Abstract
Scab, caused by the fungus Venturia inaequalis, is one of the most important diseases of apple. Although major scab resistance genes (R gene) have been widely studied, little is known about the molecular mechanisms underlying partial resistance, thought to be more durable. We used a candidate gene approach to decipher the genetic determinism of the durable partial resistance in ‘Président Roulin’, an old Belgian apple cultivar. Pathological tests using monoconidial isolates of V. inaequalis on F1 ‘Gala’ x ‘Président Roulin’ progeny suggested that partial resistance was broad spectrum but resulted from the combination of several race-specific interactions and was governed by at least five R genes. From an earlier transcript profiling study, we selected 13 pathogen-regulated genes in ‘Président Roulin’ with a known role in plant defense and characterized their expression over a time-course experiment. These candidate defense genes (CDGs) were regulated between 6 and 120 h after inoculation. Most were significantly up- or downregulated in incompatible interactions only or were induced earlier compared with compatible interactions. Among them, eight were mapped in silico within chromosomal regions containing disease resistance factors (R gene analogues, major scab R genes or quantitative trait loci). We also investigated the extent of the correlation between CDG expression data and phenotypic variation in the progeny. We estimated that the induction of nine out of 10 CDGs accounted for up to 46 % of the phenotypic variance. Our study has improved the understanding of partial apple scab resistance and could be used in developing functional molecular markers for breeding new ‘spray-free’ cultivars with durable scab resistance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Apple scab, caused by Venturia inaequalis, is one of the most severe and widespread fungal diseases, affecting most of the apple growing areas in the temperate regions of Europe, Asia, the Americas and Australasia (Dunemann and Egerer 2010). Currently, apple scab is controlled by the intensive use of pesticides in the orchard. Due to its major impact on economic cost, environment and human health, apple scab has received a lot of attention by researchers and has become the most studied plant–pathogen interaction involving a woody species (Cova et al. 2010). The growing of resistant varieties has been proposed as an effective alternative to the use of pesticides, significantly reducing the number of fungicidal treatments needed to protect susceptible cultivars (Parisi et al. 2000).
Apple genetic resistance to scab is complex, involving loci for both complete and partial resistance to the pathogen, and a ‘great deal of gray area’ between these two extremes (Poland et al. 2009). Complete resistance confers high levels of protection and fully inhibits pathogen reproduction. This resistance is governed by a single major resistance gene in the host and involves a gene-for-gene relationship (GfG) with the avirulence (Avr) gene of the pathogen (Flor 1971). It is often non-durable since it facilitates a strong directional selection on pathogen isolates presenting higher virulence, which can lead to resistance breakdown (Gessler et al. 2006), as illustrated with the Rvi6 (Vf) scab R gene (Parisi et al. 1993). In contrast, partial resistance confers a reduction rather than a lack of disease, and the disease phenotypes tend to be measured quantitatively. It is generally controlled by multiple loci of partial effects, referred to as quantitative trait loci (QTLs). Although experimental evidence for durability remains scarce and despite the fact that partial resistance could also be subjected to erosion (Caffier et al. 2014), such resistance is frequently assumed to be more durable than complete resistance (Parlevliet 2002; Kou and Wang 2010), presumably due to the smaller effects and the multiplicity of the partial resistance genes (Poland et al. 2009). Exploring partial resistance has therefore become an attractive alternative for controlling apple scab.
To date, at least 18 major resistance genes (R genes) have been identified in Malus species and domesticated apples (Bus et al. 2011; Soriano et al. 2014). Most of them have been mapped to the apple genome and their GfG relationships with Avr genes in V. inaequalis defined (Bus et al. 2011; Caffier et al. 2015), but so far only two have been cloned. The Rvi6 (Vf) resistance locus revealed the presence of a cluster of four resistance gene paralogs (called HcrVf genes), similar to the tomato Cf resistance gene, encoding leucine-rich repeat receptor-like proteins (LRR-RLP) (Vinatzer et al. 2001), and Rvi15 (Vr2) has been reported to contain three toll interleukin1 receptor-nucleotide binding site-LRR (TIR-NBS-LRR) genes (Galli et al. 2010). The function of all these genes was analyzed and only two, HcrVf2 (Belfanti et al. 2004; Joshi et al. 2011) and Vr2-C (Schouten et al. 2014) for Rvi6 and Rvi15, respectively, were shown to be functional against V. inaequalis. There is disagreement in the literature on the Vf1a (syn. HcrVf1) function (Malnoy et al. 2008; Joshi et al. 2011).
QTLs for scab resistance have been identified and mapped to 10 out of the 17 linkage groups (LGs) of apple (Durel et al. 2003, 2004; Liebhard et al. 2003; Calenge et al. 2004; Soufflet-Freslon et al. 2008). Genetic studies suggested that proteins underlying partial plant resistance were involved in diverse mechanisms related to complete resistance (defeated R gene theory), basal defense and unknown mechanisms since the function of some proteins underlying QTL has not been described yet (Ballini et al. 2008; Poland et al. 2009). Although geneticists are now equipped with the whole genome sequence assembly of Malus x domestica (Velasco et al. 2010), the nature of proteins governing partial scab resistance remains largely unknown. This is likely mainly due to the high number of candidate defense genes located inside the large confidence interval of the identified apple scab QTLs. An alternative strategy for deciphering the molecular basis of QTLs is to identify transcriptional responses of genes as key drivers of quantitative traits in experiments that combine both positional information and functional sequence tags such as cDNAs. In fact, the molecular basis underlying allelic variation at QTLs is similar to the identified variation for simple Mendelian loci, namely, alterations in gene expression or protein function (Paran and Zamir 2003). Expression gene profiling could be carried out between contrasting QTL genotypes (Wayne and McIntyre 2002; Hazen et al. 2005; Baxter et al. 2005; Steiner et al. 2009) or between each individual of a segregating population (Damerval et al. 1994; Jansen and Nap 2001; Liu et al. 2011). In this latter case, gene expression data can be analyzed in combination with molecular marker data, making possible the use of QTL analysis for the identification of influential genes and gene products (expression QTL or eQTL).
‘Président Roulin’ is an old Belgian cultivar with durable resistance and is used in apple breeding programs at the Walloon Agricultural Research Center (CRA-W) to broaden genetic apple scab resistance and thus reduce the risk of resistance breakdown. Symptoms of resistance range from chlorosis to necrosis with only slight sporulation (Chevalier class 3a) (Chevalier et al. 1991). Its main components of resistance are reduced incidence and severity compared with the susceptible cultivars ‘Jonagold’ and ‘Golden Delicious’, as well as a delay in the appearance of the first symptoms (Villette 2000). In our breeding program, the resistance in ‘Président Roulin’ has also been shown to be polygenically inherited (unpublished data), but the number and identity of resistance loci remain unknown. Previously, using cDNA Amplified Length Polymorphism (cDNA-AFLP), we analyzed the transcript profiling of ‘Président Roulin’ (partially resistant) and ‘Gala’ (susceptible) at 48 h post inoculation (hpi) (Bastiaanse et al. 2014). Among the 20,500 transcript-derived fragments (TDFs) generated during the analysis, we identified potential candidate defense genes (CDGs) that could form the basis of the partial resistance in ‘Président Roulin’. These genes were shown to be significantly (up to twofold) induced or repressed 48 h after pathogen challenge, compared with the susceptible cultivar ‘Gala’, and encoded for proteins known to act in disease resistance in other plant–pathogen interactions.
In this study, we first characterized the genetic determinism of the partial resistance of ‘Président Roulin’ by looking for differential interactions between various races of V. inaequalis and genotypes of F1 ‘Gala’ x ‘Président Roulin’. Then we proposed a candidate gene approach (Pflieger et al. 2001) to identify potential CDGs underlying the partial resistance of ‘Président Roulin’ consisting of three steps: the choice, screening, and functional assessment of CDGs. Thirteen CDGs were chosen among the set of CDGs identified previously in ‘Président Roulin’ (Bastiaanse et al. 2014). Then, these CDGs were screened by investigating their dynamic of expression across a time-course experiment during scab infection and their co-localization with resistance gene analogues (RGAs), major R genes or apple scab QTLs. Finally, contributions to the functional assessment of a subset of 10 CDGs were achieved by investigating the extent of correlation between their expression and the level of resistance/susceptibility in the segregating population.
Materials and methods
Evaluation of the partial scab resistance of ‘Président Roulin’ in unsprayed orchards
Over some 25 years (from 1984 to 2009), scab incidence on the leaves of the partially resistant ‘Président Roulin’ cultivar has been recorded at the Walloon Agricultural Research Center in Gembloux, Belgium, in various orchards under natural scab infection and in the absence of fungicide treatments. Scab incidence was scored using a 1–9 scale adapted from Lateur and Populer (1994), where 1 = no incidence and 9 = tree completely infected. Depending on the year, scab incidence was scored between 1 and 17 trees. The highly susceptible ‘Golden Delicious’ cultivar was used as the control. Typical scab resistance reactions on ‘Président Roulin’ under natural infection in the field were described.
Plant material and inoculation with V. inaequalis
Inoculation was performed on the clonal parents ‘Président Roulin’ (partially resistant) and ‘Gala’ (susceptible) and their progeny, comprising 120 seedlings. The clonal ‘Président Roulin’ and ‘Gala’ trees were grafted on M9 rootstocks and grown in 3-l plastic pots with a potting mix in the greenhouse at 20 °C under 16 h of illumination by daylight-incandescent lights. The seedlings were sown in trays with a seed-raising mix containing a general slow-release fertilizer. The trays were saturated with water, lined with plastic and stored in a cool store for a stratification period of 80 days, followed by germination in the glasshouse. The seedlings were then grown in individual plastic pots on their own roots for 2 years. After this, they were grafted in triplicate on M9 rootstocks and grown in 3-l plastic pots with a potting mix in the glasshouse, as previously described.
In this study, various monoconidial isolates of V. inaequalis obtained from the Institut National de la Recherche Agronomique (INRA) collection at Angers, France, and from the Plant Research International (PRI) collection at Wageningen, The Netherlands, were used (Additional Table 1). For the gene expression analysis, young leaves of actively growing plants were sprayed with a suspension mix of conidia from six strains of V. inaequalis belonging to race 1: EU-B04, EU-B16, EU-D49, EU-F05, EU-F11, and EU-I09 (Caffier et al. 2015; Bus, personal communication). Isolates were grown separately and mixed together to constitute the inoculum calibrated at 2.5 × 105 conidia ml−1. The leaves were sprayed in sufficient quantities to form small droplets on the leaf surface before run off. The control plants were inoculated with sterile water. In a phytopathological test, we also inoculated monoconidial isolates belonging to races 1, 2, 3, 6, 7, 8, 9, 10 and 13 (1639, EU-NL24, EU-B04, 1066, EU-D42, EU-NL05, EU-B05, EU-D49) (Caffier et al. 2015; Bus, personal communication), separately, on ‘Gala’ x ‘Président Roulin’ progeny and their parents, using the droplet inoculation technique described by Bus et al. (2005). A 130 μl droplet containing the inoculum calibrated at 1 × 105 conidia ml−1 was deposited in the small inoculation chambers clipped onto the youngest fully expanded leaf of actively growing shoots. The inoculations were performed once on each triplicate of the 120 genotypes of the segregating population.
Isolate cultivation, storage, and inoculum preparation were carried out as described by Bastiaanse et al. (2014). After inoculation, the plants were incubated for 2 days under optimal conditions for infection (at 20 °C under maximum relative humidity, RH) and were then transferred to the greenhouse (20 °C, 60–80 % RH). The small chambers used in the droplet inoculation technique were removed after the 2-day incubation stage. Symptoms were recorded 21 days after inoculation. We scored sporulation severity on a scale of 1 to 5 (1 = no sporulation, 5 = heavy sporulation). These scores were used to compute maximum sporulation severity (MSPOR) over the genotype replications, taking account of the most susceptible plant. Where appropriate, a distinction was made in the resistance symptoms between hypersensitive reactions (HR), necrosis (N), chlorosis (Chl), stellate necrosis (SN), and stellate chlorosis (SC). The plants were classified as being resistant (R) or susceptible (S) according to the MSPOR index (R from 1 to 3 and S >3) and the presence of resistance reactions.
RNA extraction
Leaf tissues challenged by the inoculum mix of race 1 of V. inaequalis were harvested from the youngest leaves of 89 individuals randomly selected from the 120 ‘Gala’ x ‘Président Roulin’ seedlings (one plant per leaf sampling) at 48 hpi and from mock-inoculated plants of each genotype. Leaf tissues were also harvested from the clonal ‘Président Roulin’ and ‘Gala’ cultivars (two plants per leaf sampling) at 6, 24, 48, 72, 96, and 120 hpi, as well as from control plants mock-inoculated with water at each of the corresponding time points. The leaves were quickly frozen in liquid nitrogen and stored at −80 °C prior to total RNA extraction. Total RNA was isolated from 100 mg of ground leaf tissue, using the extraction method described by Gasic et al. (2004), and DNase I treatment was performed (Fermentas Inc). With total RNA extracted from the leaves of the clonal ‘Président Roulin’ and ‘Gala’ trees only, a further mRNA purification step was performed starting from 250 μg total RNA, using the Qiagen Oligotex mRNA kit (Qiagen Inc.). For all samples, RNA purity and concentration were measured using Nanodrop technology (Thermo Scientific Inc.) and double stranded cDNA was finally obtained starting from 500 ng mRNA, using the Superscript Double Stranded cDNA Synthesis kit (Invitrogen Inc.) and following the manufacturer’s instructions.
Selection of partial resistance candidate genes and quantitative real-time reverse transcription PCR (qRT-PCR)
From our previous cDNA-AFLP study (Bastiaanse et al. 2014), we selected 13 TDFs as potential CDGs for partial resistance in ‘Président Roulin’ against V. inaequalis. They were specifically induced in ‘Président Roulin’ at 48 hpi and encoded for proteins reported in the literature to have a potential role in the general defense response pathway (Table 1). They were involved in the pathogen recognition, signal transduction, transcription, reactive oxygen species (ROS) production, protein modification, carbohydrate metabolism, and cell wall organization. Among them, only two CDGs (44EU122/44EU118) were also differentially regulated in the susceptible ‘Gala’ cultivar. They were selected because they encoded for a cysteine protease that could balance the action of 37DU41, a cysteine protease inhibitor that is part of the TDF selection.
Primers specific to the TDFs were designed using Primer3 software (Rozen and Skaletsky 2000). qRT-PCR was performed using the Biorad CFX96 and Maxima SYBR Green qPCR master mix (Fermentas Inc.) following the manufacturer’s instructions. Amplification of the 13 TDFs was achieved using the mRNA extracted from the clonal trees at different time points after infection, while a subset of 10 TDFs was chosen for qRT-PCR using the mRNA from the segregating populations. This later study aimed to identify the extent of correlation between gene expression of the CDGs and resistance level in each individual of the population. The subset of 10 CDGs was selected based on the diversity of their biological function and on their mapped locations in regions known to contain disease resistance factors (RGAs, QTLs, or major apple scab R genes). Two CDGs were involved in pathogen recognition (43DU149′ and 44AU9), two in gene transcription (53HU89) and signal transduction (2EU181), and five in plant defense reactions (44EU122, 44GU182, 43DU149, 37DU41, and 44GU173).
The PCR conditions were the same for all primer pairs: initial denaturation at 95 °C for 10′, followed by 40 cycles of denaturation at 95 °C for 15″, annealing at 60 °C for 30″ and extension at 72 °C for 30″. Fluorescence data were collected at the end of the annealing step. After cycling, the samples were denatured at 95 °C for 10″. The melting curve was then determined in order to differentiate between the desired amplicons and any primer dimers or DNA contaminants (in the 65–95 °C range, with a temperature increment of 0.5 °C for 5″). Each reaction was run in duplicate (technical replicate). LinRegPCR software was used to confirm that individual PCR efficiencies were about 2 for each primer pair (Ramakers et al. 2003). A list of the specific primer pairs used for each TDF is given in (Additional Table 2).
Data analysis
Severity of scab infection on the cultivars ‘Président Roulin’ and ‘Golden Delicious’ in the orchard over the 17 years of observation were subjected to the ANOVA procedure and a Pearson correlation coefficient between both cultivar disease scores over years was calculated.
In all the gene expression experiments, the relative expression ratio between scab-inoculated and water-treated plants was evaluated using the ΔΔCt method described by Applied Biosystems (fold change = 2ΔΔCt), with the glyceraldehyde 3-phosphate dehydrogenase gene (GAPDH) as the internal reference (primers sequence F-5′CAAGGTCATCCATGACAACTTTG3′, R-5′ GTCCACCACCCTGTTGCTGTAG3′). GAPDH has appeared to be stably expressed in other qRT-PCR studies conducted on apple (Aldaghi et al. 2012; Gadiou and Kundu 2012), as well as under our experimental conditions (Bastiaanse et al. 2014). Individual ΔCt values from each technical replicate of the qRT-PCR experiments were subjected to the ANOVA procedure at a statistically significant level of P < 0.05. The biological level of significance for differential gene expression was set at the minimum level of twofold change between inoculated and non-inoculated plants.
In the segregating population, the effect of the resistant vs susceptible status of the genotypes (R/S, considered as an explanatory factor) on the log-transformed relative expression of the individual CDGs (log2ΔΔCt) was tested using a single factor ANOVA. The significance level was fixed at P < 0.05, and the proportion of variation of log2ΔΔCt explained by the R/S factor (R 2) was computed. Then in order to estimate the contribution to the sporulation severity of several CDGs jointly considered, a multiple regression of MSPOR (as the explained variable) was performed on CDGs log2ΔΔCt values (multiple explanatory variables). In this regression analysis, only the CDGs significantly associated with resistance by the ANOVA test were included. All the statistical analyses were performed using Minitab 16 software.
In silico mapping and co-localization with RGAs, QTLs, or apple scab major R genes
The CDG sequences were compared with the whole genome sequence assembly v1.0 of the ‘Golden Delicious’ apple cultivar (Velasco et al. 2010) using a BLAST-N sequence similarity search and taking account of the best blast result. QTLs and major scab R genes already identified in apple (Bus et al. 2011; Gessler et al. 2006; Soufflet-Freslon et al. 2008) were also anchored on this physical map by blasting the sequences of the flanking SSR molecular markers (retrieved from http://www.hidras.unimi.it) on the apple genome sequence assembly. We added to the map only those markers with an E value ≤3e−3, a ratio of matched bases to a marker sequence equal to 100 % and a position on the expected chromosome of ‘Golden Delicious’ (according to their genetic position). Chromosomal positions of RGAs (individual or organized in clusters) were obtained from the work reported by Perazzolli et al. (2014). Only CDGs mapping inside a QTL confidence interval or mapping within 250 kb from any RGAs or a major scab R gene were considered to co-localize with genomic regions involved in resistance.
Results
Scab incidence on ‘Président Roulin’ under natural infection in the field
Since the first evaluation in 1984, scab incidence in the field has always been low for ‘Président Roulin’, with a mean incidence ranging from 1 to 3.75, depending on the year under investigation (Fig. 1). Compared with ‘Président Roulin’, the ‘Golden Delicious’ cultivar has always been significantly more susceptible (scab incidence mean ranging from 3 to 8.3) (P < 0.001). When more scab lesions were observed on ‘Président Roulin’ leaves, a tendency of higher scab incidence levels was also observed on ‘Golden Delicious’ (Pearson correlation coefficient between both cultivar disease scores of 0.285, significant at P < 0.05). Fluctuation in scab severity in both cultivars could reflect the variation in scab pressure over the years in the orchard. In the field (Additional Figure 1), typical scab resistance reactions in ‘Président Roulin’ corresponded with a mix of Chl and N reactions with no or limited sporulation (class 3a of Chevalier et al. 1991).
Scab incidence recorded on ‘Président Roulin’ (partially resistant) and ‘Golden Delicious’ (susceptible) over some 25 years in different orchards in Gembloux, Belgium. Scab incidence was evaluated under natural infection and no fungicide treatments using the scale proposed by Lateur and Populer (1994) where 1 = no incidence and 9 = tree completely infected. Bars represent the scab incidence mean recorded on between 1 and 17 trees, depending on the year and the cultivar. Associated standard deviations of each histogram are represented
Pathological tests and disease assessment in the glasshouse
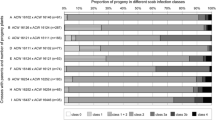
Pathological tests on ‘Gala’ x ‘Président Roulin’ progeny and their parents revealed differential interactions between the various monoconidial isolates of V. inaequalis and the genotypes. Representative symptoms for each parent, as well as the segregation ratio (R:S) observed in the progeny for each inoculum, are presented in Table 2. ‘Gala’ was highly susceptible (MSPOR 5) to all isolates tested, without any resistance reaction. In contrast, ‘Président Roulin’ was resistant to all isolates, with strong resistance reactions (Chl, N, or HR) and sporulation levels ranging from 1 (no sporulation) to 3 (50 % sporulation), depending on the isolate. Additional Figure 2 illustrates reactions observed after inoculation with the various scab isolates. Different segregation ratios between resistant and susceptible seedlings (R:S ratio) were observed for each inoculum (Table 2): R:S = 1:1 for the isolates 1639, EU-B05, EU-D49, and the inoculum mix; R:S = 1:2 for the isolates EU-D42 and EU-NL05; and R:S = 1:3 for the isolates EU-NL24, EU-B04, and 1066 (P > 0.05).
From our pathological tests (Table 3), we observed that 22 seedlings out of the 120 tested were resistant for all nine inoculums and 45 were fully susceptible. The remaining 53 seedlings showed differential interactions with V. inaequalis. The same resistance behavior was observed for some inocula: no differential interactions were observed between EU-B04 and 1066, or between EU-B05, EU-D49, and the inoculum mix. Also, some seedlings were incompatible with only one isolate among the set of isolates tested (the other isolates were all compatible with these particular seedlings). This was particularly so for 1639 and the isolates EU-B05/EU-D49/inoculum mix, which had the same resistance behavior. In contrast, more complex differential interactions were observed with the other isolates. For example, we noticed that isolate EU-NL24 showed resistance symptoms only on genotypes that were incompatible with all the isolates tested, and for eight genotypes, the resistance reactions were recorded after inoculation of all isolates, except for EU-NL24.
Co-localization of CDGs with QTLs or apple scab major R genes
Anchoring the existing apple genetic map to the whole genome sequence assembly of ‘Golden Delicious’ (Velasco et al. 2010) revealed a few discrepancies in the order of the microsatellite markers compared with the position on the genetic map produced by Gessler et al. (2006). Also, the relationship between genetic distance in centimorgans (cM) and the physical distance in mega base pairs (Mbp) was found to vary from 0.232 Mbp to 0.838 Mbp per cM, depending on the LG, with an average of 0.398 Mbp/cM.
All the CDGs could be anchored accurately on 11 out of the 17 apple LG (E value <1e-28). Of the 13 CDGs tested, eight mapped in silico on the QTL interval for apple scab resistance and/or near major apple scab R genes or RGAs (Fig. 2).
In silico mapping of the CDGs for partial apple scab resistance identified by cDNA-AFLP and co-localization with RGAs, major R genes, and QTLs for apple scab resistance. a Example of comparison of the genetic map (cM) created by Gessler et al. (2006) with the physical map (Mbp) obtained by alignment of SSR molecular markers on the whole genome sequence assembly of apple (by BLAST sequence similarity search) (Velasco et al. 2010). b Physical map obtained for the other LGs. Black bars represent the interval of confidence of apple scab resistance QTLs as given by Gessler et al. (2006). Dots represent single and clusters of RGAs according to Perazzolli et al. (2014); number of dots reflects the number of RGAs inside the clusters. Italic bold indicates the published major scab resistance genes; bold indicates our disease CGRs for partial scab resistance. Arrows indicate disease CDGs showing co-localization (<250 kb) with RGAs, QTLs, or major apple scab R genes
Investigation of the time-course expression during V. inaequalis–Malus interactions
The expression profiles of the 13 CDGs were analyzed by qRT-PCR over a time-course experiment during the V. inaequalis–Malus interaction (at 6, 24, 48, 72, 96, and 120 hpi). For 7 out of the 13 CDGs, a significant change in expression (more than twofold) was observed for ‘Président Roulin’ (partially resistant), but not for ‘Gala’ (susceptible) over the different time points tested. When a significant change was observed for ‘Gala’, it happened later than in ‘Président Roulin’, except for 43DU149 where regulation happened earlier in the susceptible cultivar but in the opposite direction (repression instead of overexpression). Finally, except for one CDG, gene expression regulation in ‘Président Roulin’ occurred between 24 hpi and 120 hpi, the latest time point tested. 44GU182 only, a gene encoding a carboxypeptidase, was regulated at the early time point of 6 hpi. Expression profiles of the CDGs are presented in Fig. 3, along with their annotation in plant defense response according to the literature (the other possible roles of these CDGs in the general plant metabolism are not presented here).
Time-course expression analysis of CDGs analyzed by qRT-PCR during V. inaequalis–Malus interaction. The possible involvement in the recognition, the transduction of the signal, and the plant defense responses of specific genes was inferred from their involvement in other plant–pathogen interactions reported in the literature. The other possible roles of these CDGs in the general plant metabolism are not presented here. Blue histograms indicate response (induction for positive values and repression for negative values) in ‘Président Roulin’ (partially resistant), red histograms in ‘Gala’ (susceptible) at 6, 24, 48, 72, 96 or 120 hpi. The relative expression of CDGs (ΔΔCt) was expressed in ‘fold change’ between infected and mock-inoculated plants at each time point of the experiment, and was based on the expression of the housekeeping gene GAPDH. Individual ΔCt values from the qRT-PCR experiments were subjected to the ANOVA procedure, at a statistically significance level of P < 0.05 (indicated by *). The bar indicates the biologically significant level of up to twofold induction or repression
Expression studies of candidate resistance genes in a segregating F1 population
A study of the regulation of expression of a subset of 10 CDGs in a segregating population (out of the 13 CDGs initially selected) was made in order to identify the extent of correlation between gene expression and resistance level in each genotype. The significance of the effect of the R/S status on the regulation of gene expression, and the proportion of variance ‘explained’ by the R/S levels conditioned by various V. inaequalis isolates are presented in Table 4. The results of EU-B04/1066 and EU-B05/EU-D49/inoculum mix were grouped because no differential interactions between those isolates were observed.
Non-significant effect (P value >0.05) was found for only one CDG (37DU41) during the ANOVA analysis. This CDG showed homology with a cysteine protease inhibitor gene. For the other CDGs tested, significant effects were obtained for at least two inocula and the R 2 varied between 4.6 and 21.3 %, depending on the inoculum tested. The largest coefficient was found for the phenotypes of EU-B05/EU-D49/inoculum mix and the expression data of 43DU149′ (CC-NBS-LRR resistance protein). When the regulation of expression of the whole set of TDFs was considered, the percentage of phenotypic variance of MSPOR accounted by the regulation of CDGs expression ranged from 16.1 % (isolate EU-NL24) to 46.0 % (EU-B05/EU-D49/inoculum mix).
Discussion
Scab resistance in ‘Président Roulin’ is partial and race specific
The resistance in ‘Président Roulin’ corresponded with partial resistance as defined by Parlevliet and van Ommeren (1975). Under natural infection in the orchard (Additional Figure 1) or when inoculated with various V. inaequalis isolates (Table 2), the resistance in ‘Président Roulin’ was incomplete, allowing limited and significantly reduced pathogen growth and reproduction as compared with a susceptible genotype. Only with the 1639 isolate did ‘Président Roulin’ exhibit strong resistance reactions of HR with no sporulation. As these symptoms have never been seen before with a spray inoculation technique using this particular isolate (data not shown) or under natural infection in the field, we believe that they were conditioned by the particular conditions in the small inoculation chambers. As already demonstrated in the apple–V. inaequalis interactions (Caffier et al. 2015), the droplet method may overexpress the resistance cascade in an incompatible reaction due to a high concentration of effectors in a limited area of the leaf. Although it would never be possible to test this thoroughly, resistance of ‘Président Roulin’ also seems to be broad spectrum, since it provided a level of resistance against all the isolates artificially inoculated as well as the isolate population present in the orchard in Belgium over 25 years of experimentation.
Pathological tests conducted on the F1 segregating population revealed differential interactions between the isolates tested and the host genotypes (Table 3). In addition, under natural infection in the field, we observed a combination of resistance reactions with various degrees of sporulation on the leaf surfaces of ‘Président Roulin’ (Additional Figure 2). These observations suggest that, along with possible basal defense response, some components of the partial resistance also show race specificity against V. inaequalis: the differential interactions would reflect the interactions between Avr genes in V. inaequalis and R genes in the host. Race specificity of QTLs has been widely demonstrated in the V. inaequalis–apple interaction (Calenge et al. 2004; Durel et al. 2003) as well as in other pathosystems (Caranta et al. 1997; Qi 1999; Dogimont et al. 2000; Chen et al. 2003; Talukder et al. 2004; González et al. 2012). One hypothesis would be that similar GfG relationships would govern complete and partial resistance. In the latter case (referred as minor gene-for-minor gene interaction), defective R genes would recognize, with low efficiency, pathogens and would trigger a weaker (partial) defense response (Parlevliet and Zadoks 1977; McHale et al. 2006). An observation supporting this hypothesis is that ‘defeated’ R genes have been shown to have a residual effect and act as a QTL against virulent strains of the pathogen. This phenomenon has been observed for the major resistance genes Rvi4 (Vh4) (Bus et al. 2011) and Rvi6 (Vf) (Durel et al. 2000) in the V. inaequalis–apple interaction and in other plant–pathogen interactions (Nass et al. 1981; Brodny et al. 1986; Li et al. 1999). So far as we know, however, evidence of a direct interaction between the product of a QTL for resistance and the matching Avr factor is still lacking. Genes controlling partial resistance to pathogens remain poorly documented, and the molecular mechanisms underlying this kind of resistance are still unknown.
A hypothetical model of GfG relationships governing the partial resistance of ‘Président Roulin’
Based on the patterns of differential interactions observed in the segregating population, and the theory that for each R gene in the host, there is an Avr gene in the pathogen (Flor 1971), we suggested that the partial resistance of ‘Président Roulin’ would be governed by at least five race-specific minor genes. The presence of the first two loci was based on the observation that some seedlings were incompatible with only one isolate among the set of isolates tested (1639 and EU-B05/EU-D49/inoculum mix having the same resistance behavior), and overcome by the other one (Table 3). A third locus was hypothesized from the observation that isolate EU-NL24 showed incompatible interactions only with seedlings resistant to the full set of isolates. This same isolate, EU-NL24, could also sporulate on genotypes for which resistance symptoms were observed for all the other isolates tested, suggesting the presence of the resistance locus 4 in our GfG relationship model. Finally, additional differential interactions, notably plants being compatible with isolates 1639, NL24, and B04/1066, and incompatible for all the other isolates tested, suggested the existence of a fifth resistance locus.
From these observations, we hypothesized a minor gene-for-minor gene relationship model in ‘Président Roulin’ (Table 5) with loci 1 and 2 providing resistance to isolate 1639 and to the EU-B05/EU-D49/inoculum mix, respectively; locus 3 providing a broad spectrum resistance to the whole set of isolates tested, as would locus 4, except for EU-NL24. Finally, locus 5 would be incompatible with EU-D42, EU-NL05, and EU-B05/EU-D49/inoculum mix. These resistance loci do not seem to be tightly linked on the same chromosome since a relatively high number of seedlings (corresponding to the recombination frequency between R genes) was observed for each pattern of differential interaction (from three to 12 plants). These loci are probably involved in the recognition of the pathogen by the host, rather than the defense-related genes acting downstream of the plant defense response. Also, we do not discard the hypothesis that other minor gene-for-minor gene interactions, not revealed by the V. inaequalis tested, could be involved in the partial resistance, or loci governing more basal resistance (non-race specific). This polygenic control seems to be durable, with a demonstrated resistance in ‘Président Roulin’ over 25 years under high disease pressure conditions (in absence of fungicides application). Pathogen isolates that overcome one of the genes would gain only a marginal advantage (Poland et al. 2009).
Segregation ratios suggest a complex genetic control of partial resistance
Interestingly, some V. inaequalis isolates showed a segregation that was not significantly different from R:S = 1:3 (EU-NL24, EU-B04, 1066) and 1:2 (EU-D42, EU-NL05). When the R:S = 1:1 segregation ratio indicates the inheritance of a dominant R gene, segregation of resistance in less than half of the progeny suggests more complex genetic interactions, as already observed in the apple–V. inaequalis interaction (Durel et al. 2003; Bastiaanse et al. 2015). For instance, a ratio of R:S = 1:3 could involve a recessive genetic control of plant resistance (Büschges et al. 1997; Deslandes et al. 2002; Iyer and McCouch 2004; Diaz-Pendon et al. 2004; Iyer-Pascuzzi and McCouch 2007; Antony et al. 2010) which, in some cases, has been demonstrated to be broad spectrum and durable (van Schie and Takken 2014). Another hypothesis would be the action of epistatic interactions among R genes in ‘Président Roulin’ that we cannot explain with the data in this study. Further genetics studies, including the creation of new segregating populations involving backcrosses, DNA molecular marker work, or the cloning of the resistance loci, could help confirm the status of the genetic control of the complex resistance of ‘Président Roulin’ against V. inaequalis.
A candidate gene approach to dissect the partial resistance of ‘Président Roulin’
In the current study, we proposed a candidate gene approach to better understand the molecular basis of the partial scab resistance of ‘Président Roulin’ consisting of three steps: the choice, screening, and functional validation of CDGs (Pflieger et al. 2001).
Choice of the CDGs
Thirteen CDGs were selected from a previous study (Bastiaanse et al. 2014) aiming to identify differentially expressed genes in ‘Président Roulin’, 48 h after V. inaequalis inoculation, as compared to the susceptible cultivar ‘Gala’. CDGs specifically induced or repressed in ‘Président Roulin’ and showing homology with plant defense genes were chosen (Table 1). In fact, numerous studies demonstrated that, beside basal host resistance, the key difference between resistant and susceptible hosts is the timely recognition of the invading pathogen and the activation of plant defense genes which is accompanied by the accumulation of corresponding gene products (Durrant et al. 2000; Alignan et al. 2006; Gabriëls et al. 2006; Wang et al. 2010; Shi et al. 2011; Li et al. 2012; Paris et al. 2012).
The selected CDGs could act at different levels of the plant defense system, from the early recognition of the pathogen, which could be at the basis of the interpretation of our scab inoculation tests, to the activation of downstream plant defense responses. In fact, signaling and defense genes could also be invoked as good candidates for explaining the resistance of ‘Président Roulin’. Among the set of selected CDGs, 43DU149′/56AU33′, and 44AU9, encoded for R proteins that are at the basis of the direct or indirect recognition of the pathogen (McHale et al. 2006). These R proteins are part of membrane-associated protein complexes, including the NBS-LRR family proteins (to date, the widest class of R genes cloned in plants) and LRR receptor kinase-like proteins. Then, external stimuli are usually transduced into an intracellular host defense response consisting of signaling proteins such as mitogen-activated protein kinase (MAPK) homologous with 2EU181 (Zhang and Klessig 2001), or serine carboxypeptidases similar to 44GU182. This later protein could also act in response to wounding (Moura et al. 2001; Liu et al. 2008). The subsequent stage of elicitation of defense-related proteins is the transcriptional activation of specific genes to redirect and alter the flow of metabolites required to sustain a pathogen attack. Transcription factors such as zinc finger homeodomain proteins (HD) (53HU89) might be a key element (Park et al. 2007). Finally, downstream events in plant defense response include the generation of highly toxic environments by massively producing reactive oxygen species (ROS) (involving peroxidase, 43DU149, and tocopherol cyclase, 51HU129′) and the activation of HR, a form of local programmed cell death restricting pathogen invasion (involving both cysteine proteinase, 44EU122/44EU118, and cysteine proteinase inhibitors, 37DU41). Plants can also secrete hydrolytic enzymes that target pathogen cell walls (e.g., chitinases, 56AU29) and trigger local cell wall fortifications (e.g., pectin methylesterase, PME) and the pectin methylesterase inhibitor (PMEis, such as 44GU173). Since most of these CDGs (apart from the typical NBS-LRR resistance proteins), encode for proteins not strictly involved in plant disease resistance, but also act in other plant physiological traits, their real involvement in the partial resistance in ‘Président Roulin’ still has to be investigated.
Screening of the CDGs: co-localization with scab major R genes, QTLs, or analogues of resistance genes
The availability of the whole genome sequence assembly of apple (Velasco et al. 2010), together with the annotation of RGAs and the mapping of numerous scab R genes and QTLs (Gessler et al. 2006; Bus et al. 2011), enabled us to investigate the co-localization of our CDGs with genomic regions known to control resistance. In an apple F1 segregating population, this approach has been previously used to identify genes whose steady-state transcript abundance was associated with the inheritance of resistance to powdery mildew disease and woolly apple aphid (Jensen et al. 2014). It has also been used as a criterion to select CDGs for partial resistance in various plant–pathogen interactions (Wang et al. 2001; González et al. 2010; Schweizer and Stein 2011). Since expressed sequence tags (ESTs) generally have a biological meaning by representing a particular gene (Matthews et al. 2001), compared with neutral markers used to map QTL, it may increase the knowledge about the genes underlying the agronomic traits.
Our CDGs were anchored to the existing ‘Golden delicious’ genetic map (Velasco et al. 2010). Eight out of the 13 pathogen-regulated CDGs co-localized with apple genomic regions known to act in scab resistance (QTLs, major scab R loci, or RGAs) (Fig. 2). We cannot discard the hypothesis that such co-localization occurred only by chance, regarding the high number of RGAs (868 RGAs) annotated in the reference apple genome (Perazzolli et al. 2014) but this could constitute a further indication of the potential role of our CDGs in the partial resistance of ‘Président Roulin’. Obviously, in our analysis, we could check only for co-localizations with R genes and QTLs that had been detected so far. In addition, information on the genomic loci that effectively regulate the expression level of the TDF of interest is still lacking from this analysis. The measured mRNA levels could be the product of regulation of the parent gene or another gene, mapping somewhere else in the genome (cis- or trans-regulatory elements) (Gilad et al. 2008). In our study, an eQTL analysis of our segregating population would help to throw some light on these issues.
Screening of the CDGs: expression regulation in ‘Président Roulin’ challenged by V. inaequalis
Since timely recognition of the pathogen could determine the resistance status of a plant, we investigated expression regulation of our CDGs in a time-course experiment.
First, our time-course experiment confirmed the expression pattern obtained at 48 hpi in our previous cDNA-AFLP study (Bastiaanse et al. 2014). Results also showed an induction or a repression of the CDGs during partially incompatible interaction with V. inaequalis, or a delay in this regulation during compatible interaction (Fig. 3). If these genes effectively act in the resistance of ‘Président Roulin’, this would suggest that, as important as the identity itself of the gene being activated, the quicker expression of some key genes could be crucial for the fate of the interaction between ‘Président Roulin’ and V. inaequalis. Sequences of the CDGs showed homologies to genes acting in the recognition of the pathogen, the transduction of the signal, or the activation of defense-related proteins. Our previous study also demonstrated that most of the transcripts that were expressed in ‘Président Roulin’ were also expressed in ‘Gala’ (Bastiaanse et al. 2014). This preponderance of quantitative and/or kinetic transcriptomic differences between resistant and susceptible responses, over the qualitative one, corroborates findings in other plant–pathogen interactions (Maleck et al. 2000; Jwa et al. 2001; Tao et al. 2003; Eulgem 2005; Li et al. 2006).
Finally, our knowledge of the dynamic of leaf infection by V. inaequalis enabled us to compare the timing of CDG expression with particular events of pathogen development. First to be activated at 24 and 48 hpi were the genes that could act in the early events of pathogen perception, signal transduction, and gene transcription. This might reflect physical and/or chemical stress experienced at the beginning of infection processes. At this phase of infection, appressoria are formed and runner hyphae spread under the cuticle in direct contact with the epidermal cells (Nusbaum and Keitt 1938; Ortega et al. 1998). At 48 hpi, and up to 120 hpi, the latest time point tested, transcriptional responses of other defense-related CDGs occurred: PR proteins (peroxidase, chitinase), a pectin methylesterase inhibitor, a cysteine protease, a tocopherol cyclase, a cysteine protease inhibitor, and a carboxypeptidase. This could lead to the appearance of the first resistance reactions in the host: collapsed epidermal cells under the subcuticular primary hyphae at 48 hpi and macroscopically visible necrosis, corresponding with large areas of collapsed epidermis cells, around 96 hpi (Nusbaum and Keitt 1938; Chevalier et al. 1991).
Based on their putative functions and their gene expression regulation during pathogen attack, we suggest in Fig. 3 a possible representation of the sequential activation of the CDGs based on their involvement during the plant–pathogen interaction: (1) genes involved in the recognition of the pathogen (43DU149′, 56AU33′, and 44AU9); (2) genes involved in the signal transduction and gene transcription (2EU181 and 53HU89); and (3) genes involved in plant defense responses (56AU29, 44GU173, 43DU149, 44EU122/44EU118, 51HU129′, 37DU41, and 44GU182).
Functional assessment
In an attempt to functionally assess the involvement of our CDGs in the partial resistance of ‘Président Roulin’, we estimated the extent of correlations (in terms of % of ‘explained variance’) between expression regulation of a subset of CDGs and level of scab resistance of individuals composing a segregating population ‘Président Roulin’ x ‘Gala’. The term ‘explained variance’ could be misleading here since correlation between two variables does not necessarily imply causation (that one causes another), but could constitute the step forward for further evaluations of the CDGs (e.g., plant transformation with the CDGs). In our study, regulation of several CDGs (considered jointly) contributed from 16 to 46 % of the sporulation severity (MSPOR) of the individuals (Table 4). This proportion of phenotypic variance ‘explained’ by the regulation of our CDGs is modest, but is of the same magnitude as the ratio found for QTL markers controlling apple scab resistance in various progenies (Durel et al. 2003; Liebhard et al. 2003; Calenge et al. 2004; Soufflet-Freslon et al. 2008). This is probably due to the minor effect of loci governing partial resistance.
Apart from 44EU122/44EU118 (encoding for the same protein), the CDGs all mapped in silico in apple genomic regions are known to contain disease resistance factors. Interestingly, the highest proportion (21.3 %) of individual CDG expression variance explained by the R/S levels was the regulation of 43DU149′, encoding a classic NBS-LRR resistance protein. Such larger effect could be due to its action upstream of plant defense reactions (recognition of the pathogen). Conversely, the effect of genes acting downstream could be diluted among the numerous defense-related genes participating in this stage of plant–pathogen interaction. In our study, an important part of the phenotypic variation of MSPOR remains ‘unexplained’ (54 %). This might result from gene actions not covered by our study, either defense-related genes identified in ‘Président Roulin’ but not investigated here (Bastiaanse et al. 2014) or genes not previously reported to be involved in disease resistance. Such genes with no similarity to any previously reported defense genes were shown to underlie some partial resistance QTL (Fukuoka et al. 2007; Zenbayashi-Sawata et al. 2007).
Conclusions and perspectives
In this study we identified potential CDGs for the polygenic (at least five R loci) and durable partial resistance in ‘Président Roulin’. These CDGs were shown to be specifically regulated in ‘Président Roulin’ or were induced earlier, compared with the susceptible ‘Gala’. They encoded for proteins that could act at various stages of plant–pathogen interaction, from pathogen perception to the activation of downstream defense reactions. Nine CDGs accounted for 46 % of the phenotypic variance of the disease severity, eight of them mapped within chromosomal regions containing disease resistance factors (RGAs, major scab R genes, or QTL). This is a modest indicator of the potential involvement of these CDGs in partial resistance against apple scab, regarding notably the small number of CDGs tested on a limited segregating population, and the absence of a genetic map for ‘Président Roulin’. Future studies, such as plant transformation with the CDGs and eQTL analysis with larger populations, will determine how strategies might be developed to incorporate these genes into breeding programs. If functional markers for partial disease resistance could be developed based on these CDGs, as illustrated by various studies (Liu et al. 2004, 2011), these markers would have greater breeding value and alleviate the recombination problems associated with the use of neutral molecular markers in genome-informed breeding programs (Andersen and Luebberstedt 2003).
References
Aldaghi M, Bertaccini A, Lepoivre P (2012) cDNA-AFLP analysis of gene expression changes in apple trees induced by phytoplasma infection during compatible interaction. Eur J Plant Pathol 134:117–130
Alignan M, Hewezi T, Petitprez M, Dechamp-Guillaume G, Gentzbittel L (2006) A cDNA microarray approach to decipher sunflower (Helianthus annuus) responses to the necrotrophic fungus Phoma macdonaldii. New Phytol 170:523–536
Almagro L, Gómez Ros LV, Belchi-Navarro S, Bru R, Ros Barceló A, Pedreño MA (2009) Class III peroxidases in plant defence reactions. J Exp Bot 60:377–390
An SH, Sohn KH, Choi HW, Hwang IS, Lee SC, Hwang BK (2008) Pepper pectin methylesterase inhibitor protein CaPMEI1 is required for antifungal activity, basal disease resistance and abiotic stress tolerance. Planta 228:61–78
Andersen JR, Luebberstedt T (2003) Functional markers in plants. Trends Plant Sci 8:554–560
Antony G, Zhou J, Huang S, Li T, Liu B, White F, Yang B (2010) Rice xa13 recessive resistance to bacterial blight is defeated by induction of the disease susceptibility gene Os-11N3. Plant Cell 22:3864–3876
Avrova AO, Stewart HE, De Jong W, Heilbronn J, Lyon GD, Birch PRJ (1999) A cysteine protease gene is expressed early in resistant potato interactions with Phytophthora infestans. Mol Plant-Microbe Interact 12:1114–1119
Ballini E, Morel JB, Droc G, Price A, Courtois B, Notteghem JL, Tharreau D (2008) A genome-wide meta-analysis of rice blast resistance genes and quantitative trait loci provides new insights into partial and complete resistance. Mol Plant-Microbe Interact 21:859–868
Bastiaanse H, Muhovski Y, Parisi O, Paris R, Mingeot D, Lateur M (2014) Gene expression profiling by cDNA-AFLP reveals potential candidate genes for partial resistance of ‘Président Roulin’ against Venturia inaequalis. BMC Genomics 15:1043
Bastiaanse H, Bassett HCM, Kirk C, Gardiner SE, Deng C, Groenworld R, Chagné D, Bus VGM (2015) Scab resistance in ‘Geneva’ apple is conditioned by a resistance gene cluster with complex genetic control. Mol Plant Pathol. doi:10.1111/mpp.12269
Baxter CJ, Sabar M, Quick WP, Sweetlove LJ (2005) Comparison of changes in fruit gene expression in tomato introgression lines provides evidence of genome-wide transcriptional changes and reveals links to mapped QTLs and described traits. J Exp Bot 56:1591–1604
Belfanti E, Silfverberg-Dilworth E, Tartarini S, Patocchi A, Barbieri M, Zhu J, Vinatzer BA, Gianfranceschi L, Gessler C, Sansavivi S (2004) The HcrVf2 gene from a wild apple confers scab resistance to a transgenic cultivated variety. Proc Natl Acad Sci 101:886–890
Brodny U, Nelson RR, Gregory LV (1986) Residual and interactive expression of ‘defeated’ wheat stem rust resistance genes. Phytopathology 76:546–549
Bus VGM, Laurens FND, van de Weg WE, Rusholme RL, Rikkerink EHA, Gardiner SE, Bassett HCM, Kodde LP, Plummer KM (2005) The Vh8 locus of a new gene-for-gene interaction between Venturia inaequalis and the wild apple Malus sieversii is closely linked to the Vh2 locus in Malus pumila R12740-7A. New Phytol 166:1035–1049
Bus VGM, Rikkerink EHA, Caffier V, Durel CE, Plummer KM (2011) Revision of the nomenclature of the differential host-pathogen interactions of Venturia inaequalis and Malus. Annu Rev Phytopathol 49:391–413
Büschges R, Hollricher K, Panstruga R, Simons G, Wolter M, Frijters A, van Daelen R, van der Lee T, Diergaarde P, Groenendijk J, Töpsch S, Vos P, Salamini F, Schulze-Lefert P (1997) The barley Mlo gene: a novel control element of plant pathogen resistance. Cell 88:695–705
Caffier V, Lasserre-Zuber P, Giraud M, Lascostes M, Stievenard R, Lemarquand A, van de Weg E, Expert P, Denancé C, Didelot F, Le Cam B, Durel CE (2014) Erosion of quantitative host resistance in the apple × Venturia inaequalis pathosystem. Infect Genet Evol 27:481–489
Caffier V, Patocchi A, Expert P, Bellanger MN, Durel CE, Hilber-Bodmer M, Broggini GAL, Groenwold R, Bus VGM (2015) Virulence characterization of Venturia inaequalis reference isolates on the differential set of Malus hosts. Plant Dis 99:370–375
Calenge F, Faure A, Goerre M, Gebhardt C, Van de Weg WE, Parisi L, Durel CE (2004) Quantitative Trait Loci (QTL) analysis reveals both broad-spectrum and isolate-specific QTL for scab resistance in an apple progeny challenged with eight isolates of Venturia inaequalis. Phytopathology 94:370–379
Caranta C, Lefebvre V, Palloix A (1997) Polygenic resistance of pepper to potyviruses consists of a combination of isolate-specific and broad-spectrum quantitative trait loci. Mol Plant-Microbe Interact 10:872–878
Chen H, Wang S, Xing Y, Xu C, Hayes PM, Zhang Q (2003) Comparative analyses of genomic locations and race specificities of loci for quantitative resistance to Pyricularia grisea in rice and barley. Proc Natl Acad Sci 100:2544–2549
Chevalier M, Lespinasse Y, Renaudin S (1991) A microscopic study of different classes of symptoms by the Vf gene in apple for resistance to scab (Venturia inaequalis). Plant Pathol 40:249–256
Collinge DB, Kragh KM, Mikkelsen JD, Nielsen KK, Rasmussen U, Vad K (1993) Plant chitinases. Plant J 3:31–40
Cova V, Paris R, Passerotti S, Zini E, Gessler C, Pertot I, Loi N, Musetti R, Komjanc M (2010) Mapping and functional analysis of four apple receptor-like protein kinases related to LRPKm1 in HcrVf2-transgenic and wild-type apple plants. Tree Genet Genomes 6:389–403
Damerval C, Maurice A, Josse JM, De Vienne D (1994) Quantitative trait loci underlying gene product variation: a novel perspective for analyzing regulation of genome expression. Genetics 137:289–301
De Gara L, de Pinto MC, Tommasi F (2003) The antioxidant systems vis-à-vis reactive oxygen species during plant–pathogen interaction. Plant Physiol Biochem 41:863–870
Deslandes L, Olivier J, Theulières F, Hirsch J, Feng DX, Bittner-Eddy P, Beynon J, Marco Y (2002) Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc Natl Acad Sci 99:2404–2409
Diaz-Pendon JA, Truniger V, Nieto C, Garcia-Mas J, Bendahmane A, Aranda MA (2004) Advances in understanding recessive resistance to plant viruses. Mol Plant Pathol 5:223–233
Dogimont C, Leconte L, Perin C, Thabuis A, Lecoq H, Pitrat M (2000) Identification of QTL contributing to resistance to different strains of cucumber mosaic cucumovirus in melon. Acta Horticult 510:391–398
D’Silva I, Poirier GG, Heath MC (1998) Activation of cysteine proteases in cowpea plants during the hypersensitive response-a form of programmed cell death. Exp Cell Res 245:389–399
Dunemann F, Egerer J (2010) A major resistance gene from Russian apple ‘Antonovka’ conferring field immunity against apple scab is closely linked to the Vf locus. Tree Genet Genomes 6:627–633
Durel CE, Parisi L, Laurens F, Venisse JS, Jourjon MF (2000) Does the Vf gene maintain a residual resistance to apple scab despite its breakdown by Venturia inaequalis race 6 strains. Acta Horticult 538:575–580
Durel CE, Parisi L, Laurens F, Van de Weg WE, Liebhard R, Jourjon MF (2003) Genetic dissection of partial resistance to race 6 of Venturia inaequalis in apple. Genome 46:224–234
Durel CE, Calenge F, Parisi L, van de Weg WE, Kodde LP, Liebhard R, Gessler C, Thiermann M, Dunemann F, Gennari F, Tartarini S (2004) An overview of the position and robustness of scab resistance QTLs and major genes by aligning genetic maps of five apple progenies. ISHS. Acta Horticult 663:135–140
Durrant WE, Rowland O, Piedras P, Hammond-Kosack KE, Jones JDG (2000) cDNA AFLP reveals a striking overlap in race-specific resistance and wound response gene expression profiles. Plant Cell 12:963–977
Eckey C, Korell M, Leib K, Biedenkopf D, Janses C, Langen G, Kogel KH (2004) Identification of powdery mildew-induced barley genes by cDNA-AFLP: functional assessment of an early expressed MAP kinase. Plant Mol Biol 55:1–15
Eulgem T (2005) Regulation of the Arabidopsis defense transcriptome. Trends Plant Sci 10:71–78
Flor HH (1971) Current status of the gene-for-gene concept. Annu Rev Phytopathol 9:275–296
Fukuoka S, Okuno K, Kawase M (2007) Rice blast disease gene Pi21, resistance gene pi21 and utilization thereof. Patent WO/2007/000880
Gabriëls SHEJ, Takken FL, Vossen JH, de Jong CF, Liu Q, Turk SCHJ, Wachowski LK, Peters J, Witsenboer HMA, de Wit PJGM, Joosten MHJ (2006) cDNA-AFLP combined with functional analysis reveals novel genes involved in the hypersensitive response. Mol Plant-Microbe Interact 19:567–576
Gadiou S, Kundu JK (2012) Evaluation of reference genes for the relative quantification of apple stem grooving virus and apple mosaic virus in apple trees. Indian J Virol 23:39–41
Galli P, Patocchi A, Broggini GA, Gessler C (2010) The Rvi15 (Vr2) apple scab resistance locus contains three TIR-NBS-LRR genes. Mol Plant-Microbe Interact 23:608–617
Gasic K, Hernandez A, Korban SS (2004) RNA extraction from different apple tissues rich in polyphenols and polysaccharides for cDNA library construction. Plant Mol Biol Report 22:437a–437g
Gessler C, Patocchi A, Sansavini S, Tartarini S, Gianfranceschi L (2006) Venturia inaequalis resistance in apple. Crit Rev Plant Sci 25:473–503
Gilad Y, Rifkin SA, Pritchard JK (2008) Revealing the architecture of gene regulation: the promise of eQTL studies. Trends Genet 24:408–415
González AM, Marcel TC, Kohutova Z, Stam P, van der Linden CG, Niks RE (2010) Peroxidase profiling reveals genetic linkage between peroxidase gene clusters and basal host and non-host resistance to rusts and mildew in barley. Plos One 5:e10495
González AM, Marcel TC, Niks RE (2012) Evidence for a minor gene-for-minor gene interaction explaining nonhypersensitive polygenic partial disease resistance. Phytopathology 102:1086–1093
Gutierrez-Campos R, Torres-Acosta JA, Saucedo-Arias LJ, Gomez-Lim MA (1999) The use of cysteine proteinase inhibitors to engineer resistance against potyviruses in transgenic tobacco plants. Nat Biotechnol 17:1223–1226
Hao L, Hsiang T, Goodwin PH (2006) Role of two cysteine proteinases in the susceptible response of Nicotiana benthamiana to Colletotrichum destructivum and the hypersensitive response to Pseudomonas syringae pv. tomato. Plant Sci 170:1001–1009
Hazen SP, Pathan MS, Sanchez A, Baxter I, Dunn M, Estes B, Chang HS, Zhu T, Kreps JA, Nguyen HT (2005) Expression profiling of rice segregating for drought tolerance QTLs using a rice genome array. Funct Integr Genomics 5:104–116
Iyer AS, McCouch SR (2004) The rice bacterial blight resistance gene xa5 encodes a novel form of disease resistance. Mol Plant-Microbe Interact 17:1348–1354
Iyer-Pascuzzi AS, McCouch SR (2007) Recessive resistance genes and the Oryza sativa-Xanthomonas oryzae pv. oryzae pathosystem. Mol Plant-Microbe Interact 20:731–739
Jansen RC, Nap JP (2001) Genetical genomics: the added value from segregation. Trends Genet 17:388–391
Jensen PJ, Fazio G, Altman N, Praul C, McNellis TW (2014) Mapping in an apple (Malus x domestica) F1 segregating population based on physical clustering of differentially expressed genes. BMC Genomics 15:261
Jongedijk E, Tigelaar H, van Roekel JSC, Bres-Vloemans SA, Dekker I, van den Elzen PJM, Cornelissen BJC, Melchers LS (1995) Synergistic activity of chitinases and β-1,3-glucanases enhances fungal resistance in transgenic tomato plants. Euphytica 85:173–180
Joshi SG, Schaart JG, Groenwold R, Jacobsen E, Schouten HJ, Krens FA (2011) Functional analysis and expression profiling of HcrVf1 and HcrVf2 for development of scab resistant cisgenic and intragenic apples. Plant Mol Biol 75:579–591
Jwa NS, Agrawal GK, Rakwal R, Park CH, Agrawal VP (2001) Molecular cloning and characterization of a novel jasmonate inducible pathogenesis-related class 10 protein gene, JIOsPR10, from rice (Oryza sativa L.) seedling leaves. Biochem Biophys Res Commun 286:973–983
Komjanc M, Festi S, Rizzotti L, Cervone F, De Lorenzo G (1999) A leucine-rich repeat receptor-like protein kinase (LRPKm1) gene is induced in Malus x domestica by Venturia inaequalis infection and salicylic acid treatment. Plant Mol Biol 40:945–957
Korfhage U, Trezzini GF, Meier I, Hahlbrock K, Somssich IE (1994) Plant homeodomain protein involved in transcriptional regulation of a pathogen defense-related gene. Plant Cell 6:695–708
Kou Y, Wang, S (2010) Broad-spectrum and durability: understanding of quantitative disease resistance. Curr Opin Plant Biol 13:181--185
Krishnaveni S, Muthukrishnan S, Liang GH, Wilde G, Manickam A (1999) Induction of chitinases and β-1,3-glucanases in resistant and susceptible cultivars of sorghum in response to insect attack, fungal infection and wounding. Plant Sci 144:9–16
Krüger J, Thomas CM, Golstein C, Dixon MS, Smoker M, Tang S, Mulder L, Jones JDG (2002) A tomato cysteine protease required for Cf-2-dependent disease resistance and suppression of autonecrosis. Science 296:744–747
Lateur M, Populer C (1994) Screening fruit tree genetic resources in Belgium for disease resistance and other desirable characters. In: Schmidt H, Kellerhals M (eds) Progress in temperate fruit breeding. Kluwer, Dordrecht, pp 425–431
Li ZK, Luo LJ, Mei HW, Paterson AH, Zhao XH, Zhong DB, Wang YP, Yu XQ, Zhu L, Tabien R, Stansel JW, Ying CS (1999) A ‘defeated’ rice resistance gene acts as a QTL against a virulent strain of Xanthomonas oryzae pv. oryzae. Mol Genet Genomics 261:58–63
Li C, Bai Y, Jacobsen E, Visser R, Lindhout P, Bonnema G (2006) Tomato defense to the powdery mildew fungus: differences in expression of genes in susceptible, monogenic- and polygenic resistance responses are mainly in timing. Plant Mol Biol 62:127–140
Li C, Faino L, Dong L, Fan J, Kiss L, De Giovanni C, Lebeda A, Scott J, Matsuda Y, Toyoda H, Lindhout P, Visser RGF, Bonnema G, Bai Y (2012) Characterization of polygenic resistance to powdery mildew in tomato at cytological, biochemical and gene expression level. Mol Plant Pathol 13:148–159
Liebhard R, Koller B, Patocchi A, Kellerhals M, Pfammatter W, Jermini M, Gessler C (2003) Mapping quantitative field resistance against apple scab in a 'Fiesta' x 'Discovery' progeny. Phytopathology 93:493–501
Lionetti V, Raiola A, Camardella L, Giovane A, Obel N, Pauly M, Favaron F, Cervone F, Bellincampi D (2007) Overexpression of pectin methylesterase inhibitors in Arabidopsis restricts fungal infection by Botrytis cinerea. Plant Physiol 143:1871–1880
Liu B, Zhang S, Zhu X, Yang Q, Wu S, Mei M, Mauleon R, Leach J, Mew T, Leung H (2004) Candidate defense genes as predictors of quantitative blast resistance in rice. Mol Plant-Microbe Interact 17:1146–1152
Liu H, Wang X, Zhang H, Yang Y, Ge X, Song F (2008) A rice serine carboxypeptidase-like gene OsBISCPL1 is involved in regulation of defense responses against biotic and oxidative stress. Gene 420:57–65
Liu R, Wang B, Guo W, Wang L, Zhang T (2011) Differential gene expression and associated QTL mapping for cotton yield based on a cDNA-AFLP transcriptome map in an immortalized F2. Theor Appl Genet 123:439–454
Maleck K, Levine A, Eulgem T, Morgan A, Schmid J, Lawton KA, Dangl JL, Dietrich RA (2000) The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat Genet 26:403–420
Malnoy M, Xu M, Borejsza-Wysocka E, Korban SS, Aldwinckle HS (2008) Two receptor-like genes, Vfa1 and Vfa2, confer resistance to the fungal pathogen Venturia inaequalis inciting apple scab disease. Mol Plant-Microbe Interact 21:448–458
Matthews BF, Devine TE, Weisemann JM, Beard HS, Lewers KS, McDonald MH, Park YB, Maiti R, Lin JJ, Kuo J, Pedroni MJ, Cregan PB, Saunders JA (2001) Incorporation of sequenced cDNA and genomic markers into the soybean genetic map. Crop Sci 41:516–521
McHale L, Tan X, Koehl P, Michelmore RW (2006) Plant NBS-LRR proteins: adaptable guards. Genome Biol 7:212
Métraux JP, Boller T (1986) Local and systemic induction of chitinase in cucumber plants in response to viral, bacterial and fungal infections. Physiol Mol Plant Pathol 28:161–169
Moura DS, Bergey DR, Ryan CA (2001) Characterization and localization of a wound-inducible type I serine-carb oxypeptidase from leaves of tomato plants (Lycopersicon esculentum Mill.). Planta 212:222–230
Nass HA, Pedersen WL, MacKenzie DR, Nelson RR (1981) The residual effect of some defeated powdery mildew Erysiphe graminis f.sp. tritici resistance genes in isolines of winter wheat. Phytopathology 71:1315–1348
Nusbaum C, Keitt GW (1938) A cytological study of host–parasite relations of Venturia inaequalis on apple leaves. J Agric Res 56:595–618
Ortega F, Steiner U, Dehne HW (1998) Induced resistance to apple scab: microscopic studies on the infection cycle of Venturia inaequalis (Cke.) Wint. J Phytopathol 146:399–405
Paran I, Zamir D (2003) Quantitative traits in plants: beyond the QTL. Trends Genet 19:303–306
Paris R, Dondini L, Zannini G, Bastia D, Marasco E, Gualdi V, Rizzi V, Piffanelli P, Mantovani V, Tartarini S (2012) dHPLC efficiency for semi-automated cDNAAFLP analyses and fragment collection in the apple scab-resistance gene model. Planta 235:1065–1080
Parisi L, Lespinasse Y, Guillaumes J, Krüger J (1993) A new race of Venturia inaequalis virulent to apples with resistance due to the Vf gene. Phytopathology 83:533–537
Parisi L, Orts R, Rivenez Damboise MO, Lefeuvre M, Lagarde MP (2000) Protection intégrée du verger de pommier de l’an Tavelure et oïdium: variétés résistantes et lutte raisonnée. Arboriculture Fruitière 486:25–29
Park HC, Kim ML, Lee SM, Bahk JD, Yun DJ, Lim CO, Hong JC, Lee SY, Cho MJ, Chung WS (2007) Pathogen-induced binding of the soybean zinc finger homeodomain proteins GmZF-HD1 and GmZF-HD2 to two repeats of ATTA homeodomain binding site in the calmodulin isoform 4 (GmCaM4) promoter. Nucleic Acids Res 35:3612–3623
Parlevliet JE (2002) Durability of resistance against fungal, bacterial and viral pathogens; present situation. Euphytica 124:147–156
Parlevliet JE, van Ommeren A (1975) Partial resistance of barley to leaf rust Puccinia hordei II. Relationship between field trials micro plot tests and latent period. Euphytica 24:293–303
Parlevliet JE, Zadoks JC (1977) The integrated concept of disease resistance: a new view including horizontal and vertical resistance in plants. Euphytica 26:5–21
Pedley KF, Martin GB (2004) Identification of MAPKs and their possible MAPK kinase activators involved in the Pto-mediated defense response of tomato. J Biol Chem 279:49229–49235
Perazzolli M, Malacarne G, Baldo A, Righetti L, Bailey A, Fontana P, Velasco R, Malnoy M (2014) Characterization of resistance gene analogues (RGAs) in apple (Malus × domestica Borkh.) and their evolutionary history of the Rosaceae family. PLoS One 9:e83844
Pflieger S, Lefebvre V, Causse M (2001) The candidate gene approach in plant genetics: a review. Mol Breed 7:275–291
Poland JA, Balint-Kurti PJ, Wisser RJ, Pratt RC, Nelson RJ (2009) Shades of gray: the world of quantitative disease resistance. Trends Plant Sci 14:21–29
Qi X (1999) Isolate-specific QTL for partial resistance to Puccinia hordei in barley. Theor Appl Genet 99:877–884
Ramakers C, Ruijter JM, Lekanne-Deprez RH, Moorman AFM (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339:62–66
Rozen S, Skaletsky HJ (2000) Primer3 on the www for general users and for biologist programmers. In: Krawetz S, Misener S (eds) Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, pp 365–386
Sahai AS, Manocha MS (1993) Chitinases of fungi and plants: their involvement in morphogenesis and host-parasite interaction. FEMS Microbiol Rev 11:317–338
Schouten HJ, Brinkhuis J, van der Burgh A, Schaart JG, Groenwold R, Broggini GA, Gessler C (2014) Cloning and functional characterization of the Rvi15 (Vr2) gene for apple scab resistance. Tree Genet Genomes 10:251–260
Schweizer P, Stein N (2011) Large-scale data integration reveals co localization of gene functional groups with meta-QTL for multiple disease resistance in barley. Mol Plant-Microbe Interact 24:1492–1501
Shi C, Chaudhary S, Yu K, Park SJ, Navabi A, McClean PE (2011) Identification of candidate genes associated with CBB resistance in common bean HR45 (Phaseolus vulgaris L.) using cDNA-AFLP. Mol Biol Rep 38:75–81
Solomon M, Belenghia B, Delledonne M, Menachema E, Levine A (1999) The involvement of cysteine proteases and protease inhibitor genes in the regulation of programmed cell death in plants. Plant Cell 11:431–443
Song WY, Wang GL, Chen LL, Kim HS, Pi LY, Holsten T, Gardner J, Wang B, Zhai WX, Zhu LH, Fauquet C, Ronald P (1995) A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270:1804–1806
Soriano JM, Madduri M, Schaart JG, van der Burgh A, van Kaauwen MP, Tomic L, Groenworld R, Velasco R, van de Weg E, Schouten HJ (2014) Fine mapping of the gene Rvi18 (V25) for broad-spectrum resistance to apple scab, and development of a linked SSR marker suitable for marker-assisted breeding. Mol Breed 34:2021–2032
Soufflet-Freslon V, Gianfranceschi L, Patocchi A, Durel CE (2008) Inheritance studies of apple scab resistance and identification of Rvi14, a new major gene that acts together with other broad-spectrum QTL. Genome 51:657–667
Steiner B, Kurz H, Lemmens M, Buerstmayr H (2009) Differential gene expression of related wheat lines with contrasting levels of head blight resistance after Fusarium graminearum inoculation. Theor Appl Genet 118:753–764
Talukder ZI, Tharreau D, Price AH (2004) Quantitative trait loci analysis suggests that partial resistance to rice blast is mostly determined by race-specific interactions. New Phytol 162:197–209
Tao Y, Xie Z, Chen W, Glazebrook J, Chang HS, Han B, Zhu T, Zou G, Katagiri F (2003) Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell 15:317–330
van Schie CC, Takken FL (2014) Susceptibility genes 101: how to be a good host. Annu Rev Phytopathol 52:551–581
Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar SK, Troggio M et al (2010) The genome of the domesticated apple (Malus x domestica Borkh.). Nat Genet 42:833–839
Villette I (2000) Contribution à la mise en évidence des composantes de la résistance partielle du pommier (Malus x domestica Borkh.) à la tavelure (Venturia inaequalis (Cke.) Wint.). Dissertation, Gembloux Agro-Bio Tech, University of Liège
Vinatzer BA, Patocchi A, Gianfranceschi L, Tartarini S, Zhang HB, Gessler C, Sansavini S (2001) Apple contains receptor-like genes homologous to the Cladiosporium fulvum resistance gene family of tomato with a cluster of genes cosegregating with Vf apple scab resistance. Mol Plant-Microbe Interact 14:508–515
Vögeli-Lange R, Hansen-Gehri A, Boller T, Meins F (1988) Induction of the defense-related glucanohydrolases, β-1,3-glucanase and chitinase, by tobacco mosaic virus infection of tobacco leaves. Plant Sci 54:171–176
Wang Z, Taramino G, Yang D, Liu G, Tingey SV, Miao GH, Wang GL (2001) Rice ESTs with disease-resistance gene-or defense-response gene-like sequences mapped to regions containing major resistance genes or QTLs. Mol Genet Genomics 265:302–310
Wang X, Liu W, Chen X, Tang C, Dong Y, Ma J, Huang X, Wei G, Han Q, Huang L, Kang Z (2010) Differential gene expression in incompatible interaction between wheat and stripe rust fungus revealed by cDNA-AFLP and comparison to compatible interaction. BMC Plant Biol 10:9
Wayne ML, McIntyre LM (2002) Combining mapping and arraying: an approach to candidate gene identification. Proc Natl Acad Sci 99:14903–14906
Zenbayashi-Sawata K, Fukuoka S, Katagiri S, Fujisawa M, Matsumoto T, Ashizawa T, Koizumi S (2007) Genetic and physical mapping of the partial resistance gene, pi34, to blast in rice. Phytopathology 97:598–602
Zhang S, Klessig DF (2001) MAPK cascades in plant defense signalling. Trends Plant Sci 6:520–527
Acknowledgments
We thank Valérie Caffier from Institut National de la Recherche Agronomique (INRA) in Angers, France, and Vincent Bus from the New Zealand Institute for Plant and Food Research (PFR) in Havelock North, New Zealand, for supplying the V. inaequalis isolates. We are also grateful to Vincent for his helpful advice on the implementation of the pathological test in small inoculation chambers. We thank Yves Brostaux (Gembloux Agro-Bio Tech, University of Liège, Belgium) for assisting in the statistical analysis of the gene expression data as well as Sébastien Massart (GxABT-University of Liège, Belgium) and Amy Watson (Queensland Alliance for Agriculture and Food Innovation, University of Queensland, Australia) for revising the manuscript draft. We thank the RGF team at CRA-W (particularly Laurent Delpierre, Thibaut Donis, Pascal Dupont, Alain Rondia, Patrick Houben and Anne Van Landschoot) for helping in the grafting and growing of the apple trees used in this experiment. This work was supported by the Moerman funds (CRA-W) within the framework of the POMINNO project entitled ‘Recherche de méthodes rapides de sélection de variétés innovantes de pommes de qualités différenciées et adaptées à une agriculture durable’.
Data archiving statement
The cDNA sequences that formed the basis of our gene expression study were deposited at DDBJ/EMBL/GenBank in the library LIBEST_028504 under the following accession numbers: 43DU149′/JZ719417, 56AU33′/JZ719506, 44AU9/JZ719419, 2EU181/JZ719320, 53HU89/JZ719483, 43DU149/JZ719416, 51HU129′/JZ719472, 44EU122/ JZ719578, 44EU118/ JZ719577, 37DU41/ JZ719360, 44GU182/JZ719427, 56AU29/ JZ719503, 44GU173/JZ719426.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by E. Dirlewanger
This article is part of the Topical Collection on Disease Resistance
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(A.) Resistance reactions observed on ‘Président Roulin’ leaves under natural infection in the orchard compared with (B.) the heavy sporulation on a susceptible ‘Gala’ leaf. (C.) close-up of the different reactions observed on ‘Président Roulin’: Necrosis (N) with limited sporulation, Chlorosis-Necrosis symptoms (Ch/N) and No Reaction (NR) with limited sporulation. (PNG 1149 kb)
ESM 2
Scab resistance reactions observed on a ‘Gala’ x ‘Président Roulin’ progeny 21 days after controlled inoculation with various monoconidial V. inaequalis isolates using the small chambers inoculation technique (Bus et al. 2005). (A.) Setting up the inoculation chambers on the leaf, a different isolate being inoculated in each well. (B.) Differential interactions obtained with the incompatible isolate 1639 (left, pin-point) and the compatible isolate EU-NL24 (right, heavy sporulation), alongside a close-up of different scab symptoms: (C) HR reactions (pin point), (D) chlorosis, (E) stellate necrosis, 33 (F) chlorosis with limited sporulation, (G) chlorosis and necrosis with limited sporulation, (H) susceptibility. (PNG 1862 kb)
ESM 3
(DOCX 14 kb)
ESM 4
(DOCX 15 kb)
Rights and permissions
About this article
Cite this article
Bastiaanse, H., Muhovski, Y., Mingeot, D. et al. Candidate defense genes as predictors of partial resistance in ‘Président Roulin’ against apple scab caused by Venturia inaequalis . Tree Genetics & Genomes 11, 125 (2015). https://doi.org/10.1007/s11295-015-0948-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-015-0948-9