent selfing and mating among related individuals. Moreover, effective pollen dispersal in Olea by wind may have further enhanced the outcrossing rate (Pinillos and Cuevas 2009). However, in rare cases, selfing occurs in olive cultivars (Mookerjee et al. 2005). Although this information cannot be directly transferred to O. europaea subsp. cuspidata and bottlenecks may have erased some of the S-alleles, a high outcrossing rate was to be expected. The outcrossing rate obtained with MLTR averaged over the whole metapopulation was high (0.967), and we obtained a moderate level of biparental inbreeding (0.170). Using individual parentages, we estimated a selfing rate of 12% for the whole metapopulation. When using the neighborhood model, the selfing rate was estimated as 14%. We argue that the latter estimates are of high biological relevance, as both the mother trees and their effective pollen sources were genotyped. In addition, the MLTR procedure does not consider the presence of mismatching loci between the mother and seed genotypes, and under this condition, it categorizes the selfed individuals into biparental inbreeding.
In contrast to the expectation (high level of selfing in small and fragmented populations; Eckert et al. 2010), outcrossing was significantly higher in the fragmented subpopulation WE in comparison to the larger and dense subpopulation TG. In most previously analyzed small fragmented populations, low outcrossing was due to a small number of effective pollen donors (Murawski and Hamrick 1991; Lee 2000). However, a higher effective number of pollen donors per mother tree (lower correlated paternity) were detected in the fragmented WE, indicating no limitation on mate availability. Higher pollen flow into the fragmented subpopulation could be a reason for the enhanced outcrossing rate at WE. The effect of canopy cover at TG could be another reason for the contrasting result of the outcrossing rate as a closed canopy cover can prevent effective pollen movement (Millerón et al. 2012).
The estimated effective number of pollen sources per mother tree in the metapopulation amounted in all three different methodical approaches to very similar estimates: 6.76 [MLTR], 6.48 [parentage analysis], and 5.56 [TWOGENER]. These estimates are within the lower range of wind-pollinated tree species (Smouse and Sork 2004). The estimate was unexpectedly high at WE compared to TG. A higher number of flowering trees at WE can be assumed as 56% of the sampled trees were involved in pollination, while at TG, this proportion was only 39%. Spatially restricted pollen dispersal at TG as indicated by high pollen pool heterogeneity might have contributed to the differences between the two subpopulations. In addition, variation in flowering synchrony and in male fertility between the subpopulations might have contributed to our observation (Dyer and Sork 2001; Robledo-Arnuncio et al. 2004; Bittencourt and Sebbenn 2007).
Pollen immigration into a studied area depends on several factors such as dispersal mechanism, phenology, size, and the level of isolation (Burczyk et al. 2004). Generally, pollen flow is very high when the studied trees are part of a continuous population. We investigated a circumscribed neighborhood of 2.53 ha in the center of a continuous olive forest. When applying the neighborhood model, we obtained a 50% immigration rate into this neighborhood; however, when the paternity was analyzed, a much lower rate (35%) was obtained, which was not affected by cryptic gene flow (0.7%). The results from the former may reflect reality better because parentage analysis was based on a relaxed paternity assignment which could overestimate the rate of non-immigrants, while this is taken into account in the probability models. A comparable result was obtained in Oenocarpus bataua where a 28% higher immigration rate was estimated using the neighborhood method compared to the parentage analysis (Ottewell et al. 2012).
Mating was non-random. Pollen movement was frequent within a small radius (< 200 m), which is in agreement with results observed in other wind-pollinated tree species (Robledo-Arnuncio et al. 2004; Bittencourt and Sebbenn 2007, 2008; Pluess et al. 2009). A pollen dusting experiment in the Mediterranean olive indicated that higher concentrations of pollen can be found within a short radius from the emitting source. However, Pinillos and Cuevas (2009) found out that pollen grains can cross more than 10 km. In our experiment, we detected pollen movement up to 3.26 km (the max. distance of all sampled trees was 3.46 km). This observation is similar to that of Besnard et al. (2009) who—also employing a paternity analysis—detected an effective pollen movement of approximately 2 km in a highly fragmented Laperrine’s olive population.
The occurrence of bidirectional long-distance pollen movement connected all parts of the metapopulation, implying that pollen movement can counteract a loss of genetic diversity in isolated forest stands. Similar findings were also reported in other studies at a fragmented landscape level (White et al. 2002; Wang et al. 2011; Mona et al. 2014). A high pollen movement from a dense and large to a fragmented and small subpopulation was expected based on previous findings (White et al. 2002; Bacles and Ennos 2008). Indeed, also our results support this general picture; however, the number of effective pollen donors (pollen limitation) detected in each subpopulation does not explain our results. The predominant wind direction in northwestern Ethiopia is from north to south (see Annual Climate Bulletin under http://www.ethiomet.gov.et), which does not support our finding of an anisotropic pollen movement. However, local wind directions might still affect the predominant gene flow from TG to WE. We suggest that major factors affecting anisotropy could be a contrasting tree density and/or canopy cover in each subpopulation. For instance, Dyer and Sork (2001) reported a significant negative correlation of the pollen pool diversity with tree density. Our estimate of more than a threefold inflated genetic differentiation of the pollen pool at TG compared to WE, even after accounting for the size of the population, adult genetic structure, and number of mother trees, supports our suggestion. Pollen pool differentiation among all subpopulations (Φ ft = 0.090) was very similar as in other wind-pollinated tree species (Dyer and Sork 2001; Bittencourt and Sebbenn 2008). Nevertheless, the lack of data on local wind direction and our non-exhaustive sampling at WE prevented a detailed comparison of pollen immigration under the two fragmentation scenarios.
Pollen dispersal was modeled at local and landscape levels using two methods (NM+ and KINDIST) which are designed for two different sampling schemes (Smouse and Sork 2004). Both scales resulted in a fat-tailed (b < 1) dispersal, which indicates a higher probability of long dispersal events in comparison to a simple exponential function. Similarly, a number of studies on tree species with different pollination behaviors and densities indicated that pollen dispersal follows from a slight to a strong fat-tailed dispersal curve (Austerlitz et al. 2004; Oddou-Muratorio et al. 2005; Robledo-Arnuncio and Gil 2005; Wang et al. 2010; Ramos et al. 2016). The dispersal kernel was more strongly fat tailed at the landscape than at the local level, although using two different approaches with different functions. This is consistent with the expectation from two contrasting sampling scales and our observation of effective pollination across the fragments using individual parentage. Slower decay of long-distance pollen dispersal in the landscape will maintain genetic connectivity of fragmented subpopulations. Only two parametric functions (Weibull and exponential power) were effective in modeling of the dispersal and have resulted in plausible estimates of the dispersal distance. These two functions are also mentioned as powerful to model dispersals with a high probability of long-distance events (Austerlitz et al. 2004).
At the landscape level, the average pollination distance using parentage analysis (276 m) was lower than that using KINDIST (417 m). The KINDIST method is sensitive to the presence of selfing and unknown pollen sources in the dataset; therefore, we removed all data resulting from selfing. However, the inflation due to unknown pollen sources remains unclear. Therefore, we consider the parentage estimate to be more reasonable. At the local level, the estimated average pollen dispersal distance (174 m) modeled with NM+ was lower than that at the landscape level. This is to be expected as modeling was based on samples taken only from the ISP as a small neighborhood area with a high rate of immigration. Moreover, our findings were according to the restricted pollen movement observed in the ISP. An indirect estimate of the gene dispersal (220 m) obtained from spatial genetic structure analysis using exclusively adult trees at the landscape level (Kassa et al. 2017) was lower than contemporal estimates at the same spatial scale using parentage analysis or KINDIST. We presume that the seed dispersal in Olea europaea subsp. cuspidata is more limited than pollen dispersal. Hence, the estimate of the historic gene flow underestimates the pollen flow and differences between historic and contemporary pollen movement are not so pronounced. Model predictions on the reproductive success using individual DBH and height as linear covariates indicated that bigger trees were more successful pollinators. This can be explained by their higher pollen production, higher point of pollen emission, and/or a shifted sexual function. Similarly, other studies found a positive association between size of tree and mating success (Oddou-Muratorio et al. 2005; Saro et al. 2014).
In conclusion, we observed long-distance pollen dispersal and high outcrossing in this subspecies. This is in accordance with previous results when studying historic gene flow (Kassa et al. 2017) which could effectively maintain genetic diversity in (sub-)populations in a fragmented landscape. Even small church forests distributed throughout middle and high elevations in Ethiopia very likely contribute to the maintenance of a diverse gene pool in Olea. The genetic system of this tree species is obviously adapted to fragmentation and occurrence at low density.
References
Abiyu HA (2012) The role of seed dispersal, exclosures, nurse shrubs and trees around churches and farms for restoration of ecosystem diversity and productivity in the Ethiopian highlands. Dissertation, University of Natural Resources and Life sciences, Vienna
Abiyu A, Teketay D, Glatzel G, Gratzer G (2015) Tree seed dispersal by African civets in the Afromontane highlands: too long a latrine to be effective for tree population dynamics. Afr J Ecol 53(4):588–591. https://doi.org/10.1111/aje.12198
Anderson CD, Epperson BK, Fortin MJ, Holderegger R, James PMA, Rosenberg MS, Scribner KT, Spear S (2010) Considering spatial and temporal scale in landscape-genetic studies of gene flow. Mol Ecol 19(17):3565–3575. https://doi.org/10.1111/j.1365-294X.2010.04757.x
Austerlitz F, Smouse PE (2001) Two-Generation analysis of pollen flow across a landscape. II. Relation between Φft, pollen dispersal and interfemale distance. Genetics 157(2):851–857
Austerlitz F, Dick CW, Dutech C, Klein EK, Oddou-Moratorio S, Smouse PE, Sork VL (2004) Using genetic markers to estimate the pollen dispersal curve. Mol Ecol 13(4):937–954. https://doi.org/10.1111/j.1365-294X.2004.02100.x
Baali-Cherif D, Besnard G (2005) High genetic diversity and clonal growth in relict populations of Olea europaea subsp. laperrinei (Oleaceae) from Hoggar, Algeria. Ann Bot 96(5):823–830. https://doi.org/10.1093/aob/mci232
Bacles CFE, Ennos RA (2008) Paternity analysis of pollen-mediated gene flow for Fraxinus Excelsior L. in a fragmented landscape. Heredity 101(4):368–380. https://doi.org/10.1038/hdy.2008.66
Bekele T (2005) Recruitment, survival and growth of Olea europaea subsp. cuspidata seedlings and juveniles in dry Afromontane forests of northern Ethiopia. Trop Ecol 46(1):113–126
Bekele-Tesemma A, Birnie A, Tengnäs B (1993) Useful trees and shrubs for Ethiopia: identification, propagation and pastoral communities. Regional soil conservation unit, Swedish international development authority, Nairobi 474p
Besnard G, Khadari B, Villemur P, Bervillé A (2000) Cytoplasmic male sterility in the olive (Olea europaea L.) Theor Appl Genet 100(7):1018–1024. https://doi.org/10.1007/s001220051383
Besnard G, García-Verdugo C, Rubio De Casas R, Treier UA, Galland N, Vargas P (2008) Polyploidy in the olive complex (Olea europaea): evidence from flow cytometry and nuclear microsatellite analyses. Ann Bot 101(1):25–30. https://doi.org/10.1093/aob/mcm275
Besnard G, Baali-Cherif D, Bettinelli-Riccardi S, Parietti D, Bouguedoura N (2009) Pollen-mediated gene flow in a highly fragmented landscape: consequences for defining a conservation strategy of the relict Laperrine’s olive. CR Biol 332(7):662–672. https://doi.org/10.1016/j.crvi.2009.02.003
Besnard G, El Bakkali A, Haouane H, Baali-Cherif D, Moukhli A, Khadari B (2013) Population genetics of Mediterranean and Saharan olives: geographic patterns of differentiation and evidence for early generation of admixture. Ann Bot 112(7):1293–1302. https://doi.org/10.1093/aob/mct196
Bittencourt JVM, Sebbenn AM (2007) Patterns of pollen and seed dispersal in small, fragmented population of the wind-pollinated tree Araucaria angustifolia in southern Brazil. Heredity 99(6):580–591. https://doi.org/10.1038/sj.hdy.6801019
Bittencourt JVM, Sebbenn AM (2008) Pollen movement within a continuous forest of a wind-pollinated Araucaria angustifolia, inferred from paternity and TWOGENER analysis. Conserv Genet 9(4):855–868. https://doi.org/10.1007/s10592-007-9411-2
Breton C, Bervillé A (2013) From the olive flower to the drupe: flower types, pollination, self and inter-compatibility and fruit set. In: Dr. Barbara S (ed.) The Mediterranean genetic code—grapevine and olive, In Tech, pp 291–213, DOI: https://doi.org/10.5772/55312
Breton CM, Farinelli D, Shafiq S, Heslop-Harrison JS, Sedgley M, Bervillé AJ (2014) The self-incompatibility mating system of the olive (Olea europaea L.) functions with dominance between S-alleles. Tree Genet Genomes 10(4):1055–1067. https://doi.org/10.1007/s11295-014-0742-0
Burczyk J, Chybicki IJ (2004) Cautions on direct gene flow estimation in plant populations. Evolution 11:956–963. https://doi.org/10.1111/j.0014-3820.2004.tb00430.x
Burczyk J, Koralewski TE (2005) Parentage versus two-generation analysis for estimating pollen-mediated gene flow in plant populations. Mol Ecol 14(8):2525–2537. https://doi.org/10.1111/j.1365-294X.2005.02593.x
Burczyk J, Adams WT, Moran JF, Griffin AR (2002) Complex patterns of mating revealed in a Eucalyptus regnans seed orchard using allozyme markers and the neighborhood model. Mol Ecol 11(11):2379–2391. https://doi.org/10.1046/j.1365-294X.2002.01603.x
Burczyk J, DiFazio SP, Adams WT (2004) Gene flow in forest trees: how far do genes really travel? Forest Genet 11(3–4):179–192
Byrne M, Elliott P, Yates C, Coates DJ (2007) Extensive pollen dispersal in a bird-pollinated shrub, Calothamnus quadrifidus, in a fragmented landscape. Mol Ecol 16(6):1303–1314. https://doi.org/10.1111/j.1365-294X.2006.03204.x
Byrne M, Elliott CP, Yates CJ, Coates DJ (2008) Maintenance of high pollen dispersal in Eucalyptus wandoo, a dominant tree of the fragmented agricultural region in Western Australia. Conserv Genet 9(1):97–105. https://doi.org/10.1007/s10592-007-9311-5
Caceres ME, Pupilli F, Sarri V, Mencuccini M, Ceccarelli M (2016) Floral biology in Olea europaea subsp. cuspidata: a comparative structural and functional characterization. Flora 222(6):27–36. https://doi.org/10.1016/j.flora.2016.03.006
Chybicki IJ, Burczyk J (2010) NM+: software implementing parentage-based models for estimating gene dispersal and mating patterns in plants. Mol Ecol Resour 10(6):1071–1075. https://doi.org/10.1111/j.1755-0998.2010.02849.x
Cuevas J, Polito VS (2004) The role of staminate flowers in the breeding system of Olea europaea (Oleaceae): an andromonocious, wind pollinated taxon. Ann Bot 93(5):547–553. https://doi.org/10.1093/aob/mch079
Cuneo P, Leishman MR (2006) African olive (Olea europeaea subsp. cuspidata) as an environmental weed in eastern Australia: a review. Cunninghamia 9(4):545–577
Darbyshire I, Lamb H, Umer M (2003) Forest clearance and regrowth in northern Ethiopia during the last 3000 years. The Holocene 13(4):537–546. https://doi.org/10.1191/0959683603hl644rp
Dow BD, Ashley MV (1996) Microsatellite analysis of seed dispersal and parentage of saplings in bur oak, Quercus macrocarpa. Mol Ecol 5(5):615–627. https://doi.org/10.1111/j.1365-294X.1996.tb00357.x
Dyer RJ, Sork VL (2001) Pollen pool heterogeneity in shortleaf pine, Pinus echinata Mill. Mol Ecol 10(4):859–866. https://doi.org/10.1046/j.1365-294X.2001.01251.x
Eckert CG, Kalisz S, Geber MA, Sargent R, Elle E, Cheptou PO, Goodwillie C, Johnston MO, Kelly JK, Moeller DA, Porcher E, Ree RH, Vallejo-Marín M, Winn AA (2010) Plant mating system in a changing world. Trends Ecol Evol 25(1):35–43. https://doi.org/10.1016/j.tree.2009.06.013
Fuchs EJ, Lobo JA, Quesada M (2003) Effects of forest fragmentation and flowering phenology on the reproductive success and mating patterns of the tropical dry forest tree Pachira quinata. Conserv Biol 17(1):149–157. https://doi.org/10.1046/j.1523-1739.2003.01140.x
García-Verdugo C, Forrest AD, Fay MF, Vergas P (2010) The relevance of gene flow in metapopulation dynamics of an oceanic island endemic, Olea europaea subsp. guanchica. Evolution 64(12):6525–3536. https://doi.org/10.1111/j.1558-5646.2010.01091.x
Goodell K, Elam DR, Nason JD, Ellstrand NC (1997) Gene flow among small populations of a self-incompatible plant: integration between demography and genetics. Am J Bot 84(10):1362–1371. https://doi.org/10.2307/2446134
Goudet J (2001) FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3). Available from http://www.unil.ch/izea/softwares/fstat.html. Updated from Goudet (1995)
Green PS (2002) A revision of Olea L. (Oleaceae). Kew Bull 57(1):91–140. https://doi.org/10.2307/4110824
Hanson TR, Brunsfeld SJ, Finegan B, Waits LP (2008) Pollen dispersal and genetic structure of the tropical tree Dipteryx panamensis in a fragmented Costa Rican landscape. Mol Ecol 17(8):2060–2073. https://doi.org/10.1111/j.1365-294X.2008.03726.x
Ismail SA, Ghazoul J, Ravikanth G, Shaanker RU, Kushalappa CG, Kettle CJ (2012) Does long-distance pollen dispersal preclude inbreeding in tropical trees? Fragmentation genetics of Dysoxylum malabaricum in an agro-forest landscape. Mol Ecol 21(22):5484–5496. https://doi.org/10.1111/mec.12054
Kalinowski ST, Tapar ML, Marshall TC (2007) Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol 16(5):1099–1106. https://doi.org/10.1111/j.1365-294X.2007.03089.x
Kamm U, Rotach P, Gugerli F, Siroky M, Edwards P, Holderegger R (2009) Frequent long-distance gene flow in a rare temperate forest tree (Sorbus domestica) at the landscape scale. Heredity 103(6):476–482. https://doi.org/10.1038/hdy.2009.70
Kassa A, Konrad H, Geburek T (2017) Landscape genetic structure of Olea europaea subsp. cuspidata in Ethiopian highland forest fragments. Conserv Genet 18(6):1463–1474. https://doi.org/10.1007/s10592-017-0993-z
Kremer A, Ronce O, Robledo-Arnuncio JJ, Guillaume F, Bohrer G, Nathan R, Bridle JR, Gomulkiewicz R, Klein EK, Ritland K, Kuparinen A, Gerber S, Schueler S (2012) Long-distance gene flow and adaptation of forest trees to rapid climate change. Eco Lett 5:378–392
Lee SL (2000) Mating system parameters of Dryobalanops aromatic Gaertn. f. (Dipterocarpaceae) in three different forest types and a seed orchard. Heredity 85(4):338–345. https://doi.org/10.1046/j.1365-2540.2000.00761.x
Marshall TC, Slate J, Kruuk LEB, Pemberton JM (1998) Statistical confidence for likelihood-based paternity inference in natural populations. Mol Ecol 7(5):639–655. https://doi.org/10.1046/j.1365-294x.1998.00374.x
Millerón M, de Haredia UL, Lorenzo Z, Perea R, Dounavi A, Alonso J, Gil L, Nanos N (2012) Effect of canopy closure on pollen dispersal in a wind-pollinated species (Fagus sylvatica). Plant Ecol 213(11):1715–1728. https://doi.org/10.1007/s11258-012-0125-2
Mona S, Ray N, Arenas M, Excoffier L (2014) Genetic consequences of habitat fragmentation during a range expansion. Heredity 112(3):291–299. https://doi.org/10.1038/hdy.2013.105
Mookerjee S, Guerin J, Collins G, Ford CM (2005) Paternity analysis using microsatellite markers to identify pollen donors in an olive grove. Theor Appl Genet 111(6):1174–1182. https://doi.org/10.1007/s00122-005-0049-5
Murawski DA, Hamrick JL (1991) The effect of density of flowering individuals on the mating system of nine tropical tree species. Heredity 67(2):167–174. https://doi.org/10.1038/hdy.1991.76
Oddou-Muratorio S, Klein EK, Austerlitz F (2005) Pollen flow in the wildservice tree, Sorbus torminalis (L.) Crantz. II. Pollen dispersal and heterogeneity in mating success inferred from parent-offspring analysis. Mol Ecol 14(14):4441–4452. https://doi.org/10.1111/j.1365-294X.2005.02720.x
Ottewell K, Grey E, Castillo F, Karubian J (2012) The pollen dispersal kernel and mating system of an insect-pollinated tropical palm, Oenocarpus bataua. Heredity 109(6):332–339. https://doi.org/10.1038/hdy.2012.40
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28(19):2537–2539. https://doi.org/10.1093/bioinformatics/bts460
Peakall R, Smouse PE (2012) GENALEX 6.5: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6(1):288–295. https://doi.org/10.1111/j.1471-8286.2005.01155.x
Petit RJ, El Mousadik A, Pons O (1998) Identifying populations for conservation on the basis of genetic markers. Conserv Biol 12(4):844–855. https://doi.org/10.1046/j.1523-1739.1998.96489.x
Pinillos V, Cuevas J (2009) Open pollination provides sufficient level of cross-pollen in Spanish monovarietal olive orchards. Hort Science 44(2):499–502
Pluess AR, Sork VL, Dolan B, Davis FW, Grivet D, Merg K, Papp J, Smouse PE (2009) Short distance pollen movement in a wind-pollinated tree, Quercus labata (Fagaceae). Forest Ecol Manag 258(5):735–744. https://doi.org/10.1016/j.foreco.2009.05.014
Ramos SLF, Dequigiovanni G, Sebbenn AM, Lopez MTG, Kageyama PY, Vasconcelos de Macédo JL, Kirst M, Veasey A (2016) Spatial genetic structure, genetic diversity and pollen dispersal a harvested population of Astrocaryum aculeatum in the Brazilian Amazon. BMC Genet 17(1):63. https://doi.org/10.1186/s12863-016-0371-8
Ritland K (2002) Extensions of models for the estimation of mating systems using n independent loci. Heredity 88(4):221–228. https://doi.org/10.1038/sj.hdy.6800029
Robledo-Arnuncio JJ, Austerlitz F (2006) Pollen dispersal in spatially aggregated populations. Am Nat 168(4):500–511. https://doi.org/10.1086/507881
Robledo-Arnuncio JJ, Gil L (2005) Patterns of pollen dispersal in a small population of Pinus syvestris L. revealed by total-exclusion paternity analysis. Heredity 94(1):13–22. https://doi.org/10.1038/sj.hdy.6800542
Robledo-Arnuncio JJ, Alía R, Gil L (2004) Increased selfing and correlated paternity in small population of a predominantly outcrossing conifer, Pinus sylvestris. Mol Ecol 13(9):2567–2577. https://doi.org/10.1111/j.1365-294X.2004.02251.x
Robledo-Arnuncio JJ, Austerlitz F, Smouse PE (2007) POLDISP: a software package for indirect estimation of contemporary pollen dispersal. Mol Ecol Notes 7(5):763–766. https://doi.org/10.1111/j.1471-8286.2007.01706.x
Saro I, Rubledo-Arnuncio JJ, González-Pérez MA, Sosa PA (2014) Patterns of pollen dispersal in a small population of the Canarian endemic palm (Phoenix canariensis). Heredity 113(3):215–223. https://doi.org/10.1038/hdy.2014.16
Smouse PE, Sork VL (2004) Measuring pollen flow in forest trees: an exposition of alternative approaches. For Ecol Manag 197(1-3):21–38. https://doi.org/10.1016/j.foreco.2004.05.049
Smouse PE, Dyer RJ, Westfall RD, Sork VL (2001) Two-generation analysis of pollen flow across a landscape. I. Male gamete heterogeneity among females. Evolution 55(2):260–271
Sokal RR, Rohlf FJ (1995) Biometry: the principles and practices of statistics in biological research, 3rd edn. WH Freeman and Company, New York
Sork VL, Smouse PE (2006) Genetic analysis of landscape connectivity in tree populations. Landsc Ecol 21(6):821–836. https://doi.org/10.1007/s10980-005-5415-9
Sork VL, Nason J, Campbell DR, Fernandez JF (1999) Landscape approaches to historical and contemporary gene flow in plants. Trends Ecol Evol 14(6):219–224. https://doi.org/10.1016/S0169-5347(98)01585-7
Wang J, Kang M, Gao P, Huang H (2010) Cotemporary pollen flow and mating pattern of a subtropical canopy tree. Forest Ecol Manag 260(12):2180–2188. https://doi.org/10.1016/j.foreco.2010.09.016
Wang R, Compton SG, Chen XY (2011) Fragmentation can increase spatial genetic structure without decreasing pollen-mediated gene flow in a wind-pollinated tree. Mol Ecol 20(21):4421–4432. https://doi.org/10.1111/j.1365-294X.2011.05293.x
White GM, Boshier DH, Powell W (2002) Increased pollen flow counteracts fragmentation in a tropical dry forest: an example from Swietenia humilis Zuccarini. PNAS 99(4):2038–2042. https://doi.org/10.1073/pnas.042649999
Young A, Boyle T, Brown T (1996) The population genetic consequences of habitat fragmentation for plants. Trends Ecol Evol 11(10):413–418. https://doi.org/10.1016/0169-5347(96)10045-8
Zegeye H, Teketay D, Kelbessa E (2011) Diversity and regeneration status of woody species in Tara Gedam and Abebaye forest, northwestern Ethiopia. J For Res 22(3):315–328. https://doi.org/10.1007/s11676-011-0176-6
Acknowledgments
We would like to acknowledge the project “Carbon storage and soil biodiversity in forest landscapes in Ethiopia: Knowledge base and participatory management (Carbo part)” funded by the Austrian Federal Ministry of Agriculture, Forestry, Environment and Water Management (Grant UW.1.3.2/0122–V/4/2013) for supporting this work. The help of Karl-Manfred Schweinzer and Thomas Thalmayr in creating the figures is gratefully acknowledged.
Data archiving statement
All data will be deposited in DRYAD upon acceptance for publication.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by G. G. Vendramin
Electronic supplementary material
Table S1
(DOCX 13 kb)
Rights and permissions
About this article
Cite this article
Kassa, A., Konrad, H. & Geburek, T. Mating pattern and pollen dispersal in the wild olive tree (Olea europaea subsp. cuspidata). Tree Genetics & Genomes 14, 3 (2018). https://doi.org/10.1007/s11295-017-1215-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-017-1215-z