Abstract
Cacopsylla pyri (pear psylla) is one of the most serious pests of pear (Pyrus spp.) in Europe. It can cause high yield losses, and its control has become difficult since it has developed resistance to a wide range of pesticides. Pear breeders are developing new cultivars resistant to pear psyllids, and Asian species, such as Pyrus ussuriensis and Pyrus × bretschneideri, are good sources of resistance. Antixenosis and antibiosis resistance to psylla were both identified in pear; they may differ in the biological mechanism and probably have different genetic backgrounds. We crossed interspecific P. × bretschneideri × Pyrus communis hybrid PEAR3, resistant to pear psylla, with the susceptible European pear cultivar ‘Moonglow’ to obtain an F1 population for the genetic mapping of the resistance. Quantitative trait locus (QTL) analysis was carried out for antibiosis by measuring the number of surviving nymphs and the nymphal development, using a novel phenotyping protocol and a saturated genetic map made of single-nucleotide polymorphism (SNP) and microsatellite (simple sequence repeats (SSR)) markers. A stable QTL was detected on linkage group (LG) 8 of PEAR3 (R 2 = 17.2–39.1 %). In addition, QTLs were detected on LG5 (R 2 = 10.8 %) of PEAR3 and on LG15 of ‘Moonglow’ (R 2 = 13.7 %).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pear psyllids (Hemiptera, Psyllidae) are one of the most serious pests of pear (Pyrus spp.). The most damaging psylla species identified are Cacopsylla pyri (Linnaeus), endemic to Europe, Cacopsylla pyricola (Fӧrster) to Europe and North America, and Cacopsylla bidens (Ŝulc) to Europe and the Middle East (Shaltiel-Harpaz et al. 2014). The pear psylla life cycle begins with the eggs, laid singly or in clusters on the host plant, which hatch into nymphs that go through five instars (Atger and Bigre 1982) (Fig. 1a). After the last molt, nymphs develop into male or female adults (Fig. 1b), which are able to reproduce sexually within a few days (Hodkinson 2009). The development rate of all immature psylla stages is highly affected by temperature: the egg stage duration ranges between 6 and 28 days, with a direct linear correlation with temperature, while the young nymphal (L1 to L3) and old nymphal (L4–L5) stages last 10–19 and 12–18 days, respectively, with an asymptotic relationship with temperature (Kapatos and Stratopoulou 1999). Both young and adult psylla feed on the plant by inserting their stylets into the phloem. However, the main damage on the host is caused by the production of honeydew by actively feeding nymphs, which in turn is a favorite substrate for sooty mould fungi. This fungi, by blocking photosynthesis, cause necrosis on the leaves of infested plants (Salvianti et al. 2008) and russet the fruits, reducing their market value (Pasqualini et al. 2006). During summer, psylla can give rise to several overlapping generations (Schaub et al. 2005), leading to high pest densities that can induce leaf and fruit drop, and reduce fruit size (Shaltiel-Harpaz et al. 2014), hence causing high yield losses. Moreover, pear psylla is the major vector of the phytoplasma (Candidatus Phytoplasma pyri) responsible for pear decline disease (Salvianti et al. 2008).
Control of pear psylla in orchards is based mainly on the use of insecticides (for example, amitraz, abamectin, organophosphates, pyrethroids) (Civolani 2012). However, the insect has developed resistance to a great number of them (Harries and Burts 1965; Buès et al. 2003; Civolani et al. 2007), while biological control strategies based on the use of natural enemies are not sufficient to prevent the damage (Berrada et al. 1995). Therefore, development of new pear cultivars with durable resistance is an effective and sustainable strategy for psylla control.
The three types of plant resistance to insects are antixenosis, antibiosis, and tolerance (Hesler and Tharp 2005; Bell 2013a). Antixenosis prevents insects from colonizing the host or sustain feeding, and antibiosis affects the pest biology, while tolerance is the ability of the plant to grow despite infestation (Hesler and Tharp 2005). Antixenosis to pear psylla is characterized by ovipositional deterrence and feeding inhibition, whereas antibiosis is expressed by nymphal mortality and delayed development (Bell and Stuart 1990). These types of resistance may not share a common molecular and biological mechanism, because some pear genotypes only show one or the other (Pasqualini et al. 2006). Antixenosis and antibiosis resistances to C. pyri in Europe and C. pyricola in North America have been characterized, and cultivars with different levels of resistance were identified among European and Asian pears and interspecific hybrids (Bell and Stuart 1990; Bell 1992, 2013a; Robert et al. 2004; Robert and Raimbault 2004). Most of the Pyrus communis-resistant varieties originate from Eastern Europe and were found, or supposed, to be triploid, which means that they might not be pure P. communis (Bell 2013a). Asian pear cultivars have long been used as sources of resistance to psylla (Westigard et al. 1970); Harris and Lamb (1973) showed that Pyrus ussuriensis resistance, based on counts of nymphs on the seedlings, was heritable and dominant when crossed with P. communis. Also, Pasqualini et al. (2006) showed that the Asian species P. ussuriensis and Pyrus pyrifolia were able to transmit psylla resistance to their progeny, although some variability was observed, depending on the parent combinations. They evaluated the resistance in terms of settling of adults, ovipositional antixenosis, and nymphal antibiosis and concluded that the last one was the most important type of resistance in the observed crosses. On the other hand, resistant cultivars of East European origin did not appear able to transmit high degrees of resistance to nymphal feeding to their progeny, except for ‘Erabasma.’ Moreover, in crosses involving European pear cultivars, susceptibility was dominant (Bell 2013b). It is important to underline that the studies of Harris and Lamb (1973) and of Bell (2013b), other than using different sources of resistance, evaluated two distinct modes of resistance. Therefore, it is clear that the investigation of all types of resistance to pear psylla is fundamental to determine whether a cultivar will be useful for breeding programs.
Pear psylla resistance is considered to be a polygenic trait (Pasqualini et al. 2006; Lespinasse et al. 2008), and to date, only one quantitative trait locus (QTL) has been detected on pear linkage group (LG) 17 using the interspecific population ‘Angélys’ (P. communis) × NY10355 (P. ussuriensis × P. communis) (Bouvier et al. 2011). To our knowledge, only one other study focused on mapping pest resistance loci in pear: Evans et al. (2008) mapped a major gene for resistance to Dysaphis pyri to LG17 of the snow pear (Pyrus nivalis). In contrast, in the apple (Malus × domestica) genome, several loci linked to insect resistance, especially to aphids, have been mapped: a resistance gene and a QTL for the leaf-curling aphid (Dysaphis devecta (Walker)) on LG7 (Roche et al. 1997; Cevik and King 2002; Stoeckli et al. 2008b); a QTL for the rosy apple aphid (Dysaphis plantaginea (Passerini)) resistance on LG17 (Stoeckli et al. 2008b); a QTL for antibiosis resistance to the green apple aphid (Aphis pomi De Geer) on LG11 (Stoeckli et al. 2008a); four major genes conferring resistance to woolly apple aphid (Eriosoma lanigerum (Hausmann)) on LGs 7, 8, and 17 (Bus et al. 2008, 2010); and a QTL associated with the carpophagous codling moth (Cydia pomonella L.) susceptibility on LG10 (Stoeckli et al. 2009).
We investigated a new source of resistance to pear psylla derived from the Asian species Pyrus × bretschneideri. Interspecific hybrid PEAR3 (P. × bretschneideri ‘Xuehuali’Footnote 1 × P. communis ‘Max Red Bartlett’) was crossed with the European cultivar ‘Moonglow’ to develop a segregating population for QTL mapping. PEAR3 was previously proven to be moderately resistant to psylla (unpublished data), while ‘Moonglow’ was reported as moderately to highly susceptible (Bell 1984; Berrada et al. 1995). In a mono-varietal pear orchard, the insect is closer to a no-choice situation (Pasqualini et al. 2006). Therefore, we focused on antibiosis resistance, predominantly expressed as a reduced development rate of the insects. A novel phenotyping protocol was developed to screen large numbers of plants simultaneously, and its repeatability was tested over 2 years. Montanari et al. (2013) scanned 220 progeny of the PEAR3 × ‘Moonglow’ segregating population with single-nucleotide polymorphism (SNP) and simple sequence repeats (SSR) markers and constructed two high density parental genetic maps. PEAR3 map consisted of 208 markers spanning 979.8 cM (with a density of one marker every 4.7 cM and a LG average length of 57.6 cM), and ‘Moonglow’ map consisted of 464 markers spanning 1016.6 cM (with a density of one marker every 2.2 cM and a LG average length of 59.8 cM). These parental maps were used to detect QTLs for C. pyri resistance.
Materials and methods
Plant material
An F1 population derived from PEAR3 × ‘Moonglow’ was screened for pear psylla resistance in 2013 and 2014 at the INRA site of Angers (France), testing, respectively, 96 and 98 progeny, along with the two parents and five controls: P. communis cultivars ‘Angélys,’ ‘Harrow Sweet,’ Michigan-US 437, and ‘Williams Bon Chrétien’ and the interspecific P. ussuriensis × P. communis hybrid NY10355. All genotypes were grafted on ‘Kirchensaller’ rootstocks and randomized in the greenhouse with an average of 5 and 7 replicates in 2013 and 2014, respectively. Potted plants were placed on benches, and the climatic conditions in the greenhouse were controlled in order to keep an average temperature of 22/18 °C day/night and 55 % of relative humidity (RH). Pots were fertilized with a nutrient solution (N17–P10–K30) one to seven times per week, depending on growth condition of the plants. Irrigation was applied manually when needed.
At the infestation dates, the shoots were at least 15 cm tall and actively growing.
Infestation and assessments
C. pyri was reared on ‘Williams Bon Chrétien’ pear in insect-proof cages placed in a climatic chamber (16/8 h day/night photoperiod, 100 μM/m2/s minimum photosynthetic photon flux density, 22/18 °C temperature, and 70–90 % of RH) (Fig. 2a). Adults were collected for infestation 1 week after the last molt, when males and females were visually recognizable, and put in separate tubes (Fig. 2b). In order to perform a no-choice test and guarantee oviposition on all genotypes, the upper three to four leaves of each shoot were covered with light net bags, and one male and one female were introduced in each bag (Fig. 2c). After 8 days, the bags were removed, making sure not to leave any live adults in the greenhouse, and the numbers of eggs were counted with the aid of binocular magnifiers, using six classes (0 = no eggs, 1 = 1–10 eggs, 2 = 11–30 eggs, 3 = 31–50 eggs, 4 = 51–100 eggs, and 5 = more than 100 eggs). From this moment on, plants were monitored constantly to determine when all the eggs had hatched, but no new adults had appeared, which was when the nymph assessment was performed (22 and 26 days after infestation, in 2013 and 2014 respectively). Nymph assessment was the most crucial part of the experiment: in order to introduce as little variability as possible to the phenotypic traits, the right balance between time (the assessment had to be completed within very few days) and the unavoidable subjectivity of the scorer (more observers, more variability) had to be achieved. Nymphal mortality was measured by counting the numbers of living young (L1, L2, and L3 instars) and old (L4 and L5 instars) nymphs (Fig. 1a) with the use of a stereomicroscope.
Method for psylla infestation in an interspecific pear population. a Cacopsylla pyri was reared on ‘Williams Bon Chrétien’ trees placed in insect-proof cages. b On the infestation date, male and female adults were captured into separate tubes. c The main shoot for each genotype grown in the greenhouse was covered with a light net bag, and one male and one female were introduced inside each bag
Statistical analysis and QTL mapping
R studio (http://www.rstudio.com) was used for statistical analyses. Shoots that stopped growing were excluded from the analyses.
Raw data (eggs, total nymphs, young nymphs, and old nymphs) were tested for normality using the Lilliefors and Shapiro–Francia tests (Thode 2002), in which the null hypothesis is that the data were normally distributed.
In both years, the nymph counting on all plants took 3 days, during which the insects continued to develop, and involved ten scorers. Therefore, the significance of the “scoring date” and “scorer” effects, considered as fixed effects, on the number of nymphs at different stages was tested, using ANOVA. The model of the analysis of variance was considered reliable when the residual errors were normal, which was verified with “residual versus fitted” and “normal quantile–quantile” plots. For each year, averages were adjusted according to the significant (ρ < 0.05) fixed effects and the distributions of the adjusted means were again tested for normality with the Lilliefors and Shapiro–Francia tests. The egg phenotypic data were treated both as a factor affecting the number of nymphs and a trait for QTL mapping. In the first case, the ordered “factor eggs” (obtained from the variable “eggs” by applying the function factor in R with the argument ordered = TRUE) was added to the ANOVA model for the means adjustment.
For each year of phenotyping, the correlations between the adjusted means of the different traits were tested, in particular, “eggs versus total nymphs,” “young versus total nymphs,” “old versus total nymphs,” and “young versus old nymphs.” The correlation coefficients were calculated using the Pearson formula when the traits were normally distributed or Spearman formula (for ranked data) when at least one of the two traits was not normal. The statistical significance of the correlations was also evaluated.
The traits considered for QTL mapping were as follows: (i) class of eggs (eggs), (ii) number of total nymphs (“total nymphs”), and (iii) the ratio of the number of old/number of total nymphs (“old/total nymphs”). The broad-sense heritability (H 2) of genotypic means within each progeny for all these traits was estimated using the formula \( {H}^2={\sigma}_g^2\div \left({\sigma}_g^2+\raisebox{1ex}{${\sigma}_e^2$}\!\left/ \!\raisebox{-1ex}{$n$}\right.\right) \), where n is the mean number of replicates per genotype, σ 2 g is the genetic variance (i.e., inter-genotype variance), and σ 2 e is the residual error variance (Calenge et al. 2005; Durel et al. 2009).
QTL mapping was performed with the MapQTL 5.0 software (Van Ooijen 2004), using interval mapping (IM) (Lander and Botstein 1989) and, when multiple QTLs were detected, restricted multiple QTL mapping (rMQM) (Jansen 1993; Jansen and Stam 1994), selecting the closest marker to the QTL peak as cofactor. QTLs were also detected using the Haley–Knott (HK) regression method (Haley and Knott 1992), with the package “qtl” of R (Rqtl) (http://www.rqtl.org). The significant logarithm of odds (LOD) threshold (ρ = 0.05) for each trait was determined after genome-wide permutation tests (Churchill and Doerge 1994) using 1000 permutations. The genetic maps of the parents PEAR3 and ‘Moonglow’ used for QTL mapping were the ones published by Montanari et al. (2013), with minor modifications: (i) eight new markers were added to LG5 of PEAR3 (ss475882774, ss475883501, ss475878404, ss475879604) and LG15 of ‘Moonglow’ (ss475881341, ss475881255, NB129a, ss527789616) and (ii) the SNPs heterozygous with the same alleles in both parents were removed prior to analyses.
Possible epistatic interactions between detected QTLs were tested using ANOVA with the formula Y k = μ + M1 + M2 + (M1 * M2) + ε k , where Y k is the phenotypic value of the genotype k, μ is the phenotypic mean of the population, M1 and M2 are the actual effects of markers M1 (the closest to the peak of QTL1) and M2 (the closest to the peak of QTL2), (M1 * M2) is the interaction effect between the markers M1 and M2, and ε k is the residual effect. The normality of the residual errors was verified as explained before. The percentage of the phenotypic variation explained by all the significant (ρ < 0.05) QTLs and epistatic interactions (R 2 or coefficient of multiple determination) was estimated using the formula \( \mathrm{global}\;{R}^2=1-\left(\raisebox{1ex}{${\mathrm{SS}}_{\mathrm{res}}$}\!\left/ \!\raisebox{-1ex}{${\mathrm{SS}}_{\mathrm{tot}}$}\right.\right) \), where SSres is the residual sum of squares and SStot is the total sum of squares.
Comparison between the 2 years
The data collected in 2013 and 2014 were compared in two ways: first, they were pooled together and the significance of the effect “year” was tested; then, the adjusted means from 2013 and 2014 were compared in R, and the correlation coefficients were estimated. As for the comparisons between different traits, the Pearson formula was used when the two compared sets of data followed a normal distribution; otherwise, the Spearman rank correlation coefficient was calculated. In order to verify if our interpretation of the correlation coefficient was correct, we also tested the statistical significance of the correlations.
Results
Egg and nymph assessments
In total, 405 and 504 trees from 96 and 98 replicated seedlings were screened for host resistance to pear psylla in 2013 and 2014, respectively. When the infestations were carried out, plant shoots were actively growing and psylla females were ready for oviposition. By the time the bags were removed, 8 days later, the plants were just a little withered, and the oviposition was well advanced. The nymph assessment started 22 and 26 days after infestation in 2013 and 2014, respectively. When looking at the response of the controls, ‘Williams Bon Chrétien’ and NY10355 were always highly susceptible and highly resistant, respectively (Fig. 3). For the total number of nymphs in 2013, the parent ‘Moonglow’ was comparable to ‘Williams Bon Chrétien’ and PEAR3 to NY10355, and both parents were not significantly different from each other (according to Tukey test); no significant difference was observed between any of the controls for old/total nymphs. Also, in 2014, the PEAR3 response was similar to that of NY10355, while the total number of nymphs for this parent, although not significantly different from ‘Williams Bon Chrétien,’ was lower than Michigan-US 437. This year, like in 2013, the total number of nymphs and the old/total nymph ratio in ‘Moonglow’ were not significantly different from those in PEAR3 (Fig. 3). It is worth mentioning that while the number of replicates for ‘Moonglow’ in 2013 and 2014 was comparable (four and three), in 2013, PEAR3 had only one replicate and six in 2014.
Comparison between the different pear accessions used as controls in a psylla resistance phenotyping in 2013 and 2014. Significantly different genotypes, according to Tukey test, are identified by different letters. PEAR3 and ‘Moonglow’ (Moon) are the parents of the interspecific pear population tested; ‘Williams Bon Chrétien’ (WBC), ‘Harrow Sweet’ (HS), ‘Angélys’ (Angel), and Michigan-US 437 (Mich) the susceptible controls; and NY10355 (NY) the resistant control
The number of eggs was expected to be consistent amongst all plants, because the insects were in a no-choice situation. However, only few eggs were counted on the antixenotic and antibiotic NY10355 (Salvianti et al. 2008) (Fig. 3), which demonstrates that antixenosis was possible. Several transgressive seedlings were observed amongst the progeny when looking at the arithmetic means for each genotype. This was consistent with the polygenic nature of the trait, and it was probably emphasized by the level of resistance of the two parents, which was not as different as expected.
Phenotypic distribution, environmental effects, and heritability
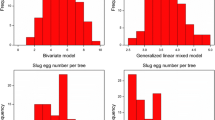
The results of the Lilliefors and Shapiro–Francia tests performed on the raw data were consistent, with only the latter reported here. The tests indicated that none of the measured variables had a normal distribution (ρ < 0.05) (Table 1) and all were biased toward resistance, except for eggs (Online Resource 1). Nevertheless, the residual errors turned out to be normally distributed, and no transformation or non-parametric analysis was needed. Consequently, ANOVA was used to evaluate the significance of the effects factor eggs (i.e., the number of laid eggs considered as an ordered factor affecting the number of nymphs), year, scoring date, and scorer on the phenotypic traits. A higher infestation was observed in the second year (2014), since significantly higher numbers of eggs and total nymphs were scored. Moreover, nymphal development was faster in 2014 than in 2013, as revealed by the quicker evolution of the numbers of young and old nymphs over the 3 days of assessment. Indeed, in 2014, the decrease of young nymphs and the increase of the old ones from the first to the third day of assessment were significantly greater than in 2013 (Δ(young) = −6.62 and Δ(old) = +2.12 in 2013 and −14.58 and +25.5 in 2014), with the number of old nymphs largely surpassing the number of young in 2014 (Fig. 4). The genotype significantly affected all the traits in both years. The environmental effects factor eggs, scoring date, and scorer were also significant in both years, and the phenotypic means were adjusted according to them. The distribution of the adjusted means was normal for the traits total nymphs and “young nymphs” in 2013 and, for eggs, young nymphs (although only according to Lilliefors test and not to Shapiro–Francia) and old/total nymphs in 2014, while it was not normal for the other traits (Table 2, Online Resource 2). The number of eggs and the total number of nymphs were significantly (ρ < 0.05) positively correlated and had Spearman coefficients r s = 0.747 and 0.672 in 2013 and 2014, respectively (Fig. 5). The total number of nymphs was very highly correlated with the number of young nymphs in 2013 (r s = 0.958) and to the number of old nymphs in 2014 (r s = 0.946) (Fig. 5).
Effect of the three nymph scoring dates on the number of young and old nymphs counted in an interspecific pear population in 2013 and 2014. The different letters represent significance difference (according to Tukey test) within each category: young nymphs in 2013, old nymphs in 2013, young nymphs in 2014, and old nymphs in 2014. Young nymphs are represented by the dark grey bars and old nymphs by the light grey bars. For each year, the difference between the third and the first days of assessments for the numbers of young and of old nymphs (delta) was calculated
Correlation between different psylla resistance phenotypic traits measured in a pear segregating population in 2013 and 2014. For each comparison, correlation coefficients and their significance level (*** = <0.001; ** = ρ < 0.01; * = ρ < 0.05; ns not significative) are shown. The Pearson formula (r) was used when both traits were normally distributed; otherwise, the Spearman formula (r s) was used
The estimated broad-sense heritability (H 2) was generally high for each trait (Table 3), with the highest values observed for total nymphs (0.63 in 2013 and 0.68 in 2014). The heritability for eggs was lower than the other traits in 2013 (0.45), but in the same range as for total nymphs in 2014 (0.58), thus indicating that antixenosis was significantly contributing to psylla resistance in this experiment. In contrast, the ratio old/total nymphs had a high heritability in 2013 (0.65), but it was lower in 2014 (0.45). The H 2 was lower when considering the factor eggs in ANOVA (data not presented).
Phenotypic correlation between years
A significant (ρ < 0.05) linear correlation was observed for the trait total nymphs between 2013 and 2014, with a Spearman coefficient of r s = 0.474 (Fig. 6a). Conversely, the traits eggs and old/total nymphs showed very weak or no correlation (Fig. 6b). For the trait eggs, the Spearman coefficient was r s = 0.249.
Phenotypic variability between years for psylla resistance in a pear segregating population. For each trait, the means adjusted for the environmental factors were used. Correlation coefficients, calculated using Spearman formula (r s), and their significance level (*** = <0.001; ** = ρ < 0.01; * = ρ < 0.01; ns not significative) are also shown. a For the total number of nymphs (“Total nymphs”), a linear correlation was observed between 2013 and 2014. b The number of eggs (“Eggs”) and the ratio of old/total nymphs (“Old/tot nymphs”) showed weak or absence of correlation between 2013 and 2014
QTL detection
QTLs were detected for all measured traits except the old/total nymphs in 2013 by IM or rMQM using MapQTL and by HK regression using Rqtl, with the significance of genome-wide LOD thresholds ranging between 3.1 and 3.3 after permutation tests (Table 4). The phenotypic variation explained by each QTL (R 2) is reported in Table 4, as well as the global R 2 estimated for each trait taking into account possible epistatic interactions between QTLs (global R 2), when several QTLs were present. The detected QTLs were the same whether or not the “factor eggs” was added into the model. However, the LOD scores were more significant without the factor eggs. Therefore, the results obtained with the factor eggs were not reported.
QTL detection in 2013
A QTL on LG8 of PEAR3 was detected with both methods for eggs and total nymphs, while for old/total nymphs, no QTL was found. The marker closest to the QTL peak was SSR CH05a02, with its 130 bp allele associated with resistance. This QTL on LG8 explained between 17.2 % (calculated with HK for the eggs) and 39.1 % (calculated with rMQM for total nymphs) of the phenotypic variation. When using MapQTL, two other putative QTLs, just a little below the threshold, were mapped to LG5 of PEAR3 for eggs (R 2 = 9.9 %), associated with SNP ss475875754, and to LG11 of PEAR3 for total nymphs (R 2 = 8.4 %), associated with SNP ss475877524. However, neither of these QTLs was detected when using the HK method. The residual errors calculated post-QTL analysis were normally distributed for the trait total nymphs, but not for eggs.
QTL detection in 2014
The QTL on LG8 of PEAR3 was confirmed in 2014 for eggs (just below the threshold) and total nymphs using both methods. Its peak was located on the upper part of the LG, at the same position as in 2013 or above it. By looking at marker CH05a02, the favorable allele was 130 bp, as in 2013. Moreover, a QTL just below the threshold was found on the same location also for old/total nymphs. The R 2 of the LG8 QTL ranged between 10.9 and 29.5 %, again with the highest value for the “total number of nymphs.” The QTL on LG5 of PEAR3 for eggs (marker ss475875754) was not detected in 2014, but a QTL in a very close position (marker ss475878404) was found for the trait total nymphs, although not confirmed when using the regression method. For total nymphs, another putative QTL was found on LG11 of PEAR3 using MapQTL, close to the one detected in 2013; however, its peak was below the threshold. Furthermore, a QTL was mapped to LG15 of ‘Moonglow’ for the old/total nymph ratio, significant only with the HK method. LG5, LG11, and LG15 QTLs had usually smaller effects than the one on LG8, with R 2 values ranging between 7.7 and 13.7 %. Both in 2014 and 2013, the resistance was associated to allele “G” of SNP ss475875754 and to allele “0” (null allele) of ss475878404 on LG5, and to allele G of ss475877524 and to allele 0 (null allele) of ss475882338 on LG11. On LG15, the QTL peak was close to SNP ss475883269 and “T” was the favorable allele. The global R 2 was estimated for the total number of nymphs (global R 2 = 50.5 %), and a significant interaction effect (epistasis) was detected between the LG8 and LG5 QTLs. The residual errors were normally distributed for all the traits except for eggs, like in 2013.
The positions of all the QTLs detected in 2013 and 2014 are shown on the genetic map (Fig. 7).
When R 2 was compared with the estimated broad-sense heritability (Table 3), it was noticed that it was always lower than H 2.
Discussion
A robust, repeatable, and high-throughput phenotyping protocol for psylla resistance genetic analysis
Collecting quantitative and reproducible phenotypic data with minimal environmental effect over large numbers of segregating seedlings is crucial for QTL mapping. Multiple protocols have been developed previously for phenotyping antibiosis to psylla (Berrada et al. 1995; Pasqualini et al. 2006; Bell 2013a, b), but none of them was suitable for the purpose of assessing resistance in a large segregating population. The logistical and reproducible challenges were exacerbated by the necessity of a strict phenological synchronization between the plant and the pest and the creation of an environment with optimal growing conditions for both of them. The phenotyping protocol that we developed employed about ten people for only 5 days each year to study the antibiosis resistance of pear to psylla and to collect quantitative data from hundreds of plants. The egg and nymph assessments were carried out over a short time frame in order to minimize non-genetic factors, such as the influence of temperature and RH on insect development, and allowed the detection of robust QTLs. Furthermore, this protocol has proved to be repeatable across years.
Sensitivity of the assessment period
When considering both tests performed in 2013 and 2014, it was interesting to notice that the date of the nymph assessment (scoring date) was a very sensitive parameter. Firstly, considering each year separately, this effect turned out to be significant (Fig. 4), despite that the scoring had been performed on three consecutive days with a complete randomization of the assessed genotypes and a stable involvement. Consequently, the number of young and old nymphs quickly evolved in a short period of time, which aspect was corrected year-per-year by adjusting the phenotypic means of the seedlings according to the scoring date effect. Secondly, when comparing 2013 and 2014 tests, the nymph assessment was performed with a small discrepancy regarding the number of days after infestation (22 and 26 days, respectively). Moreover, nymph development was faster in 2014 than in 2013 (Fig. 4), even though the temperature and the RH measured inside the greenhouse were comparable. The later assessment and quicker nymphal development in 2014 with respect to 2013 explain the stronger correlation between young nymphs and total nymphs in 2013 and between “old nymphs” and total nymphs in 2014 (Fig. 5). As total nymphs = young nymphs + old nymphs, a later and faster (respectively, earlier and slower) assessment gave higher emphasis to old relative to nymphs in agreement with the nymph developmental process.
Discrimination between antibiosis and antixenosis
In this experiment, we initially wanted to evaluate the antibiosis resistance to pear psylla, putting insects in a no-choice situation in order to reduce as much as possible the variability in the oviposition rate among the different genotypes. The reason for that was to mimic a mono-varietal pear orchard where the insect has no choice for the variety on which to lay its eggs. Nevertheless, in practice, ovipositional antixenosis and antibiosis resistance could not be completely separated, and we also measured significant variability among the genotypes for the eggs trait. An extreme situation was observed for NY10355, where the number of eggs laid was particularly small, thus indicating that this genotype exhibits a strong ovipositional antixenosis resistance. Basically, there is a (chronological) dependency between the final number of total nymphs and the initial number of eggs. A small number of laid eggs prevents one from observing a large number of nymphs, whereas a large number of laid eggs allows the observation of small, medium, or large number of nymphs according to the subsequent antibiosis resistance of the genotypes. This was clearly shown in the “triangle” relationship between total nymphs and eggs (Fig. 5), with less variation in total nymphs for small values of eggs and larger variation for high values of eggs. We tried to correct for the number of nymphs according to the number of eggs in order to focus on antibiosis, but we came across the imprecision of our initial egg assessment protocol, as an ordinal scale with large intervals and only six classes was used. Moreover, as antixenosis applies earlier in the parasitic process than antibiosis, it can hamper the correct detection of antibiosis resistance by hiding its genetic variation, especially for those genotypes with strong ovipositional antixenosis resistance. Such a chronological dependency creates a bias in the accuracy of the antibiosis assessment, which cannot be simply corrected by statistical approaches. Thus, ovipositional antixenosis can generate seeming antibiosis. Here, the consistency of low numbers of eggs across the replicates of several genotypes generated a moderate but significant heritability for this trait, demonstrating that there was an important contribution of antixenosis to psylla resistance in our experiment. Consistently, we were able to detect QTLs for the eggs trait. For the number of nymphs, the detected QTLs were the same whether or not the “factor eggs” was added into the ANOVA model, indicating that antibiosis was also most probably contributing to psylla resistance. The lower LOD score significance for the QTLs detected with factor eggs could be interpreted as a signature of the antixenosis impact on the antibiosis assessment. We nevertheless considered that antibiosis resistance was present and correctly mapped in the present experiment, since the number of laid eggs was rather high for many of the genotypes, with a majority of 4 or 5 scoring at the egg assessment. Thus, the new phenotyping protocol allowed an incomplete, but acceptable control of the antixenosis mechanism of resistance and a correct examination of the antibiosis.
New QTLs for pear resistance to psylla
A large effect and stable QTL inherited from the resistant parent PEAR3 was detected on LG8 for all the traits (Fig. 7). The position of this QTL was confirmed after 2 years of experiments and by using two QTL mapping methods, IM and HK regression. QTL detection by regression is more robust for non-normally distributed data (Feenstra et al. 2006), which was the case for the trait eggs (non-normal distribution of the residual errors after the QTL analysis). In 2014, when the infestation was higher, a QTL was also detected on LG15 of ‘Moonglow’ for the ratio old/total nymphs. No QTL was found for this trait in 2013, probably because of the very low numbers of old nymphs scored. Furthermore, two small-effect QTLs were detected on LG5 (for eggs in 2013 and for total nymphs in 2014) and on LG11 of PEAR3 (for total nymphs both in 2013 and 2014). However, their LOD scores were low and neither of them was confirmed using HK regression, indicating that they could be spurious QTLs (Table 4).
The number of total nymphs was the measure less prone to error. In fact, the distinction between young and old nymphs can be difficult, especially between the L3 and L4 instars (Fig. 1a); hence, some nymphs could have been allocated to the wrong class. This may explain why we found the strongest QTLs for the trait total nymphs, with the highest LOD score and R 2 (Table 4). For the trait eggs we also found a QTL on LG8 of PEAR3 in 2013 and in 2014 (just below the threshold), whose position was consistent with the QTLs found for the other traits (Fig. 7). The broad-sense heritability was usually high for all the traits (Table 3), indicating that the phenotypic variance was mostly attributable to differences in genotypes and less to the environment and that the results of our QTL mapping were reliable. However, the R 2 explained by the QTLs were always lower than the H 2, which indicates that we were not able to detect all the loci linked to psylla resistance. The reasons were imputable to the type and size of the mapping population that we used. Being an interspecific F1 population, all individuals were supposedly highly heterozygous; hence, the progeny was highly variable with possible complex genetic architecture of the studied traits involving gene interactions, which are more difficult to map. Moreover, our population consisted of just fewer than 100 genotypes, which is sufficient to detect only the largest effect QTLs. The strong QTL that we discovered on LG8 of PEAR3 also has epistatic relationships with other loci. Therefore, a larger number of genotypes would be necessary for the detection of further smaller effect QTLs in this family, if present (Collard et al. 2005). Since the parental genetic maps, especially the one of PEAR3, were not saturated, it is also possible that some QTLs are located in genomic regions not covered by markers, hence could not be detected in this experiment.
From these results, we can assume that a locus responsible for a strong antibiosis resistance, but also for ovipositional antixenosis, was located on LG8 of PEAR3. Since the confidence intervals of the QTL detected for the different traits were quite large (one-LOD support interval ranging from 6 to 25 cM, but usually higher than 18 cM) (Fig. 7), two different, but closely linked loci, one for antibiosis and one for antixenosis, could be located in the same interval. Here again, the population size (∼100 progeny) was not large enough to discriminate between both hypotheses: closely linked QTLs (approximately 20 cM or less) are not distinguishable with population size lower than 500 (Collard et al. 2005). On the other hand, the QTL for old/total nymphs on LG15 of the “susceptible” parent ‘Moonglow,’ even if its presence should be confirmed with other tests, may be more strictly linked to the antibiosis mechanism (i.e., delayed nymphal development). The observation of several transgressive seedlings amongst the progenies had already predicted the possible presence of resistance factors in both parents. Therefore, unknown sources of psylla resistance may be present among the P. communis cultivars in the ‘Moonglow’ pedigree (Montanari et al. 2013): ‘Seckel’ and ‘Bartlett’ are known to be susceptible to psylla species (Butt et al. 1988; Bell and Stuart 1990), Michigan-US 437 was tested in our experiments and resulted to be susceptible (Fig. 3), while there is no information available about ‘Roi Charles de Würtemberg,’ which could thus be more resistant. Concerning the LG8 QTL, while we do not have any information about the genotypes at this locus for the PEAR3 parents, with psylla resistance mostly found in Asian pear species (Westigard et al. 1970; Bell and Stuart 1990; Bell 2013a), it is most likely to have been inherited from ‘Xuehuali’. Previously, the cultivar ‘Xuehuali’ was reported as having good field resistance to psylla (Stanica 2002).
Co-localization with genes and QTLs for aphid resistance
The genetic map of PEAR3 could be compared with other pear and apple maps generated for the detection of QTLs and major loci for pest and disease resistance through common microsatellite markers. Two major genes for woolly apple aphid resistance were mapped to the upper part of LG8 in apple (Bus et al. 2008, 2010), the same region where we detected the major QTL for psylla resistance. Moreover, the putative QTL for the resistance of apple to A. pomi found on LG11 by Stoeckli et al. (2008a) co-localizes with the small-effect QTL that we detected on this LG for total nymphs. This is not the first time that loci associated to psylla and aphid resistance are found in chromosomal regions orthologous between species: aphid resistance genes were mapped to LG17 in both pear (Evans et al. 2008) and apple (Stoeckli et al. 2008b; Bus et al. 2008, 2010), where Bouvier et al. (2011) also detected a QTL for pear psylla resistance in the P. ussuriensis × P. communis hybrid NY10355. Aphids and psylla are both phloem feeders; therefore, finding orthologous regions linked to antibiosis resistance to these insects may indicate some common molecular resistance mechanisms. Civolani et al. (2013) conducted experiments on the probing behavior of C. pyri, and they introduced the hypothesis that strong resistance factors are present in the phloem of resistant pear accessions.
Conclusion
The results of our experiment confirmed pear psylla resistance to be a polygenic trait. Although the parents PEAR3 and ‘Moonglow’ turned out to have a much more similar response to psylla infestation than we expected, we were able to detect a stable QTL on LG8 of PEAR3. Until now, only Bouvier et al. (2011) had published results from a QTL mapping study for pear psylla resistance, but they used a different source of resistance (P. ussuriensis), and the major QTL that they found was located on LG17. Pyramiding these two sources of resistance (P. × bretschneideri and P. ussuriensis) could be an effective breeding strategy for the development of pear cultivar highly resistant to psylla.
Other experiments will be necessary to reduce the confidence interval of the QTL on LG8 and to confirm the significance of the minor QTLs on LGs 5, 11, and 15, with the final purpose of identifying markers useful for marker-assisted selection (MAS). Moreover, scoring the same population more accurately for oviposition could be useful to verify the hypothesis of the presence of two distinct loci on LG8, one for antibiosis and one for antixenosis. Pear cultivars bringing both the QTLs responsible for antixenosis and antibiosis would have a more durable resistance, more difficult to be overcome by newly evolved psylla races. Indeed, the experiment carried out by Puterka (1997) with different C. pyricola biotypes (originating from different regions in the USA) on susceptible and resistant pear varieties suggested the ability of this pest to adapt to the host resistance.
Finally, it would be interesting to study the possible localization on apple and pear LG8 and LG17 of genes responsible for the production of phloem resistance factors, which act in response to psylla and aphid infestation.
The recent publication of P. bretschneideri (Wu et al. 2013) and P. communis (Chagné et al. 2014) genome sequences will facilitate studies on the molecular determinism of agronomic traits of interest in pear, such as pest and disease resistance. The development of new markers and the functional analysis of genes in the genomic regions linked to psylla resistance will lead to a better understanding of this important, but complex trait.
References
Atger P, Bigre JP (1982) Le psylle du poirier. Centre Technique Interprofessionnel des Fruits et Légumes, Paris
Bell RL (1984) Evaluation of Pyrus germplasm for resistance to the pear psylla. Acta Horticult 161:234–237
Bell RL (1992) Additional East European Pyrus germplasm with resistance to pear psylla nymphal feeding. HortSci 27:412–413
Bell RL (2013a) Host resistance to pear psylla of breeding program selections and cultivars. HortSci 48:143–145
Bell RL (2013b) Inheritance of resistance to pear psylla nymphal feeding in pear (Pyrus communis L.) of European origin. HortSci 48:425–427
Bell RL, Stuart LC (1990) Resistance in Eastern European Pyrus germplasm to pear psylla nymphal feeding. HortSci 25:789–791
Berrada S, Nguyen TX, Lemoine J et al (1995) Thirteen pear species and cultivars evaluated for resistance to Cacopsylla pyri (Homoptera: Psyllidae). Environ Entomol 24:1604–1607
Bouvier L, Bourcy M, Boulay M et al (2011) European pear cultivar resistance to bio-pests: scab (Venturia pirina) and psylla (Cacopsylla pyri). Acta Horticult 909:459–470
Buès R, Boudinhon L, Toubon JF (2003) Resistance of pear psylla (Cacopsylla pyri L.; Hom., Psyllidae) to deltamethrin and synergism with piperonyl butoxide. J Appl Entomol 127:305–312
Bus VGM, Chagné D, Bassett HCM et al (2008) Genome mapping of three major resistance genes to woolly apple aphid (Eriosoma lanigerum Hausm.). Tree Genet Genomes 4:223–236. doi:10.1007/s11295-007-0103-3
Bus VGM, Bassett HCM, Bowatte D et al (2010) Genome mapping of an apple scab, a powdery mildew and a woolly apple aphid resistance gene from open-pollinated Mildew Immune Selection. Tree Genet Genomes 6:477–487
Butt BA, Stuart LC, Bell RL (1988) Feeding behavior of pear psylla (Homoptra: Psyllidae) nymphs on susceptible and resistant Pyrus germplasm. J Econ Entomol 81:1394–1397
Calenge F, Drouet D, Denancé C et al (2005) Identification of a major QTL together with several minor additive or epistatic QTLs for resistance to fire blight in apple in two related progenies. Theor Appl Genet 111:128–35. doi:10.1007/s00122-005-2002-z
Cevik V, King G (2002) High-resolution genetic analysis of the Sd-1 aphid resistance locus in Malus spp. Theor Appl Genet 105:346–354
Chagné D, Crowhurst RN, Pindo M et al (2014) The draft genome sequence of European pear (Pyrus communis L. ‘Bartlett’). PLoS One. doi:10.1371/journal.pone.0092644
Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138:963–71
Civolani S (2012) The past and present of pear protection against the pear psylla, Cacopsylla pyri L. Insecticides - Pest Engineering. INTECH Open Access Publisher, pp 385–408
Civolani S, Peretto R, Caroli L et al (2007) Preliminary resistance screening on abamectin in pear psylla (Hemiptera: Psyllidae ) in Northern Italy. J Econ Entomol 100:1637–1641
Civolani S, Grandi G, Chicca M et al (2013) Probing behaviour of Cacopsylla pyri on a resistant pear selection. J Appl Entomol 137:365–375. doi:10.1111/jen.12003
Collard BCY, Jahufer MZZ, Brouwer JB, Pang ECK (2005) An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: the basic concepts. Euphytica 142:169–196. doi:10.1007/s10681-005-1681-5
Durel C-E, Denancé C, Brisset MN (2009) Two distinct major QTL for resistance to fire blight co-localize on linkage group 12 in apple genotypes ‘Evereste’ and Malus floribunda clone 821. Genome 52:139–147. doi:10.1139/G08-111
Evans KM, Govan CL, Fernández-Fernández F (2008) A new gene for resistance to Dysaphis pyri in pear and identification of flanking microsatellite markers. Genome 51:1026–31. doi:10.1139/G08-093
Feenstra B, Skovgaard IM, Broman KW (2006) Mapping quantitative trait loci by an extension of the Haley-Knott regression method using estimating equations. Genetics 173:2269–82. doi:10.1534/genetics.106.058537
Haley CS, Knott SA (1992) A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity (Edinb) 69:315–24
Harries FH, Burts EC (1965) Insecticide resistance in the pear psylla. J Econ Entomol 58:172–173
Harris MK, Lamb RC (1973) Resistance to the pear psylla in pears with Pyrus ussuriensis lineage. J Am Soc Hortic Sci 98:378–381
Hesler LS, Tharp CI (2005) Antibiosis and antixenosis to Rhopalosiphum padi among triticale accessions. Euphytica 143:153–160. doi:10.1007/s10681-005-3060-7
Hodkinson ID (2009) Life cycle variation and adaptation in jumping plant lice (Insecta: Hemiptera: Psylloidea): a global synthesis. J Nat Hist 43:65–179. doi:10.1080/00222930802354167
Jansen RC (1993) Interval mapping of multiple quantitative trait loci. Genetics 135:205–211
Jansen RC, Stam P (1994) High resolution of quantitative traits into multiple loci via interval mapping. Genetics 136:1447–1455
Kapatos ET, Stratopoulou ET (1999) Duration times of the immature stages of Cacopsylla pyri L. (Hom., Psyllidae), estimated under field conditions, and their relationship to ambient temperature. J Appl Entomol 123:555–559. doi:10.1046/j.1439-0418.1999.00417.x
Lander ES, Botstein D (1989) Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121:185–99
Lespinasse Y, Chevalier M, Durel C-E et al (2008) Pear breeding for scab and psylla resistance. Acta Horticult 800:475–482
Montanari S, Saeed M, Knäbel M et al (2013) Identification of Pyrus single nucleotide polymorphisms (SNPs) and evaluation for genetic mapping in European pear and interspecific Pyrus hybrids. PLoS One. doi:10.1371/journal.pone.0077022
Pasqualini E, Civolani S, Musacchi S et al (2006) Cacopsylla pyri behaviour on new pear selections for host resistance programs. Bull Insectology 59:27–37
Puterka GJ (1997) Intraspecific variation in pear psylla (Psyllidae: Homoptera) nymphal survival and development on resistant and susceptible pear. Environ Entomol 26:552–557
Robert P, Raimbault T (2004) Resistance of some Pyrus communis cultivars and Pyrus hybrids to the pear psylla Cacopsylla pyri (Homoptera, Psyllidae). Acta Horticult 671:571–575
Robert P, Guérif P, Lemoine J, Le Lézec M (2004) Criblage de génotypes de Pyrus vis-à-vis de la résistance au psylle du poirier Cacopsylla pyri (L.). Chaiers Agric 13:349–354
Roche P, Alston FH, Maliepaard C et al (1997) RFLP and RAPD markers linked to the rosy leaf curling aphid resistance gene (Sd 1) in apple. Theor Appl Genet 94:528–533
Salvianti F, Bettini PP, Giordani E et al (2008) Identification by suppression subtractive hybridization of genes expressed in pear (Pyrus spp.) upon infestation with Cacopsylla pyri (Homoptera: Psyllidae). J Plant Physiol 165:1808–16. doi:10.1016/j.jplph.2007.12.010
Schaub L, Graf B, Butturini A (2005) Phenological model of pear psylla Cacopsylla pyri. Entomol Exp Appl 117:105–111. doi:10.1111/j.1570-7458.2005.00339.x
Shaltiel-Harpaz L, Soroker V, Kedoshim R et al (2014) Two pear accessions evaluated for susceptibility to pear psylla Cacopsylla bidens (Šulc) in Israel. Pest Manag Sci 70:234–9. doi:10.1002/ps.3543
Stanica F (2002) Behaviour of four over grafted Chinese pear varieties (Pyrus serotina) in Bucuresti area. Acta Horticult 596:405–409
Stoeckli S, Mody K, Dorn S (2008a) Aphis pomi (Hemiptera: Aphididae) population development, shoot characteristics, and antibiosis resistance in different apple genotypes. J Econ Entomol 101:1341–8
Stoeckli S, Mody K, Gessler C et al (2008b) QTL analysis for aphid resistance and growth traits in apple. Tree Genet Genomes 4:833–847
Stoeckli S, Mody K, Gessler C et al (2009) Quantitative trait locus mapping of resistance in apple to Cydia pomonella and Lyonetia clerkella and of two selected fruit traits. Ann Appl Biol 154:377–387
Thode HC (2002) Testing for normality. CRC Press, New York
Van Ooijen JW (2004) MapQTL 5, Software for the mapping of quantitative trait loci in experimental population. Kyazma BV, Wageningen
Wang Y (2002) Genetic resources of deciduous fruit and nut crops in China. China Agricultural Science and Technology Press, Beijing
Westigard PH, Westwood MN, Lombard PB (1970) Host preference and resistance of Pyrus species to the pear psylla, Psylla pyricola Foerster. J Am Soc Hortic Sci 95:34–6
Wu J, Wang Z, Shi Z et al (2013) The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res 23:396–408. doi:10.1101/gr.144311.112
Acknowledgments
SM was funded by the Fondazione Edmund Mach (FEM) PhD School. We greatly thank Déborah Renault, Jasmine McCarthy, Ferreol Braud, Sophie Aligon, Medhi Al-Rifai, Clement Joffrion, Sylvain Hanteville, Valérie Le Mignon, Arnaud Guyader, and Christine Boursier (UMR1345 IRHS, Angers, France) for their precious help in the scoring on C. pyri eggs and nymphs. We also thank Lester Brewer at PFR for generating the PEAR3 × ‘Moonglow’ cross and the INEM team of IRHS, especially Michel Boucourt, and the INRA Experimental Unit (UE Horti, Angers, France), especially Christian Cattanéo, for taking care of the seedling population.
Data archiving statement
QTL data are reported in Table 4 and will be made publicly available through the Genome Database for Rosaceae (www.rosaceae.org).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by E. Dirlewanger
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM_1
Psylla resistance phenotypic data distributions in a segregating interspecific pear population in 2013 and 2014. (PDF 189 kb)
ESM_2
Distribution of the phenotypic means of psylla resistance adjusted for environmental factors in a pear segregating population in 2013 and 2014. (PDF 321 kb)
Rights and permissions
About this article
Cite this article
Montanari, S., Guérif, P., Ravon, E. et al. Genetic mapping of Cacopsylla pyri resistance in an interspecific pear (Pyrus spp.) population. Tree Genetics & Genomes 11, 74 (2015). https://doi.org/10.1007/s11295-015-0901-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-015-0901-y