Abstract
Oaks of Quercus Group Ilex are emblematic components of the Mediterranean landscapes and the full extent of their diversity in a geographic context is still poorly assessed. In order to detail differentiation patterns within Group Ilex and to illuminate causes and circumstances that underlie the distribution of its lineages, we examined plastome differentiation in the four species currently representing this oak group across the Mediterranean Basin (Quercus ilex, Quercus coccifera, Quercus aucheri, Quercus alnifolia). Plastid DNA variation was explored with three markers on 124 individuals and spatial analyses were performed with phylogenetic and landscape genetics approaches. A strong geographic sorting, entirely decoupled from species boundaries, was found. Spatial coincidences with phylogenetic splits highlights the presence of four geographic regions (the Middle East, the Aegean region, the central and the western Mediterranean) characterised by different degrees of isolation. Distinct sub-regional structures were identified. Inter- and intra-lineage divergence patterns appear to reflect the complex orogeny of the Mediterranean region and Pleistocene climatic oscillations, while a few widespread haplotypes might represent the remnants of the ancestral Group Ilex range. Regional differentiation in this group is recognised to be triggered by a number of factors, including disruption of the original range, incomplete lineage sorting, repeated phases of asymmetrical introgression and isolation. The key role played by some Mediterranean regions on the range establishment and dynamics of this oak group is discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Oaks (Quercus, Fagaceae) are among the most economically important and ecologically diverse woody angiosperms of the northern hemisphere. The genus comprises ca. 500–600 species occurring in temperate, subtropical and tropical forests, as well as in steppes, scrublands and open woodlands across Eurasia, North Africa and North and Central America (Govaerts and Frodin 1998, Menitsky 2005). As evergreen to deciduous shrubs or large trees, with many transitional forms, they play major ecological roles in numerous plant and animal communities (Tovar-Sanchez and Oyama 2004, Holz and Gradstein 2005, Blondel et al. 2010). Many oak species are currently threatened with extinction (IUCN 2016) mainly because of changes in land use, livestock grazing and unsustainable exploitation of wood resources (Johnson et al. 2002). Ongoing climate change constitutes an additional factor for reshaping species distributions and for diminishing genetic resources, especially in some highly vulnerable regions such as the Mediterranean Basin (Giorgi 2006, Klausmeyer and Shaw 2009, Lefèvre et al., 2013) and California (McIntyre et al. 2015).
A major research question in such a prominent group is to understand the evolutionary processes behind speciation and diversification. Oaks have been studied with respect to species phylogenies and intra-generic relationships (Denk and Grimm 2010, Hipp et al. 2014, Eaton et al. 2015), species genetic diversity/biogeography (e.g. Magri et al. 2007, Cavender-Bares et al. 2015, Bagnoli et al. 2016), historical range dynamics (Vessella et al. 2015) and ecological adaptation (e.g. Modesto et al. 2014). The ultimate objective is to correlate species history, diversity and the distribution of biological lineages to past and present ecological frameworks, in order to assess the genus’ biodiversity, anticipate the response of its ecosystems to predicted climate settings and highlight potential areas which deserve particular conservation measures (Hampe and Jump 2011, Hampe et al. 2013, Gavin et al. 2014).
The Mediterranean Basin is a world biodiversity hotspot (Myers 2000), whose vast species richness is generally attributed to biogeographical patterns associated with high landscape heterogeneity, complex geologic and climatic history and long human activity (Blondel et al. 2010, Nieto Feliner 2014). Despite numerous recent investigations, the conservation and management issues for this biota still need an improvement of our knowledge on the governing forces, main features and true extent of its biodiversity to further examine the impact of environmental features on species evolution and diversification (Médail and Diadema 2009, Conord et al. 2012, Nieto Feliner 2014).
Phylogeographical studies in this area have mostly interpreted species distribution and genetic structures against the backdrop of Quaternary glaciations. In continental Europe, numerous investigations identified latitudinal patterns of plant genetic diversity resulting from the Holocene post-glacial recolonisation of continental Europe from refugia typically located in the southern Mediterranean peninsulas (e.g. Brewer et al. 2002, Petit et al. 2003, Liepelt et al. 2009). In the Mediterranean area, Conord et al. (2012) found major longitudinal patterns of genetic diversity, generally characterised by a west to east increase attributable to recolonisation following substantial population size contraction after the Last Glacial Maximum (LGM). However, the Mediterranean area was less affected by the latest glaciations (Médail and Diadema 2009), and several lines of evidence have demonstrated that the genetic structure of some Mediterranean, long-lived woody species may also be the result of older processes (e.g. Magri et al. 2007, Désamoré et al. 2011, Migliore et al. 2012). In addition, confusing imprints of biological factors such as large-scale hybridisation have been also detected (Papageorgiou et al. 2008). Nonetheless, the genetic structures of many species and groups in the Mediterranean are still unknown, or not yet fully understood. Key palaeobiogeographical regions, particularly in the eastern and southern parts of the Mediterranean Basin, are severely understudied (Nieto Feliner 2014).
Sclerophyllous oaks of Quercus Group Ilex (Denk and Grimm 2010) are an emblematic feature of many relevant Mediterranean forest habitats (Blondel and Aronson 1999). In this region, the group currently includes four accepted species that can be sympatric and share ecological preferences: the narrow endemics Quercus alnifolia Poech. and Quercus aucheri Jaub. et Spach., and the widespread Quercus ilex L. and Quercus coccifera L. (including Quercus calliprinos (Webb) Holmboe).
Few extensive molecular studies have been conducted on this species group in the past, generally with heterogeneous markers and mostly focused on the central and western parts of the Mediterranean area. The currently accepted phylogeographic model for Q. ilex is based on the restriction length-polymorphic data of the plastid DNA (RFLPs; Lumaret et al. 2002). A sample of 174 populations covering a large part of the species range (but not extending further east than Crete) revealed the existence of five main genetic clusters, corresponding to the Balkan area, the Italian peninsula, North Africa, the eastern Iberian Peninsula and the European Atlantic coasts. The proposed genetic differentiation model outlined a Miocene (23–5.3 Ma) south-east (Aegean Sea) to south-west (Morocco) gradual range expansion, persistence in distinct southern glacial refugia and subsequent northward migration after the last glaciation (Petit et al. 2005).
Conversely, previous studies on Q. coccifera, based on nuclear markers such as the ribosomal internal transcribed spacer (nrITS), amplified fragment length polymorphisms (AFLPs) and allozymes (López de Heredia et al. 2007, Toumi and Lumaret 2010) showed very homogeneous genetic patterns and a consistent lack of geographical structure in the central-western part of the Mediterranean Basin, explained by recent (Holocene) range expansion. A comparative analysis of PCR-RFLPs retrieved three mixed ilex-coccifera chloroplast DNA lineages in the central and western parts of the Basin (López de Heredia et al. 2007), and indicated the complexity of the evolutionary history of these two oaks.
No population-scale data are available for Q. aucheri, whereas high interspecific differentiation and low admixture of nuclear microsatellite haplotypes were detected in Q. alnifolia and Q. coccifera populations from Cyprus; on the contrary, their chloroplast DNA haplotypes were highly shared, pointing towards ancient (pre-speciation) cytoplasmic introgression (Neophytou et al. 2011a, b).
In a recent work that aimed at elucidating the origin of Group Ilex, Simeone et al. (2016), analysed 20 Eurasian species (81 accessions) and found a major geographic sorting highly decoupled from species boundaries. The haplotypes of the 59 investigated Mediterranean samples formed or were part of three distinct lineages termed ‘Euro-Med’ (Euro-Mediterranean), ‘Cerris-Ilex’ and ‘WAHEA’ (West Asian-Himalayan-East Asian); haplotypes of the latter two lineages were also found in Group Cerris and a number of central and eastern Asian members of Groups Ilex and Cyclobalanopsis, respectively. Formation of these lineages likely predated the appearance of modern taxa and pointed towards a multiple origin of Mediterranean Group Ilex oaks. Overall, the surveyed data clearly indicate that the processes involved in the formation and diversification of Group Ilex are complex, strictly bound to a geographic context and yet to be assessed. In this work, we delved deeper into the genetic structures of this group in the Mediterranean by increasing the number of investigated specimens, including all areas previously not covered, and carrying out spatial analyses of the plastid DNA sequence variation. Our objectives were the following: (i) to detail diversity patterns in Group Ilex oaks across the entire Mediterranean; (ii) to assess the distribution of the three plastid lineages, evaluating the role of key areas in shaping diversification and (iii) to provide new data to better understand the establishment of Quercus Group Ilex in the Mediterranean region.
Material and methods
Plant material and DNA analyses
Holm oak (Q. ilex L.) is a prominent tree distributed throughout the Mediterranean region, from Pontic Turkey to the Moroccan Anti-Atlas and along the Atlantic coasts of the Iberian Peninsula and France (Fig. 1a). In terms of spatial distribution, it is more important in the central and western part of the Mediterranean Basin, where holm oak forests have enormous economic and ecological importance (Romane and Terradas 1992), than in the eastern part, where it is rare and nowhere dominant (Menitsky 2005). The species, provided with a good resprouting ability, has a large ecological amplitude, usually associated with marked morphological variation (heterophylly, shrubby forms). It grows in four bioclimates (sensu Emberger 1930): semiarid, sub-humid, humid and per-humid. However, it is essentially in the sub-humid bioclimate that the species occupies the widest range of situations, from cold to warm conditions and on a variety of substrata from 0 to 2500 m a.s.l., depending on the latitude and region (Barbero et al. 1992). Accordingly, and in agreement with a still contrasting taxonomy (c.f. Lumaret et al. 2002, Denk and Grimm 2010), the Iberian Q. ilex subsp. ballota (Desf.) Samp. (syn. Q. rotundifolia Lam.), differing in leaf shape (rounded vs. elongated) and some physiological characteristics such as drought resistance, was considered part of the species’ adaptive variation and included in the Q. ilex dataset.
Holly or Kermes Oak (Q. coccifera L./Q. calliprinos (Webb) Holmboe), is the only oak possessing a truly circum-Mediterranean range (Barbero et al. 1992; Fig. 1b). It is considered an indicator of the meso- and thermo-Mediterranean climax forests and maquis formations in the Basin (together with Q. ilex) and becomes dominant under more arid, xerophytic conditions and disturbance-prone environments due to characteristics such as marked frugality, resprouting capacity and strong adaptation to unfavourable soil conditions (Menitsky 2005). It grows in a wide variety of often-contrasting conditions and morphs (Balaguer et al. 2001). The type ‘Q. calliprinos’, cited by several authors, includes large individuals (up to 20 m high) and occurs in the eastern Mediterranean. The ‘Q. coccifera s.str.’ type grows in the western part of the Mediterranean region and mostly attains shrubby dimensions. However, clear-cut geographic restrictions, diagnostic morphological (Zohary 1961, Greuter et al. 1986) or molecular traits (Denk and Grimm 2010; this study) do not support this separation.
By contrast, Q. alnifolia Poech. and Q. aucheri Jaub. et Spach. are two narrow Eastern Mediterranean endemics (Fig. 1b). The former is a shrub or a small tree up to 10 m. It grows exclusively on the Troodos Mountains (Cyprus), confined to ultrabasic igneous rocks from 400 to 1800 m a.s.l. and forming pure or mixed forests and maquis. The latter occurs on some south-eastern Aegean islands (e.g. Rhodes) and along the coasts of south-western Anatolia (Browicz 1986). Formerly described as a subspecies or a variety within the broad concept of Q. coccifera (Zohary 1961), Q. aucheri is now considered a distinct species (Govaerts and Frodin 1998).
We utilised a combination of recently published and new material (59 and 65 individuals, respectively) to develop a sampling design with 124 samples covering the whole distribution range of the four investigated species. The final dataset included 116 samples of the two widespread species, Q. ilex and Q. coccifera (58 each), and eight samples of the narrow endemics Q. aucheri and Q. alnifolia (five and three, respectively). Leaves were collected from geo-referenced trees and preserved in silica gel. DNA extractions, primers and PCR protocols for each investigated plastid region (rbcL, trnK-matK, trnH-psbA) were the same as in Simeone et al. (2016). Sequencing was performed at Macrogen (http://www.macrogen.com); electropherograms were edited with CHROMAS 2.3 (http://www.technelysium.com.au) and checked visually. A set of sequences of Asian members of Group Ilex oaks (Quercus semecarpifolia, Quercus baloot, Quercus floribunda) were retrieved from the GenBank and used as outgroups. GenBank accession number, vouchers and other relevant information are reported in File S1.
Statistical and phylogeographical tools
We used MEGA 5.2 (Tamura et al. 2011) to build multiple alignments and calculate pairwise uncorrected p-genetic distances at the intra-/interspecific, intra- and intergroup levels. Haplotype lists, the main diversity parameters of the investigated markers and Tajima’s D and Fu’s Fs tests for population expansions were computed with DNASP 5.1 (Librado and Rozas 2009); the statistical significance was evaluated with coalescent simulations (1000 replicates). A planar phylogenetic network was produced with the Neighbor-Net (NN) algorithm implemented in SplitsTree4 (Bryant and Moulton 2004, Huson and Bryant 2006). The NN network was inferred from a matrix of pairwise K2P distances (Kimura 1980) calculated based on the concatenated sequence data; edge support was established using non-parametric bootstrapping and 1000 replicates. A principal coordinate analysis (PCoA) was performed using GenAlEx 6.5 (Peakall and Smouse 2012). The covariance matrix of the genetic distance after standardisation was used to explore and visualise data dissimilarities in a low-dimensional space. Initial species-wise haplotype analysis relied on median-joining (MJ) haplotype networks (Bandelt et al. 1999) inferred with NETWORK 4.6.1.1 (http://www.fluxus-engineering.com) based on the complete alignments and treating gaps as fifth state. MJ algorithm was invoked with default parameters (equal weight of transversion/transition) in order to handle large datasets and multistate characters, and the star contraction option was set in order to simplify the obtained networks by shrinking the star-like clusters of nodes around a founder population node.
To further understand the geographic differentiation patterns and discuss putative migration scenarios, we inferred full median networks for each of the recently recognised main plastid lineages (‘Euro-Med’, ‘Cerris-Ilex’, ‘WAHEA’; Simeone et al. 2016), i.e. in-depth parsimony-based haplotype networks that can reconstruct ancestor-descendant relationships (Bandelt et al. 1995). Whereas haplotype diversity in the rbcL, trnK and matK partial gene regions is mostly limited to single-nucleotide site variation, the intergenic spacer trnH-psbA differs mainly by sequence variation in length-polymorphic regions. These mutation patterns would either be over- or underrepresented in direct analysis using available software; hence, we extracted the variable sites from each sub-alignment (File S1) using MESQUITE (Maddison and Maddison 2011) and reconstructed by hand full median networks using the guidelines provided by Bandelt et al. (2000).
Spatial genetic structure analysis
The correlation between geographic and genetic distances was examined with the Mantel test (Mantel 1967) implemented with the pairwise linearised genetic distances of the three joined markers of each sample and the natural logarithm of geographic distances (straight-line distances in kilometres) among the sampling sites of each species (XLSTAT package implemented in Microsoft Excel).
The Genetic Landscape GIS Toolbox (Vandergast et al. 2011), integrated in ArcMap 9.3.1, was used to map a virtual genetic landscape from the residual values of a regression of geographic distance on genetic divergence. As input, we used the genetic distance matrices produced by MEGA 5.2, the coordinates of sampled individuals and a raster grid 30 arc-seconds resolution to clip the study area. We defined output cell size based on the raster and the number of the surrounding points to control the map layouts. The output map was generated using the Inverse Distance Weight (IDW) algorithm of spatial interpolation. Genetic divergence values were scaled between zero and one, so that regions of unusually high or low divergence, regardless of distance, could be detected and compared.
The presence of genetic barriers, corresponding to geographic zones of largest genetic differentiation among oak samples, was investigated using Monmonier’s maximum difference algorithm as implemented in BARRIER 2.2 (Manni et al. 2004). The geographic coordinates of each sampling site were connected by the Delaunay triangulation, and the corresponding Voronoï tessellation was derived. Once the network was obtained, each edge of the polygons was associated to its pairwise genetic (K2P) distance. We tested one to 10 genetic barriers, and their support was evaluated by means of 100 resampled bootstrap distance matrices. Only genetic barriers with arbitrary bootstrap support of P > 0.50 (robustness >50%) were retained.
Results
Genetic diversity and phylogeographic relationships
The dataset included 124 sequences for each of the three investigated plastid regions: 743 bp of the rbcL gene, a 692- to 695-bp-long (690 bp in Q. baloot) strand including the 3′-trnK intron and adjacent part of the matK gene, and the complete trnH-psbA intergenic spacer (547–579 bp). The multiple alignment of the three concatenated markers resulted in a matrix with 2035 characters. A 24-bp inversion found in almost 50% of the trnH-psbA sequences was reverse-complemented. Forty-two substitutions and 10 indels (1–14 bp in length) were found, resulting in high numbers of variable sites (S) and parsimony informative characters (PICs), and overall high levels of nucleotide polymorphism (θw; Table 1). Most of the variation was exhibited by the trnH-psbA fragment, where also the largest number of indels was encountered (9); Q. alnifolia and Q. aucheri alignments showed no indels. Site variation resulted in 44 total haplotypes (H), scaling-up to 62 with gaps considered, and leading to high degrees of both haplotype (h) and nucleotide (π) diversity. The complete data set showed uncorrected p-distances ranging from zero to 0.008. Quercus coccifera was the most variable; its intraspecific genetic diversity (0.008) equalled the maximum interspecific genetic distance seen in the complete data set. Conversely, all the diversity parameters exhibited by Q. ilex were lower. Tajima’s D tests were not significant; large negative and significant Fs values were recorded in the total dataset and in Q. coccifera.
We detected 42 haplotypes in Q. coccifera, 26 in Q. ilex, two in Q. aucheri and two in Q. alnifolia. Fifty-three of these either were found in single accessions (29 in Q. coccifera, 16 in Q. ilex and one in Q. alnifolia) or were shared by individuals of the same species (three haplotypes in Q. coccifera and Q. ilex, one in Q. alnifolia; File S1). Nine haplotypes (14.5% of total) were shared among members of different species (seven between Q. coccifera and Q. ilex, two between Q. coccifera and Q. aucheri). The total number of individuals sharing haplotypes at the interspecific level was high (61, corresponding to 49% of total) and ranged from one (in each species) to 10 (in Q. ilex). The relative numbers of the individuals of each species sharing the interspecific haplotypes ranged from a 1:1 to a 10:1 ratio.
As shown in Fig. 2a–b (see also File S1), unique haplotypes appear across the entire Mediterranean region, whereas species-exclusive shared haplotypes are restricted to certain areas (Cyprus, Iberian Peninsula, North Africa, Greece, western Turkey, Lebanon; haplotype codes H03, H10, H18, H34, H44, H48, H60). Generally shared (between Q. coccifera and Q. ilex or Q. aucheri) haplotypes have different spatial extensions (Fig. 2c). Some are limited to small islands (e.g. Mallorca, Spain, H11; Ikaria, Greece, H14) while others include samples distributed over large geographic distances (e.g. from Southwest Sardinia and Italy to the Black Sea, H6; from West Sicily and Sardinia to the Balkans, H28; from East Algeria and North Morocco to South and West France, H12).
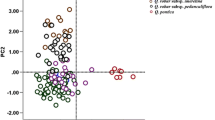
The two main coordinates of the PCoA analysis of the total dataset (Fig. 3a) explained 67.2% of variation among the data; four main groups (I, II, IV, V) and two pairs of isolated specimens (groups III, VI) can be recognised. Samples clustered according to their provenance, independent of taxonomy affiliations (Fig. 3b, c; File S1). Group I collected Q. baloot, Q. floribunda and 22 samples of Q. coccifera, Q. aucheri, Q. alnifolia from the Middle East (southern Jordan to western Turkey, including Cyprus) with ‘WAHEA’ haplotypes (Simeone et al. 2016); group II included 20 members of Q. ilex, Q. coccifera and Q. aucheri from the Aegean region (western Turkey to Greece, including Aegean islands) with ‘Cerris-Ilex’ haplotypes (Simeone et al. 2016). The remaining groups (groups III–VI) reflect haplotype diversity of the elusive ‘Euro-Med’ lineage as defined by Simeone et al. (2016). The two group III samples (Q. ilex and Q. coccifera from Crete and western Turkey) appear to be linked with the ‘Cerris-Ilex’ haplotypes; group VI samples include two close-by Q. ilex and Q. coccifera samples from north-eastern Greece that are nearly equidistant to all other samples. Thirty samples of Q. coccifera and Q. ilex from the central Mediterranean were placed in group IV (Malta, Balkan countries, Italy, Sicily, Sardinia, Corse, South France, Tunisia, Algeria) together with two Q. ilex samples from the Turkish Black Sea coast; group V was the most widespread, collecting 48 Q. coccifera and Q. ilex samples spanning from the Balkans to North Africa and the Iberian Peninsula.
PCoA analysis of the investigated dataset (a), geographic distribution of the identified clusters (b) and neighbour-net analysis showing their phylogenetic relationships (c). Colouration and roman numerals refer to distinct clusters; species, number of samples in each cluster and the three main lineages identified in Quercus group Ilex (Simeone et al. 2016) are reported
The PCoA groups can be traced in the unrooted phylogenetic network based on the concatenated sequence data (Fig. 3b). The pronounced box-like parts in the centre of the graph reflect a high level of incompatible signal in the underlying distance matrix, but also the general split between the ‘Euro-Med’ (groups III–VI) and the other two main lineages (groups I and II). It furthermore highlights the unique signal of the four, somewhat isolated samples of the ‘Euro-Med’ lineage from the Aegean region (groups III and VI), appearing intermediate between the ‘Cerris-Ilex’ and the ‘Euro-Med’ lineages. The graph further shows the high similarity of the ‘Cerris-Ilex’ s.str. haplotypes mostly confined to the Aegean region (group II, Fig. 3c) and the close relationship between the Eastern Mediterranean Q. alnifolia, Q. aucheri and Q. coccifera specimens, all showing ‘WAHEA’ haplotypes (group I), with the central Asian Ilex oaks (Q. baloot, Q. floribunda, Q. semecarpifolia). The intragroup divergences of the ‘WAHEA’ (group I) and ‘Cerris-Ilex’ (group II) lineages were lower than divergence measured within the ‘Euro-Med’ lineage (groups III–VI; 0.001 and 0.002, respectively, data not shown). The ‘Euro-Med’ lineage also was more distinct from the ‘WAHEA’ than the ‘Cerris-Ilex’ group (Table 2; 0.006 and 0.005, respectively). Within the ‘Euro-Med’ lineage, groups III and VI were more similar to ‘Cerris-Ilex’ and group V (0.003).
Spatial genetic structure
The Mantel test showed that the pairwise linearised genetic distances and the natural logarithm of geographic distances (straight-line distances in kilometres) between sampling sites of each species (Q. coccifera and Q. ilex) were not correlated (r 2 = 0.191 and 0.1335, respectively; P = 0.0002). Genetic divergence in both species cannot be linked to isolation by distance. The Genetic Landscape pattern of Q. coccifera (Fig. 4a) pinpointed regions with moderate (yellow) and sharp (red) discontinuities in species genetic diversity. Southern Anatolia and the Middle East appear to be rather uniform but strongly separated (i.e. divergent) from the rest. The same applies to the regions surrounding the Aegean Sea and North Anatolia. Populations in the Balkans and central and western Mediterranean are less differentiated relative to distance. Monmonier’s maximum difference algorithm implemented in BARRIER identified five statistically significant genetic barriers (robustness >50%). These coincide with the genetic divergence patterns (e.g. with the areas of genetic discontinuity) seen in the Genetic Landscape analysis. The main barriers (robustness =100%) run across northern and western Anatolia (roughly following the North Anatolian Fault and the contact of the Aegean Archipelago and Asia Minor), and from the south-eastern Balkan Peninsula (conjunction of the Rhodope Mts, Dinaric and Pindus Mts) along the west coast of the Aegean and the Cretan Sea. The identified barriers primarily reflect the deep divergence between the three main plastid lineages evidenced by the corresponding MJ network (see File S2a). Additional minor barriers (robustness =52–58%) run between the Balearics and Sardinia, and divide the South Anatolian and Cypriote from the Middle Eastern samples. These barriers reflect intra-lineage differentiation within the ‘Euro-Med’ (western Mediterranean) and ‘WAHEA’ lineages (eastern Mediterranean; File S2a), respectively.
The Genetic Landscape analysis for Q. ilex (Fig. 4b; see Fig. S2b for the corresponding MJ network) showed very similar results, delineating three main diverging areas: western, central and eastern Mediterranean. The eastern Mediterranean area is highly diverse in the southern Balkans and the Black Sea region, with homogeneity being restricted to the area around the Aegean Sea. BARRIER identified five statistically significant (robustness >60%) genetic barriers. Also in this case, the main genetic boundaries coincide with the patterns of genetic divergence resulting from the Genetic Landscape analysis. Such barriers run across the Bosporus (north-western Turkey), the southern Balkan Peninsula, north-south through Greece (involving the southern Dinaric Mts, Pindus, Rhodope and Peloponnese Mts), and north-south through the Balearic Sea and the Sicilian Strait.
Both inferred median networks for the ‘WAHEA’ lineage haplotypes calculated for the low (rbcL + trnK-matK) and the high (trnH-psbA) divergent marker regions essentially showed the same results, corresponding to the MJ network, genetic landscape and BARRIER data: a putatively more derived haplotype group characterizing Q. aucheri and Q. coccifera in southern Turkey (AU01-04, CO04, C051) and Rhodes (Co10), and a putatively ancestral haplotype group characteristic for the Near East, showing some variation in the trnH-psbA spacer (Fig. 5). Both lineages occur sympatrically in Cyprus (CO31, CO51), with the endemic Q. alnifolia samples showing the Near East haplotype. Reticulation is indicated by the ambiguous or recombinant signal from three disjunct individuals of Q. alnifolia (AL03) and Q. coccifera (CO42, Petra, South Jordan; CO53, Manavgat, south-western Turkey; highlighted in red). The typical Aegean ‘Cerris-Ilex’ haplotypes show very little variation (Fig. 6), but a biogeographic pattern. The most common haplotype involves most individuals and representatives of Q. ilex, Q. coccifera and Q. aucheri, covering the south-western Black Sea region and the northern Aegean islands and adjacent coastal regions of Greece and Turkey. The most common haplotype is also the most ancestral one, when compared to haplotypes of the other two main lineages (see File S2a, b). Genetic drift likely occurred in some islands (Crete: CO14, CO57, IX53; Ikaria: CO16, IX18; Gökceada: CO54).
Differentiation patterns and phylogenetic relationships between haplotypes comprising the ‘Euro-Med’ lineage are more complex, particularly when comparing the mutation patterns of the low and high divergent marker regions (Fig. 7, File S2c). However, both reconstructions are largely congruent in the recognition of the two main PCoA and NN groups (IV and V), although a higher number of haplotypes is obviously identified by the high divergent region. A striking feature is the sequence of the two group III samples (IX15, CO50). Their sequenced gene regions show four unique mutational patterns separating them from all ‘Euro-Med’ haplotypes: the thymine-dominated, length-polymorphic motif in the trnK intron links them to the ‘Cerris-Ilex’ and ‘WAHEA’ haplotypes, the unique site mutations are located in the matK gene and shared with ‘Cerris-Ilex’ (Table 3). Their trnH-psbA sequence matches the consensus within the lineage, and may actually represent the ancestral variant. In contrast, the group-specific mutations in the trnH-psbA in the two samples of group VI of the ‘Euro-Med’ lineage (IX44, CO32) are shared with the haplotypes of ‘Cerris-Ilex’, while the sequence in the other gene regions is inconspicuous. Within the two large groups (IV and V), the median networks indicate that group V, including the western Mediterranean and some Balkan samples, originated in a region spanning from northern Africa to the Iberian Peninsula, with the most ancestral haplotypes found in North Morocco and North Spain (Fig. 7, sub-lineage V2; File S2c, sub-lineages A0-A2). Morocco and Spain also host the highest diversity of haplotypes. One relatively homogenous lineage of group V, mostly composed of Q. coccifera and including three Greek Q. ilex, extends into the eastern Mediterranean (Fig. 7, sub-lineage V1; File S2c, sub-lineage A0). Group IV shows less genetic drift and occupies the central Mediterranean region (south-eastern France, Italy, north-western Africa, some parts of the Balkans). It is dominated by Q. ilex samples and notably includes three samples of Q. ilex from the humid-subtropical climate of north-western Turkey (Bosporus and Black Sea region; Fig. 7, sub-lineage IV1; File S2c, sub-lineage G1). The haplotypes of the North African Q. coccifera can be directly derived by one mutational event from those of their Italian relatives (Fig. 7, sub-lineage IV0, File S2c, sub-lineage G1-G2). The same holds for the north-westernmost samples included in this group, Q. ilex from south-eastern France.
Inferred haplotype network of the rbcL + trnK-matK marker region in Mediterranean Group Ilex oaks assigned to the ‘Euro-Med’ lineage; the occurrence of the thymine-dominated, length-polymorphic motif in the trnK intron is indicated. Species (with colours), accessions and geographic origins are reported
Discussion
In previous studies, the restricted sampling in the eastern Mediterranean region hampered the assessment of the genetic structure of Q. ilex because substantial information was lacking (Lumaret et al. 2002); at the same time, the low resolution of the markers used (allozymes; Toumi and Lumaret 2010) obscured the geographic patterning of Q. coccifera. The present work fills these gaps providing resolved phylogeographic patterns for both species. Plastome ‘non-monophyly’ (López de Heredia et al. 2007) and a complex geographic structure consisting of three major lineages with different origin (Simeone et al. 2016) are further reinforced, taxonomically and geographically expanded, and spatially detailed. New, rare haplotypes are recovered, bridging the major lineages. Finally, we identified distinct sub-regional structures, leading to a revision of the proposed models of gradual range expansion, either in ancient (Petit et al. 2005) or in recent times (Toumi and Lumaret 2010), for both Q. ilex and Q. coccifera.
Genetic diversity in Mediterranean Group Ilex oaks
The overall intra- and interspecific genetic distances (<0.01) might suggest low evolutionary rates for the chloroplast genomes of Mediterranean Group Ilex oaks, at least at the examined loci. However, within Quercus and the Fagales in general, this oak group is characterised by unusually high haplotype number and genetic diversity (Simeone et al. 2013, 2016). The genetic variation we found was sufficient to enable the detection of geographic patterns largely congruent with previous studies (Lumaret et al. 2002, López de Heredia et al. 2007). Other circum-Mediterranean woody taxa show low(er) overall diversity levels based on cpDNA sequence (e.g. Besnard et al. 2007, Rodriguez-Sanchez et al. 2009, Désamoré et al. 2011, Migliore et al. 2012, Chen et al. 2014, Mateu-Andrés et al. 2015). Standard explanations for low cpDNA variation in trees include low mutation rates, long generation times and reiterated bottleneck events promoted by ecological changes during range establishment. Although possibly affected by the limited sampling size, the low genetic diversity displayed by Q. alnifolia is in line with its status of a rare endemic relict. The non-significant Tajima’s D tests were consistent with no population growth. However, Fu’s Fs is more sensitive than Tajima’s D in detecting population expansion (Ramos-Onsins and Rozas 2002), which generally leads to largely negative Fs values. These were significantly recorded in the total dataset and in Q. coccifera but not in Q. ilex. The lower range expansion inferred for Q. ilex may correspond to the species’ demographic decrease, possibly caused by recurring climatic oscillations in the Mediterranean, with increasing aridity since the Pliocene (ca. 5.3–2.6 Mya). In contrast to the well drought-adapted Q. coccifera, Q. ilex is essentially meso-Mediterranean (Barbero et al. 1992; Quézel and Médail 2003). Menitsky (2005) also notes that Q. ilex was probably present in the Middle East until the last century (Syria, Lebanon), and it has now become sporadic in the Aegean region. The effect of natural genetic depletion in the course of range reduction could have been eventually enforced by high rates of clonal propagation following historical man-made environmental changes including over-exploitation of its forests (Blondel 2006). Quercus coccifera instead would have been less affected due to its stronger xeromorphy and higher resistance to disturbed environments. In addition, it has been noted (Lumaret et al. 2002) that in the hybridisation process, introgression is usually unidirectional, with Q. ilex predominantly acting as the maternal species and Q. coccifera as the pollen donor; therefore, asymmetric introgression and acquisition of Q. ilex haplotypes might have increased Q. coccifera plastid diversity. For instance, based on the relative number of the individuals involved, the spatial and in-depth haplotype analyses (Figs 2, 3, 4, 5, 6, and 7), it appears likely that the Sardinian and Sicilian populations of Q. coccifera picked up group IV haplotypes (H6, H28; CO02, CO39) from the local Q. ilex populations. Likewise, the Greek Q. ilex specimens with a group V haplotype (H5; IX35, IX47; otherwise restricted to Q. coccifera, File S1) may indicate mutual introgression. Alternatively, a Greek Q. ilex haplotype could have been introgressed by local Q. coccifera populations and propagated into the Adriatic region and south-eastern Italy. In general, a higher capacity of Q. coccifera to intrograde and propagate haplotypes throughout the hot and dry summer climates of the Mediterranean region seems likely. In any case, the higher diversity values of Q. coccifera vs. Q. ilex agree with the differences in their geographic range (circum-Mediterranean vs. missing from the eastern parts), historical habitat disturbance and the number of potential contact zones with related oak species.
Patterns of large haplotype sharing
Haplotype sharing among oak species is a well-established phenomenon (Petit et al. 2002). Explanations for the evident plastid ‘non-monophyly’ of oaks (e.g. incomplete sorting of ancestral lineages, introgression, recent speciation) and the extent of chlorotype exchange among interfertile species, including Q. ilex/Q. coccifera and Q. alnifolia/Q. coccifera, have been discussed in Simeone et al. (2016 and references therein). Therefore, the complex plastid differentiation patterns found in Q. coccifera and Q. ilex are not surprising (cf. López de Heredia et al. 2007). Generally, the geographic circumscription of interspecifically shared haplotypes is promoted by local introgression rather than by incomplete sorting (Hare and Avise 1998, Masta et al. 2002). Considering the distribution of the shared haplotypes, those occurring in Sardinia and Sicily (H6, H28) and in eastern Greece (H5) likely result from introgression of Q. coccifera into Q. ilex. However, it is more difficult to explain the shared haplotypes uniquely found in Ikaria and Mallorca islands and those shared across larger geographic ranges by Q. ilex/Q. coccifera and Q. coccifera/Q. aucheri (e.g. H12). The latter may result from incomplete lineage sorting (cf. Simeone et al. 2016), although occasional introgression following long-dispersal events (Lumaret et al. 2002; Toumi and Lumaret 2010) cannot be ruled out. On the other hand, recent speciation or intraspecific variation within Q. coccifera (cf. Zohary 1961) might also be invoked to explain the absence of specific haplotypes in Q. aucheri. Clearly, a more extensive population sampling and multiple markers would be needed to provide a more comprehensive characterisation of this taxon. In any case, the relative distributions of the single, intra- and interspecifically shared haplotypes appeared rather homogenous across the entire group range, and potential hybrid zones between the three ‘polyphyletic’ oaks could be inferred (besides Sardinia, Sicily, the Balkans and the western Anatolian coast) nearly all around the Mediterranean.
Phylogeographic relationships
The strong correlation between plastid haplotypes and geography is a well-known phenomenon, in both oaks (Gugger and Cavender-Bares 2013, Simeone et al. 2013) and other tree species (e.g. Acosta and Premoli 2010; Lei et al. 2012). Nevertheless, our study is the first to detail such a phenomenon in an all-inclusive taxonomic group across its entire Mediterranean range. Four geographic areas hosting distinct lineages were delineated (Fig. 3): (1) the Middle East and southern Anatolia, (2) the Aegean region, (3) the central Mediterranean (south-eastern France, Italian peninsula and north-eastern Africa) and (4) the western Mediterranean (south-western France, Iberian Peninsula, and north-western Africa).
The clusters identified in the present and other studies coincide with the major Mediterranean regions recognised as glacial refugia (Médail and Diadema 2009). This would suggest that although Pleistocene climatic oscillations (ca. 2.6–0.01 Mya) stimulated local intra-lineage diversification, the formation of the genetic lineages started earlier. This is highlighted in our data set by the exceptions from the rule, such as the general haplotype shared by the disjunct western or central Mediterranean and the southern Adriatic populations, which is also found in a few individuals of the Aegean and Euxinian (north-western Turkey) regions. Sclerophyllous oaks of Group Ilex were present in the eastern Mediterranean region since the early Miocene based on the macrofossil record (Paicheler and Blanc 1981, Velitzelos et al. 2014). It is therefore highly likely that some of the complex genetic signatures reflect older (pre-Quaternary) speciation and differentiation processes. Mesic members of Group Ilex apparently withstood the onset of the Mediterranean climate (3.2 Mya), and could be one phylogenetic source of Q. ilex (Figs 6 and 7). Simultaneously, new species evolved/migrated into the Mediterranean (Q. coccifera lineage; Figs 5, 6 and 7), where they participated in the establishment and subsequent expansion of sclerophyllous plant communities and xerophyllous taxa (Suc 1984), thanks to their xerophytic adaptations and other features such as high phenotypic plasticity, long life span, sprouting ability and bird-dispersed seeds (Herrera 1992).
Similar conclusions were made in previous works, together with general models of late Miocene gradual range expansion (Petit et al. 2005) and climate-induced Pliocene vicariance of the western populations in Q. ilex (López de Heredia et al. 2007), or recent genetic isolation of the Middle Eastern populations of Q. coccifera (Toumi and Lumaret 2010). Our study adds to this body of knowledge by providing novel regional data and explicit spatial analyses showing that the genetic structures of these species are closely linked, interdependent and can be related to the complex formation of the Mediterranean biome.
Based on the distribution of the PCoA groups and the spatial analyses (Figs 3, 4, 5, 6 and 7), the three major haplotype lineages within Quercus Group Ilex (‘WAHEA’, ‘Cerris-Ilex’, ‘Euro-Med’) are confirmed and detailed across the entire Mediterranean. A strong fragmentation in the eastern Mediterranean gene pool is evident. The central and western Mediterranean regions are characterised by a less marked fragmentation and possibly reflect to some degree higher instances of recent introgression of Q. ilex and Q. coccifera. In particular, the region including South Anatolia and the Middle East acted as a shelter and a diversification centre for species and haplotypes more tightly linked to the original ancestral stocks of the Ilex oaks, matching the ‘WAHEA’ lineage. The Aegean and adjacent Black Sea region, the location of the ‘Cerris-Ilex’ lineage but also several distinct variants of Euro-Med haplotypes, acted as a diversification centre and a crossroad (via the Balkans or the Ionian Sea) towards the central or western Mediterranean regions. In turn, the western (including Iberian Peninsula, western France and western North Africa) and central Mediterranean (including northern Balkans, south-eastern France, Italy and eastern North Africa) were home of various and more or less derived haplotypes of the ‘Euro-Med’ lineage. Active contact areas among the three major lineages are centred around the Aegean and the Black Sea coasts; contact areas among phylogroups of the ‘Euro-Med’ lineage are the Tyrrhenian region (Q. ilex: southern France, Q. coccifera: Sicily, Sardinia) and northern Africa (Q. coccifera: north-eastern Algeria).
Regional substructuring and range establishment
The Mediterranean region has been reported as home of multiple vicariant processes pre-dating the Quaternary in various Mediterranean tree species. Fragmentation of a Tethyan ancestral range during the Miocene (e.g. Magri et al. 2007), late Miocene-Pliocene climate changes (e.g. Chen et al. 2014) and isolation by distance during range establishment (e.g. Rodriguez-Sanchez et al. 2009) have been suggested as potential driving forces for gene flow interruptions and population divergence in other circum-Mediterranean woody species (see Nieto Feliner 2014). These processes produced clear phylogeographic patterns, resembling those detected in this work, although the high divergence of the Group Ilex plastome and the intense relationships shared among species and with other oak groups point to a scenario where incomplete lineage sorting and asymmetrical introgression among ancestral lineages likely played a key role. In addition, reiterated west- and eastward waves of colonisation prompted by Pleistocene climate changes contributing to an accumulation of genotypic diversity should also be considered (e.g. Migliore et al. 2012).
The Landscape Genetics and the BARRIER analyses (Fig. 4) showed that the main interruptions to haplotype migration coincide with the mountain ranges of Anatolia, Greece and the Balkans, the central Mediterranean Sea (Sea of Sardinia) and the South Aegean Sea (Libyan Sea). The Mediterranean Basin is framed by a series of mountain ranges (Rif, Maghrebides, Baetic Cordillera, Pyrenees, Alps, Dinarids, Taurus, Anatolian chain and Mt. Lebanon) that acted as centres of diversification or as barriers to plant and gene flow since their origin (Thompson 2005). Their formation was related to the Alpine orogeny, and was completed in the Pliocene (Popov et al. 2004). The strong split between the Aegean and Euxinian haplotypes of Q. aucheri, Q. coccifera and Q. ilex and the eastern Mediterranean lineage of Q. coccifera-aucheri-alnifolia fit with the so-called ‘Anatolian Diagonal’ (Davis 1971). This is a composite line of mountain ranges that runs across Anatolia, from the eastern Pontic Range to the western Taurus. It has been proposed as a significant geographic barrier shaping the phylogeography and assemblage of various animal and plant species across Turkey (e.g. Bilgin 2011; Kapli et al. 2013) since its geological origin in the Eocene (Popov et al. 2004). During some phases of the Miocene, Cyprus was connected to southern Anatolia (Robertson 1998). The distinct genetic split also fits with another important floristic division: the so-called Rechinger’s line (Rechinger 1943). This line separates Greece and the Aegean islands from Turkey, Rhodes and Western Asia, acting as a barrier to plant migration and gene flow (Bittkau and Comes 2005, Rodriguez-Sanchez et al. 2009, Gaudeul et al. 2016), and has been linked to the geographic history through the Miocene and early Pliocene (Greuter et al. 1986; Bittkau and Comes 2005, and references therein). The Aegean area (including Crete), connected to mainland Greece by numerous land bridges during the Messinian Salinity Crisis (5.93–5.33 Mya), became isolated after the re-flooding of the (western and central) Mediterranean Basin in the Pliocene (Steininger and Rögl 1984). The genetic discontinuities identified north and west of Greece separate the Aegean lineages of Q. ilex-coccifera (‘Cerris-Ilex’ group II, and groups III and VI of the ‘Euro-Med’ lineage) from the main groups within the ‘Euro-Med’ lineage (groups IV and V) and are linked to mountain ranges (the Balkan and the Hellenic arc) that were in existence since the Oligocene and acquired their present configuration in the Pleistocene (Krijgsman 2002). Accordingly, isolation of the three lineages could have started in the Miocene/Pliocene.
Inferring a colonisation sequence of the western and central Mediterranean is more difficult. Haplotype relationships across the Adriatic Sea, the Sicilian Channel and between Corsica, Sardinia and north-western Italy-south-western France constitute well-known connections for Q. ilex and Q. coccifera (Petit et al. 2005; Toumi and Lumaret 2010). The emergence of land bridges facilitated by reiterated sea-level shifts during the Pleistocene (Nieto Feliner 2014) would have allowed for genetic homogenisation and prevented increasing genetic drift in this region. At the westernmost side of the Basin, the Strait of Gibraltar also constituted a permeable barrier for both species, since the haplotypes observed in the Iberian Peninsula and North Africa constitute a unique phylogroup for both species, and one haplotype (H12) was identified on both sides of the Strait. A number of studies found that this strait was not interrupting gene flow between African and Iberian populations in both directions, despite the increased genetic drift detected in both regions (Hewitt 2011). Petit et al. (2005) concluded that Q. ilex colonised the Iberian Peninsula from North Africa, via the Italian peninsula. In this study, Q. ilex showed four distinct ‘Euro-Med’ phylogroups (i.e. groups III–VI), all with haplotypes scattered in the Eastern Mediterranean. The group IV haplotypes have a disjunct distribution centred in Italy, the Adriatic Sea and north-western Turkey (Black Sea and Marmara Sea coastal regions). No group IV haplotypes of Q. ilex are found west of southern France and Sicily; the only group V plastid signatures collected in the east are represented by one haplotype (H05; IX35, IX47) shared with a group of Balkan and Greek Q. coccifera samples. Intermediate types are not known (Fig. 7, File S2c). Our data therefore indicate that the Q. ilex haplotypes found in the Italian peninsula (group IV) are clearly distinct from the western phylogroup (group V), and both represent lineages independently evolved from the ‘Euro-Med’ common ancestor/ancestral population (Fig. 4, S2b). Considering all available evidence (Petit et al. 2005, this study), the more likely explanation is that group IV phylogroup of Q. ilex may have evolved from a now extinct progenitor; its main refuge was the Black Sea region and southern Italy, from where they colonised the rest of Italy, south-eastern France and the Adriatic Region but not the Iberian Peninsula. The Iberian Peninsula was more likely recolonised by genetically polymorphic populations from older refuges in North Africa and southern Spain. With respect to Q. coccifera, the clearly structured genetic landscape (Fig. 4a) primarily reflects its differentiation into the three main lineages characteristic of the Mediterranean Ilex oaks (Fig. 3), linked to the pre-Pleistocene topographical history of western Eurasia and to the high hybridisation ability of oaks. No barriers to gene migration are visible in the central-western Mediterranean, possibly mirroring the species’ high adaptability to the xerophytic conditions and disturbed environments that increased since the Pleistocene. Inferences of the recent biogeographic history of Q. coccifera would need to evaluate the molecular signals derived from co-occurring sibling species (Q. alnifolia, Q. aucheri and Q. ilex) throughout its range.
Conclusion
Available fossil evidence (discussed in Simeone et al. 2016) allows the interpretation of the most likely past range dynamics of evergreen oaks through the Neogene. During the Miocene and Pliocene, two distinct species complexes of Quercus Group Ilex, including morphotypes found today in Q. ilex and Q. coccifera, were prominently represented in western Eurasian and Paratethyan plant assemblages, Q. drymeja Unger and Q. mediterranea Unger (e.g. Kovar-Eder et al. 2004, Velitzelos et al. 2014). The central-eastern Mediterranean-Paratethys region therefore appears as a putative radiation centre for this oak group (Denk et al. 2012, Velitzelos et al. 2014). All gathered data point towards the Mediterranean Group Ilex plastome genepools likely originated by lineage sorting and past introgression of the early forms of these oaks into ancestral taxa, and coeval isolation favoured by the complex Miocene-Pliocene Mediterranean orogeny. The hypothesis of a southward migration from an ancestral Paratethyan range would be supported by our phylogeographic reconstruction and by a few widespread haplotypes that might represent remnants of the original range. In the Pliocene, the evergreen Mediterranean oaks became widespread after the onset of the Mediterranean climate (Suc 1984), and three divergent lineages developed gradually in the different sectors of a moulding Mediterranean. Within each sector, migration was unrestricted until the Holocene, including the crossing of major sea straits, and introgression likely reiterated until recent times. Divergence of the Mediterranean lineages in the different macro-regions is still actual today. Relevant issues are constituted by the strong isolation of the eastern region, the extremely rich gene diversity of the Aegean, the marked connection between the Balkan and the western genepools as opposed to the central Mediterranean. The key role played by North Africa still needs to be uncovered. The traditional model of gradual range expansion from single (eastern) gene pool sources should therefore be updated. Large-scale comparative nucleome investigations are required to complement the partially disentangled, puzzling phylogeography of Ilex oaks in the Mediterranean, and definitely understand the evolutionary processes behind their speciation and diversification.
References
Acosta MC, Premoli AC (2010) Evidence of chloroplast capture in South American Nothofagus (subgenus Nothofagus, Nothofagaceae). Mol Phylogenet Evol 54:235–242
Bagnoli F, Tsuda Y, Fineschi S, Bruschi P, Magri D, Zehlev P, Paule L, Simeone MC, González-Martínez SC, Vendramin GG (2016) Combining molecular and fossil data to infer demographic history of Quercus cerris: insights on European eastern glacial refugia. J Biogeogr 43:679–690
Balaguer L, Martinez-Ferri E, Valladares F, Perez-Corona ME, Baquedano FJ, Castillo FJ, Manrique E (2001) Population divergence in the plasticity of the response of Quercus coccifera to the light environment. Funct Ecol 15:124–135
Bandelt H-J, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48
Bandelt H-J, Forster P, Sykes BC, Richards MB (1995) Mitochondrial portraits of human populations using median networks. Genetics 141:743–753
Bandelt H-J, Macaulay V, Richards M (2000) Median networks: speedy construction and greedy reduction, one simulation, and two case studies from human mtDNA. Mol Phylogenet Evol 16:8–28
Barbero M, Loisel R, Quezel P (1992) Biogeography, ecology and history of Mediterranean Quercus ilex ecosystems. Vegetatio 100:19–34
Besnard G, Rubio de Casas R, Vargas P (2007) Nuclear and plastid DNA polymorphism reveals large-scale reticulation in the olive tree complex (Olea europaea L.). J Biogeogr 34:736–752
Bilgin R (2011) Back to the suture: the distribution of intraspecific genetic diversity in and around Anatolia. Int J Mol Sci 12:4080–4103
Bittkau C, Comes HP (2005) Evolutionary processes in a continental island system: Molecular phylogeography of the Aegean Nigella arvensis alliance (Ranunculaceae) inferred from chloroplast DNA. Mol Ecol 14:4065–4083
Blondel J (2006) The ‘design’ of Mediterranean landscapes: a millennial story of humans and ecological systems during the historic period. Hum Ecol 34:713–729
Blondel J, Aronson J (1999) Biology and wildlife of the Mediterranean region. Oxford University Press, Oxford
Blondel J, Aronson J, Bodiou JY, Boeuf G (2010) The Mediterranean region: biological diversity in space and time. Oxford University Press, Oxford
Brewer S, Cheddadi R, de Beaulieu JL, Reille M (2002) The spread of deciduous Quercus throughout Europe since the last glacial period. Forest Ecol Manag 156:27–48
Browicz K (1986) Chorology of trees and shrubs in South-West Asia and adjacent regions. Polish Scientific Publishers, Warsaw
Bryant D, Moulton V (2004) Neighbor-net: an agglomerative method for the construction of phylogenetic networks. Mol Biol Evol 21:255–265
Cavender-Bares J, Gonzalez-Rodriguez A, Eaton DAR, Hipp AAL, Beulke A, Manos PS (2015) Phylogeny and biogeography of the American live oaks (Quercus subsection Virentes): a genomic and population genetics approach. Mol Ecol 24:3668–3687
Chen C, Qi ZC, Xu XH, Comes HP, Koch MA, Jin XJ, Fu CX, Qiu YX (2014) Understanding the formation of Mediterranean-African-Asian disjunctions: evidence for Miocene climate-driven vicariance and recent long-distance dispersal in the Tertiary relict Smilax aspera (Smilacaceae). New Phytol 204:243–255
Conord C, Gurevitch J, Fady B (2012) Large-scale longitudinal gradients of genetic diversity: a meta-analysis across six phyla in the Mediterranean basin. Ecol Evol 2:2600–2614
Davis PH (1971) Distribution patterns in Anatolia with particular reference to endemism. In: Davis PH, Harper PC, Hedge IC (eds) Plant Life of South-West Asia. Edinburgh Botanical Society of Edinburgh, Edinburgh, pp. 15–27
Denk T, Grimm GW (2010) The oaks of western Eurasia: traditional classifications and evidence from two nuclear markers. Taxon 59:351–366
Denk T, Grímsson F, Zetter R (2012) Fagaceae from the early Oligocene of Central Europe: persisting New World and emerging Old World biogeographic links. Rev Palaeobot Palynol 169:7–20
Désamoré A, Laenen B, Devos N, Popp M (2011) Out of Africa: north-westwards Pleistocene expansions of the heather Erica arborea. J Biogeogr 38:164–176
Eaton DAR, Hipp AL, Gonzalez-Rodriguez A, Cavender-Bares J (2015) Historical introgression among the American live oaks and the comparative nature of tests for introgression. Evolution 69:2587–2601
Emberger L (1930) La végétation de la région méditerranéenne: essai d’une classification des groupements végétaux. Rev Gen Bot 42:641–662 et 705–721
Gaudeul M, Véla E, Rouhan G (2016) Eastward colonization of the Mediterranean Basin by two geographically structured clades: the case of Odontites Ludw.(Orobanchaceae). Mol Phylogenet Evol 96:140–149
Gavin DG, Fitzpatrick MC, Gugger PF, Heath KD, Rodriguez-Sanchez F et al (2014) Climate refugia: joint inference from fossil records, species distribution models and phylogeography. New Phytol 204:37–54
Giorgi F (2006) Climate change hot-spots. Geophys Res Lett 33
Govaerts R, Frodin DG (1998) World checklist and bibliography of Fagales (Betulaceae, Corylaceae, Fagaceae and Ticodendraceae). Kew Royal Botanic Gardens, Kew
Greuter W, Burdet HM, Long G (1986) Med-Checklist: a critical inventory of vascular plants of the circum-Mediterranean countries, Vol 3, Dicotyledones (Convolvulaceae-Labiatae). Conservatoire et Jardin botaniques, Ville de Genève
Gugger PF, Cavender-Bares J (2013) Molecular and morphological support for a Florida origin of the Cuban oak. J Biogeogr 40:632–645
Hampe A, Jump AS (2011) Climate relicts: past, present, future. Annl Rev Ecol Evol Syst 42:313–333
Hampe A, Rodríguez-Sánchez F, Dobrowski S, Hu FS, Gavin DG (2013) Climate refugia: from the last glacial maximum to the twenty-first century. New Phytol 197:16–18
Hare MP, Avise JC (1998) Population structure in the American oyster as inferred by nuclear gene genealogies. Mol Biol Evol 15:119–128
Herrera C (1992) Historical effects and sorting processes as explanations for contemporary ecological patterns: character syndromes in mediterranean woody plants. Am Nat 140:421–446
Hewitt GM (2011) Mediterranean peninsulas: the evolution of hotspots. In: Zachos FE, Habel JC (eds) Biodiversity hotspots. Springer, Berlin, Heidelberg, pp. 123–147
Hipp AL, Eaton DAR, Cavender-Bares J, Fitzek E, Nipper R, Manos PS (2014) A framework phylogeny of the American oak clade based on sequenced RAD data. PLoS One 9:e93975
Holz I, Gradstein RS (2005) Cryptogamic epiphytes in primary and recovering upper montane oak forests of Costa Rica—species richness, community composition and ecology. Plant Ecol 178:89–109
Huson DH, Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267
IUCN (2016) The Red List of Threatened Species. Version 2015–4; www.iucnredlist.org. Downloaded on 21 March 2016
Johnson PS, Shifley RS, Rogers R (2002) The ecology and silviculture of oaks. CABI Publishing, New York
Kapli P, Botoni D, Ilgaz Ç, Kumlutas Y, Avcı A, Rastegar-Pouyani N, Fathinia B, Lymberakis P, Ahmadzadeh F, Poulakakis N (2013) Molecular phylogeny and historical biogeography of the Anatolian lizard Apathya (Squamata, Lacertidae). Mol Phylogenet Evol 66:992–1001
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Klausmeyer KR, Shaw MR (2009) Climate change, habitat loss, protected areas and the climate adaptation potential of species in Mediterranean ecosystems worldwide. PLoS One 4:e6392
Kovar-Eder J, Kvaček Z, Ströbitzer-Hermann M (2004) The Miocene flora of Parschlug (Styria, Austria)—revision and synthesis. Ann Naturhist Mus Wien 105:45–159
Krijgsman W (2002) The Mediterranean: Mare Nostrum of Earth sciences. Earth Planet Sci Lett 205:1–12
Lefèvre F, Koskela J, Hubert J, Kraigher H, Longauer R, Olrik DC, Schueler S et al (2013) Dynamic conservation of forest genetic resources in 33 European countries. Conserv Biol 27:373–384
Lei M, Wang Q, Wu ZJ, Lopez-Pujol J, Li DZ, Zhang ZY (2012) Molecular phylogeography of Fagus engleriana (Fagaceae) in subtropical China: limited admixture among multiple refugia. Tree Genet Genomes 8:1203–1212
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Liepelt S, Cheddadi R, de Beaulieu JL, Fady B, Gömöry D, Hussendörfer E et al (2009) Postglacial range expansion and its genetic imprints in Abies alba (Mill.)—a synthesis from palaeobotanic and genetic data. Rev Palaeobot Palynol 153:139–149
López de Heredia U, Jiménez P, Collada C, Simeone MC, Bellarosa R, Schirone B, Cervera MT, Gil L (2007) Multi-marker phylogeny of three evergreen oaks reveals vicariant patterns in the Western Mediterranean. Taxon 56:1209–1209
Lumaret R, Mir C, Michaux H, Raynal V (2002) Phylogeographic variation of chloroplast DNA in holm oak (Q. ilex L.). Mol Ecol 11:2327–2336
Maddison WP, Maddison DR (2011) Mesquite: a modular system for evolutionary analysis. Version 2.75. http://mesquiteproject.org
Magri D, Fineschi S, Bellarosa R, Buonamici A, Sebastiani F, Schirone B, Simeone MC, Vendramin GG (2007) The distribution of Quercus suber chloroplast haplotypes matches the palaeogeographical history of the western Mediterranean. Mol Ecol 16:5259–5266
Manni F, Guerard E, Heyer E (2004) Geographic patterns of (genetic, morphologic, linguistic) variation: how barriers can be detected by using Monmonier’s algorithm. Hum Biol 76:173–190
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
Masta SE, Sullivan B, Lamb T, Routman EJ (2002) Phylogeography, species boundaries, and hybridization among toads of the Bufoamericanus group. Mol Phylogenet Evol 24:302–314
Mateu-Andrés I, Ciurana M-J, Aguilella A, Boisset F, Guara M, Laguna E, Currás R, Ferrer P, Vela E, Puche MF, Pedrola-Monfort J (2015) Plastid DNA homogeneity in Celtis australis L. (Cannabaceae) and Nerium oleander L. (Apocynaceae) throughout the Mediterranean Basin. Int J Plant Sci 176:421–432
McIntyre PJ, Thorne JH, Dolanc CR, Flint AL, Flint LE, Kelly M, Ackerly DD (2015) Twentieth-century shifts in forest structure in California: denser forests, smaller trees, and increased dominance of oaks. Proc Natl Acad Sci U S A 112:1458–1463
Médail F, Diadema K (2009) Glacial refugia influence plant diversity patterns in the Mediterranean Basin. J Biogeogr 36:1333–1345
Menitsky YL (2005) Oaks of Asia. Science Publishers, Enfield, New Hampshire
Migliore J, Baumel A, Juin M, Médail F (2012) From Mediterranean shores to central Saharan mountains: key phylogeographical insights from the genus Myrtus. J Biogeogr 39:942–956
Modesto IS, Miguel C, Pina-Martins F, Glushkova M, Veloso M, Paulo OS, Batista D (2014) Identifying signatures of natural selection in cork oak (Quercus suber L.) genes through SNP analysis. Tree Genet Genomes 10:1645–1660
Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J (2000) Biodiversity hotspots for conservation priorities. Nature 403:853–858
Neophytou C, Aravanopoulos FA, Fink S, Dounavi A (2011a) Interfertile oaks in an island environment. II. Limited hybridization between Quercus alnifolia Poech and Q. coccifera L. in a mixed stand. Eur J For Res 130:623–635
Neophytou C, Dounavi A, Fink S, Aravanopoulos FA (2011b) Interfertile oaks in an island environment: I. High nuclear genetic differentiation and high degree of chloroplast DNA sharing between Q. alnifolia and Q. coccifera in Cyprus. A multipopulation study. Eur J For Res 130:543–555
Nieto Feliner G (2014) Patterns and processes in plant phylogeography in the Mediterranean Basin. A review. Perspect Plant Ecol Evol Syst 16:265–278
Paicheler JC, Blanc C (1981) La flore du bassin lacustre miocène de Bes-Konak (Anatolie septentrionale, Turquie). Géol Médit 8:19–60
Papageorgiou AC, Vidalis A, Gailing O, Tsiripidis I, Hatziskakis S, Boutsios S et al (2008) Genetic variation of beech (Fagus sylvatica L.) in Rodopi (N.E. Greece). Eur J For Res 127:81–88
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in excel. Population genetic software for teaching and research—an update. Bioinformatics 28:2537–2539
Petit RJ, Csaikl UM, Bordács S, Burg K, Coart E, Cottrell J, Van Dam B, Deans JD, Dumolin-Lapègue S, Fineschi S (2002) Chloroplast DNA variation in European white oaks: phylogeography and patterns of diversity based on data from over 2600 populations. Forest Ecol Manag 156:5–26
Petit RJ, Aguinagalde I, de Beaulieu J-L, Bittkau C, Brewer S, Cheddadi R, Ennos R, Fineschi S, Grivet D, Lascoux M (2003) Glacial refugia: hotspots but not melting pots of genetic diversity. Science 300:1563
Petit RJ, Hampe A, Cheddadi R (2005) Climate changes and tree phylogeography in the Mediterranean. Taxon 54:877–885
Popov SV, Rögl F, Rozanov AY, Steininger FF, Shcherba IG, Kovac M (2004) Lithological-Paleogeographic maps of Paratethys. 10 maps. Late Eocene to Pliocene. Courier Forschungsinstitut Senckenberg 250:1–46
Quézel P, Médail F (2003) Ecologie et biogéographie des forêts du basin méditerranéen. Elsevier, Collection Environnement, Paris
Ramos-Onsins SE, Rozas J (2002) Statistical properties of new neutrality tests against population growth. Mol Biol Evol 19:2092–2100
Robertson AH (1998) Mesozoic-Tertiary tectonic evolution of the easternmost Mediterranean area: integration of marine and land evidence. Proceedings of the Ocean Drilling Program, Scientific Results 160
Rodriguez-Sanchez F, Guzman B, Valido A, Vargas P, Arroyo J (2009) Late Neogene history of the laurel tree (Laurus L., Lauraceae) based on phylogeographical analyses of Mediterranean and Macaronesian populations. J Biogeogr 36:1270–1281
Rechinger KH (1943) Flora Aegaea. Flora der Inseln und Halbinseln des ägäischen Meeres. Österreichische Akademie der Wissenschaften Wien, Mathematisch-Naturwissenschaftliche Klasse, Denkschriften,105/1
Romane F, Terradas J (1992) Quercus ilex ecosystems: function, dynamics and management. Kluver, Dordrecht
Simeone MC, Grimm GW, Papini A, Vessella F, Cardoni S, Tordoni E, Piredda R, Franc A, Denk T (2016) Plastome data reveal multiple geographic origins of Quercus Group Ilex. PeerJ 4:e1897
Simeone MC, Piredda R, Papini A, Vessella F, Schirone B (2013) Application of plastid and nuclear markers to DNA barcoding of Euro–Mediterranean oaks (Quercus, Fagaceae): problems, prospects and phylogenetic implications. Bot J Linn Soc 172:478–499
Steininger FF, Rögl F (1984) Paleogeography and palinspastic reconstruction of the Neogene of the Mediterranean and Paratethys. Geol Soc Lond, Spec Publ 17:659–668
Suc J-P (1984) Origin and evolution of the Mediterranean vegetation and climate in Europe. Nature 307:429–432
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thompson JD (2005) Plant Evolution in the Mediterranean. Oxford Univ. Press, Oxford
Toumi L, Lumaret R (2010) Genetic variation and evolutionary history of holly oak: a circum-Mediterranean species-complex [Quercus coccifera L./Q. calliprinos (Webb) Holmboe, Fagaceae]. Plant Syst Evol 290:159–171
Tovar-Sanchez E, Oyama K (2004) Natural hybridization and hybrid zones between Quercus crassifolia and Quercus crassipes (Fagaceae) in Mexico: morphological and molecular evidence. Am J Bot 91:1352–1363
Vandergast AG, Perry WM, Lugo RV, Hathaway SA (2011) Genetic landscapes GIS toolbox: tools to map patterns of genetic divergence and diversity. Mol Ecol Resour 11:158–161
Velitzelos D, Bouchal JM, Denk T (2014) Review of the Cenozoic floras and vegetation of Greece. Rev Palaeobot Palynol 204:56–117
Vessella F, Simeone MC, Schirone B (2015) Quercus suber range dynamics by ecological niche modelling: from the last interglacial to present time. Quat Sci Rev 119:85–93
Zohary M (1961) On the oak species of the Middle East. Bull Res Counc Isr 9:161–186
Acknowledgements
We warmly thank all the friends and colleagues that participated in the sample collection: Nisreen Al-Qadi, Farag Bakkar, Fotios Bekris, Laura Genco, Tunçay Güner, Magda Bou Dagher-Kharrat, Fazia Krouchi, Angeliki Laiou, Faouzi Maamouri, Maria Mayol, Karalambos Neophytou, Sergio Ortu, Antonello Salis, Leonardo Scuderi, Avra Schirone, Martina Temunovic, Maria Paola Tomasino, Enara Otaegi Veslin, Valasia Yakovoglou and Petar Zhelev. This work represents part of the PhD thesis of MV granted by Sabina Universitas (P.N. 566). GWG is financed by the Austrian Science Fund (FWF): Grant M-1751-B16. TD acknowledges funding by the Swedish Research Council (VR).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Data archiving statement
All sequence data generated as part of this study are available on GenBank (http://www.ncbi.nlm.nih.gov/genbank/) under accession numbers LM222072–LM222201 and LM222232-LM222296; other relevant data are within the paper and its Supplementary File S1.
Additional information
Communicated by F. Gugerli
Rights and permissions
About this article
Cite this article
Vitelli, M., Vessella, F., Cardoni, S. et al. Phylogeographic structuring of plastome diversity in Mediterranean oaks (Quercus Group Ilex, Fagaceae). Tree Genetics & Genomes 13, 3 (2017). https://doi.org/10.1007/s11295-016-1086-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-016-1086-8