Abstract
Apple is host to a wide range of pests and diseases, with several of these, such as apple scab, powdery mildew and woolly apple aphid, being major causes of damage in most areas around the world. Resistance breeding is an effective way of controlling pests and diseases, provided that the resistance is durable. As the gene pyramiding strategy for increasing durability requires a sufficient supply of resistance genes with different modes of action, the identification and mapping of new resistance genes is an ongoing process in breeding. In this paper, we describe the mapping of an apple scab, a powdery mildew and a woolly apple aphid gene from progeny of open-pollinated mildew immune selection. The scab resistance gene Rvi16 was identified in progeny 93.051 G07-098 and mapped to linkage group 3 of apple. The mildew and woolly aphid genes were identified in accession 93.051 G02-054. The woolly aphid resistance gene Er4 mapped to linkage group 7 to a region close to where previously the genes Sd1 and Sd2, for resistance to the rosy apple leaf-curling aphid, had been mapped. The mildew resistance gene Pl-m mapped to the same region on linkage group 11 where Pl2 had been mapped previously. Flanking markers useful for marker-assisted selection have been identified for each gene.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Apple (Malus × domestica Borkh.) is host to a wide range of pests and diseases, many of which are present in all apple-producing regions in the world (Way et al. 1989). The most common diseases that can devastate apple crops if not controlled are apple scab (Venturia inaequalis), powdery mildew (Podosphaera leucotricha) and fire blight (Erwinia amylovora). Extensive germplasm evaluations have shown that many useful sources of resistance are available for the breeding of pest and disease-resistant cultivars as an alternative to pesticides, the use of which is increasingly being disapproved of by consumers.
Resistance breeding has long been applied to apple as a means to control scab and powdery mildew. Since the earliest germplasm evaluations for scab resistance by Aderhold (1902), who recognised the value of resistant cultivars in the absence of effective fungicides, many sources of resistance have been identified. Genetic studies have shown the presence of both major gene and polygenic resistances in apple germplasm and breeders have discussed the merits and disadvantages of each type since resistance breeding commenced (Williams and Kuć 1969). In spite of the range of resistances available to breeders, most breeding programmes have for a long time relied on the Rvi6 (Vf) gene only for the development of new cultivars with scab resistance (Crosby et al. 1992; Laurens 1999). Today, other major gene and polygenic resistances take a more prominent place in most breeding programmes (Gessler et al. 2006). The same applies for mildew, where the Pl1 and Pl2 genes (Alston 1977) have long been used as the main sources of resistance in a number of programmes.
Resistance breeding is an economical option also for insects for which good sources of resistance are available, such as woolly apple aphid (Eriosoma lanigerum), a major pest in many Southern Hemisphere countries. Biological control by the parasitic wasp Aphelinus mali has shown promise (Shaw and Walker 1996), but its efficacy can be diminished by non-target effects of pesticides (Bradley et al. 1997). Also, the options for chemical control are increasingly restricted, due to a reduction in the number of pesticides available. For controlling the pest underground, breeding of resistant rootstocks is the preferred approach, as biological control is not effective and chemical control is not sustainable. The Malling–Merton rootstocks, which were specifically bred from ‘Northern Spy’ (Crane 1937) for the Southern Hemisphere countries of the British Commonwealth (Hatton 1937), have proved to be a very effective means to prevent the pest infesting the roots of apple trees.
Even though many sources of resistance to these pests have been identified, the search for new sources of new resistances is ongoing in order to provide a diverse range of resistance genes as a base for the continued development of new cultivars with pyramided resistances. The use of single gene resistances has been shown to be an ineffective approach to achieving long-lasting resistance, as most of the resistance genes used in apple to date have been overcome by the pathogen or pest at some stage, e.g. Er1 (Giliomee et al. 1968; Rock and Zeiger 1974; Sen Gupta and Miles 1975) and Er3 (Sandanayaka et al. 2003) for woolly apple aphid resistance, Rvi2 (Vh2) (Shay and Williams 1956; Bus et al. 2005b) and Rvi6 (Parisi et al. 1993) for scab resistance and Pl-m (Lespinasse 1983) and Pl2 (Caffier and Laurens 2005; Caffier and Parisi 2007) for mildew resistance. However, the seemingly general durability of gene pyramids found in Malus germplasm, e.g. scab resistance in Russian apple R12740-7A and Malus micromalus, suggests that combining resistance genes is a valid approach to durable resistance, even if they individually condition differential interactions with the pathogen (Bus 2006b). Over the last 10 years, many of these resistance genes have been mapped to the apple genome (Gardiner et al. 2007) to facilitate the application of marker-assisted selection (MAS) in the development of multiple and durably resistant cultivars.
Today, sourcing new resistances is still an important activity of the New Zealand apple-breeding programme. Plant and Food Research maintains an Apple Genetics Population commenced with seed imported from germplasm collections around the world in order to increase the genetic diversity of apple, a crop not native to New Zealand, and to ensure the long-term development of novel apple cultivars (Noiton and Shelbourne 1992). Some of this germplasm has been evaluated to identify new sources of resistance to pests and diseases (Alspach and Bus 1999; Bus et al. 2000a, 2002; Luby et al. 2002). In this paper, we present the mapping of the apple scab Rvi16, powdery mildew Pl-m and woolly apple aphid Er4 resistance genes identified in progenies of an open-pollinated (OP) family derived from ‘Mildew Immune Selection’ (MIS), which itself is an OP selection of ‘Delicious’ (Dayton 1977). While named for its mildew resistance, there are no reports on its resistance to apple scab and woolly aphid, which could not be determined either since this accession is not present in New Zealand.
Materials and methods
Plant material and phenotyping
Two progeny of the MIS OP family were used to study the genetics of resistance to one pest and two diseases. The genetics of the Rvi16 scab resistance was studied on three families across two generations, where the first generation family (173 seedlings) was raised from seed of a ‘Splendour’ × MIS OP 93.051 G07-098 cross made in 1998. A ‘fast-breeding’ approach was applied to two selections from this family, AK617 and AK653, in order to accelerate flowering (Austin et al. 2006; Volz et al. 2009), and both were crossed in 2000 with ‘Scired’ to develop second-generation families consisting of 244 and 210 seedlings, respectively. Both ‘Splendour’ and ‘Scired’ are susceptible to apple scab. The screening for scab was performed in spring (September–October) by inoculating the seedlings with V. inaequalis at the three to five true-leaf stage in the glasshouse under optimal conditions for scab infection (Gardiner et al. 1996). The F1 family was inoculated in 1999 with conidia from a mixture of isolates used in the breeding programme (‘breeding mix’), while in 2001, the F2 families were divided into two, when 105 and 71 seedlings of the AK617 and AK653 families, respectively, were inoculated with the breeding mix and the remainder with single-spore isolate J222, an isolate collected in 1996 from a leaf of an unknown cultivar at the Plant and Food Research orchard at Nelson (Bus et al. 2000b, 2005b). In the third week after inoculation, the seedlings were scored into classes using the scale according to Chevalier et al. (1991). Leaf samples for microscopic observations were collected, cleared in a chloral hydrate solution (Bruzzese and Hasan 1983) and mounted in an arabic gum solution (Cunningham 1972). The leaf sections were examined under brightfield conditions, as well as for autofluorescence of the resistance reactions in the interference blue range (excitation filter 450–490 nm, dichroic mirror 505 nm and barrier filter 515 nm) on a Nikon Optiphot microscope equipped with epi-fluorescence (Nikon, Tokyo, Japan). Digital images were taken with a CoolSnap camera (Coherent Scientific, Adelaide, Australia) and digitally adjusted for contrast in Photoshop version 6.2 (Adobe, San Jose, CA, USA). The glasshouse assessment was followed by field evaluation of the seedlings for scab resistance over 2 years when the trees were in their fourth and fifth leaf (seasons 2004/2005 and 2005/2006, respectively). The seedlings were scored in late spring/early summer on a six-point scale, where 0 = no symptoms; 1 = occasional lesions on less than 5% of the leaves; 2 = lesions on 5–15% of the leaves; 3 = lesions on 16–30% of the leaves; 4 = lesions on 31–50% of the leaves; to 5 = lesions on more than 50% of the leaves.
Accession MIS OP 93.051 G02-054, erroneously identified as MIS OP 93.051 G7-062 by Gardiner et al. (2007), was used to study the genetics of the resistance to woolly apple aphid (Er4) and powdery mildew (Pl-m). Approximately half of the seedlings from a ‘Fuji’ × MIS OP 93.051 G02-054 cross made in 1997 were evaluated for resistance to woolly apple aphid (153 seedlings) and the other half for resistance to powdery mildew (176 seedlings). ‘Fuji’ is susceptible to both powdery mildew and woolly apple aphid. Woolly aphid screening was performed in the 1998/1999 season by repeatedly inoculating the 4-month-old seedlings with infested shoot pieces in the shadehouse (Bus et al. 2008). After about 3 months, the seedlings were scored on a six-point scale according to Bus et al. (2008), where 0 = no aphids or galls and 5 = large aphid colony and more than 10 galls. The seedlings in the mildew resistance study were screened in the field without fungicide applications, to allow the uninhibited development of disease. The seedlings were scored on a six-point scale, where 0 = no symptoms; 1 = occasional small to moderate lesions on leaves; 2 = large sporulating areas on leaves and/or occasional infected shoot tip; 3 = up to 20% of shoot tips infected; 4 = 20–50% of shoot tips infected; and 5 = more than 50% of the shoots and leaves infected in the 2000/2001 and 2001/2002 seasons, when the trees were in their third and fourth leaf, respectively.
DNA isolation, amplification and genetic marker analysis
Genomic DNA was isolated from young immature leaves using the method described by Gardiner et al. (1996). Rvi16 was mapped in the ‘Scired’ × AK617 family, while both Er4 and Pl-m were mapped in the divided ‘Fuji’ × MIS OP 93.051 G02-054 family. Bulked segregant analysis (BSA) using randomly amplified polymorphic DNA (RAPDs) was used to develop markers linked to the three resistance genes. DNA bulks made of phenotypic extremes, two for resistant and two for susceptible individuals, were screened using 240, 200 and 200 Operon primers for Rvi16, Er4 and Pl-m, respectively. RAPD band amplification fragments co-segregating with the resistance were converted into sequence characterised amplified region (SCAR) markers as described by Bus et al. (2008). The SCAR markers developed were checked for co-segregation with the phenotype and then mapped when possible in a reference map developed from a ‘Malling 9’ × ‘Robusta 5’ (M.9 × R5) cross (Celton et al. 2009), in order to assign each resistance gene to a linkage group. Then simple sequence repeat (SSR) markers from the corresponding linkage group, as well as the SCAR markers, were screened over each whole segregating population to construct genetic maps.
Markers developed by Plant and Food Research are those beginning with NZms for the SSR markers, and NZsc for the SCAR markers (Table 1), some of which have been reported previously: NZmsCN943818 (LG3), NZmsDR033893 (LG11) and NZmsPal8 (Celton et al. 2009), and NZscAC20, NZscAY17/AB16 and NZscN18 (Gardiner et al. 2003). Other published SSR markers used were CH02d12 (LG11) and CH04e05 (LG7) (Liebhard et al. 2002); NB109a and NH030a (LG3) (Yamamoto et al. 2002); EMPc111a (LG7) (Fernández-Fernández et al. 2006); Hi02c06 (LG11), Hi05b09 (LG7), and Hi07d12b (LG7) (Silfverberg-Dilworth et al. 2006); and CH-Sd1 (Khan et al. 2007).
The polymerase chain reaction conditions for the SCAR and SSR markers were carried out as described by Bus et al. (2008). The mapping strategy used was that of the double pseudo testcross (Grattapaglia and Sederoff 1994). Linkage analysis and genetic map construction were performed using JoinMap v3.0 software (http://www.kyazma.nl) with the Kosambi mapping function and the critical logarithm of the odds score for marker grouping set at 5. After an initial mapping round, the genotype data was examined for the presence of double recombinants that possibly corresponded to genotype-phenotype incongruent (GPI) individuals (Gygax et al. 2004). Double recombinants were removed from the analysis to generate the final maps.
Genetic markers linked to each of the resistance genes were then evaluated on DNA from MIS (Germplasm Resources Information Network (GRIN) accession PI 589818; GMAL 2451) itself to determine the origin of the alleles.
Results
Genetics and symptoms of resistance
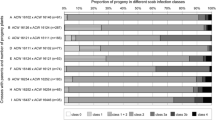
The phenotypic data for the families showed strong bimodal segregations for all three resistance genes. The ‘Splendour’ × MIS OP 93.051 G07-098 family inoculated with the V. inaequalis breeding mix in the glasshouse segregated into three main phenotypic groups: hypersensitive response (HR; disease classes 0 and 1), chlorotic resistance reaction (Chl; class 3A) and susceptibility (S; classes 3B and 4), and a minor group: (stellate) necrosis (class 2) (Table 2). The distinct phenotypic resistance classes, combined with resistance to susceptible (R:S) segregation ratios that were skewed towards resistance, suggested the presence of two scab resistance genes in accession MIS OP 93.051 G07-098. Therefore, two progeny from this family, AK617 from the HR class and AK653 from class 3A, were crossed with the same susceptible parent, ‘Scired’. However, the F2 families showed very similar segregation ratios depending on the inoculum applied. They segregated into the same three major classes as the F1 family when inoculated with the isolate mixture (Table 2). In contrast, both F2 families inoculated with the single isolate J222 showed very strong bimodal distributions into HR and S classes that did not differ significantly from R:S = 1:1 ratios expected from a major gene for the AK617 family, but not for the AK653 family (Table 2). Microscopic observations of the resistance symptoms in the glasshouse phenotyping confirmed the high scab resistance conditioned to isolate J222 by the Rvi16 gene. Many conidia on the class 0 seedlings germinated and produced appressoria but were unable to establish infections (Fig. 1a), while the development of other spores that did achieve infection were rapidly restricted. The HR responses varied from macroscopically invisible single cell (Fig. 1b) to multiple cell (Fig. 1d) necrosis of the epidermis, which involved the production of autofluorescent compounds in the resistance reaction (Fig. 1c, e) that was distinctly independent of the background autofluorescence from the vascular bundles and cell walls of the leaves. The breeding mix induced resistance reactions in the form of stellate necrosis (Fig. 1f) as well as multiple cell HR (Fig. 1g, h), sometimes combined with fungal stroma development showing limited sporulation that was not necessarily associated with autofluorescence of the underlying leaf tissues (Fig. 1i, j). The fungal stroma itself also showed autofluorescence in incompatible interactions as an indication of stress (Fig. 1f, h), but not in what appears to have been a compatible interaction on a class 3A seedling (Fig. 1j). All the resistance reactions were very superficial as they were only expressed in the epidermis and rarely involved the palisade mesophyll (infection on the adaxial side of the leaf) or the spongy mesophyll (infection on the abaxial side of the leaf). The phenotypic scores from the scab screening in the glasshouse were largely confirmed in the field, although a considerable number of the HR seedlings showed low levels of sporulation and a few seedlings could be considered susceptible since they showed higher levels of sporulation (Table 3). However, the correlation between the glasshouse and field scores was low for the seedlings in the intermediate resistance classes 2 to 3B for both the isolate mixture and isolate J222 from the glasshouse screen (Table 3). Therefore, a preliminary map around the scab resistance gene was developed first involving only the HR (classes 0 and 1) and susceptible (class 4) seedlings before the final mapping of the gene based on all seedlings for which marker data were generated.
Microscopic observations of scab resistance reactions on leaves of ‘Scired’ × AK617 progeny. a–c Arrested development of Venturia inaequalis isolate J222 on seedling BD228, which was assigned to class 0 (no visible symptoms) in the glasshouse screen. a Brightfield observation of germinated conidia that have developed appressoria but have not invoked a resistance reaction in the host. b, c Brightfield (b) and interference blue autofluorescence (c) observations of a hypersensitive response (HR) involving two epidermal cells on seedling BD228. d, e Brightfield (d) and interference blue autofluorescence (e) observations of an HR involving multiple epidermal cells on seedling BD266 that was inoculated with a mixture of V. inaequalis isolates. f Interference blue autofluorescence observation of a stellate necrotic (SN) reaction on seedling BD175 inoculated with a mixture of isolates. g–j Differential interactions of the isolate mixture on seedling BD280, which was assigned to classed 3A according to Chevalier et al. (1991). Brightfield (g) and interference blue autofluorescence (h) observations of an HR/SN reaction confirm the presence of a resistance gene in BD280. However, the limited sporulation that is visible in the brightfield (i) observation is not associated with a resistance reaction in the host as it does not show autofluorescence (j). (a = appressorium, c = conidium, m = mycelium, e = necrotic epidermis)
The major gene nature of both the powdery mildew and woolly apple aphid resistance genes were confirmed in the MIS OP 93.051 G02-054 family. The mildew sub-family segregated into 89 resistant (class 0) and 87 susceptible (classes 1–5) seedlings (P(χ 2 > 0.02) = 0.89) and the woolly aphid sub-family into 80 resistant (class 0) and 69 susceptible (classes 1–5) seedlings (P(χ 2 > 1.47) = 0.23).
Resistance gene mapping
Apple scab resistance gene Rvi16
BSA resulted in the identification of six RAPD markers (OPAJ12/1,400, OPAJ18/500, OPAN01/1,800, OPAS07/700, OPAX6/1,400 and OPAX18/800 bp) linked to apple scab resistance. Two of these (NZscAS07 and NZscAJ12) were successfully converted into SCARs, and a local map around the gene was developed, showing that NZscAJ12 was located closest to the gene. Since NZscAN01 had previously been mapped to linkage group 3 (LG3) of ‘Robusta 5’ (Peil et al. 2008), Rvi16 was assumed to be located on this LG. Therefore, SSRs from LG3 were screened over the population and the influence of the inoculum on the phenotypes of the surviving seedlings in the field, which by then was skewed towards resistance as many susceptible seedlings had died, and the mapping of the scab resistance gene was investigated further. We found that a relative large proportion of the seedlings that were susceptible to the breeding mix in the glasshouse actually exhibited the markers NZmsCN943818 and NZscAS07 that had been mapped close to the gene (Table 4). Most of these seedlings indeed were resistant in the field, which suggested that the field evaluation provided a better assessment of scab resistance than the glasshouse screen. An example of this was seedling BD280 (Fig. 1g–j), which was classed 3A in the glasshouse and therefore assumed susceptible in the preliminary assessment (see above) but exhibited both markers, while showing partial resistance in the field. The correlation between the glasshouse screen with V. inaequalis isolate J222 and the field screen was much higher than with the breeding mix (Table 4), hence the sub-population of ‘Scired’ × AK617 screened with J222 was used to map the resistance. Local genetic maps of Rvi16 based on the glasshouse and field evaluations, respectively, were constructed using four SSRs and two SCARs. As the map positions of each of the markers and the gene did not differ more than 1 cM between both maps, only the map based on the glasshouse phenotyping is presented. Rvi16 maps at a distance of about 5 cM below NZmsCN943818 towards the lower end of LG3 (Fig. 2). The alleles of this marker and SSR marker NH030a, which maps with NB109a to the same region on LG3 (Celton et al. 2009) just above the gene (Fig. 2), linked to the Rvi16 resistance gene were amplified with the DNA from the original MIS accession (Table 5).
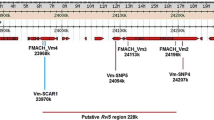
Genetic maps of the apple scab resistance gene Rvi16, the woolly apple aphid resistance gene Er4 and the powdery mildew resistance gene Pl-m identified in accessions derived from open-pollinated ‘Mildew Immune Selection’. The underlined microsatellite markers were used to trace the origins of the three resistance genes (see Table 5)
Woolly apple aphid resistance gene Er4
BSA resulted in the identification of a RAPD band obtained from the OPA04 primer co-segregating with Er4. The band was converted into a SCAR marker (NZscA04F3R3), which was mapped to LG7 in the M.9 × R5 map. More framework SSRs from this LG were then screened over the whole population, and the position of Er4 was located between the SSR markers Hi07d12a and CH04e05, at about a third down the linkage group, after seven potential GPIs were removed. The alleles of the markers CH-Sd1 (Khan et al. 2007), which mapped 17.9 cM above the gene (Fig. 2) and CH04e05 linked to Er4, were only present in the MIS OP 93.051 G02-054 accession and not in MIS itself (Table 5).
Powdery mildew resistance gene Pl-m
A 1,800-bp product of RAPD marker OPAC20 was found to be distantly (14 cM) linked to Pl-m. It was converted into a SCAR marker that mapped close to Pl-m. However, it did not segregate in the M.9 × R5 reference population. A set of SCARs previously found to be linked to other powdery mildew resistance genes was tested, and five SCARS linked to Pl2 were also linked to Pl-m (Gardiner et al. 2003). As Pl2 had been assigned to LG11 (Liebhard et al. 2002) with the aid of a marker for a major mildew resistance quantitative trait locus in A679-2 (Seglias and Gessler 1997), SSRs from this group were screened in the MIS population to confirm the linkage. The final genetic map of LG11 was constructed using five SSRs and six SCARs to locate Pl-m after seven potential GPIs were removed (Fig. 2). With the gene mapping between NZscAC20 and NZscDR033888 in the upper half of LG11, the linkage of Pl-m with Pl2 was confirmed (Gardiner et al. 2007). The marker allele of CH02d12 linked to Pl-m was amplified in both MIS OP 93.051 G02-054 and MIS itself (Table 5).
Discussion
Apple scab resistance gene Rvi16
The new major gene for scab resistance, Rvi16, which previously was reported under its ‘working name’ Vmis (Gardiner et al. 2007), was mapped towards the lower end of LG3, at some distance below the region where a scab QTL from ‘Fiesta’ × ‘Discovery’ was located (Durel et al. 2004). Its independent segregation from other scab resistance genes and its somewhat distinctive resistance symptoms indicate that the gene confers a different resistance mechanism than the other genes and QTLs. The gene also maps below the region where a major QTL for fire blight resistance from R5 was mapped recently (Peil et al. 2007, 2008). Interestingly, a large effect pear scab resistance QTL co-locates with Rvi16 on LG3 of ‘Abbé Fétel’ (Pierantoni et al. 2007), suggesting the existence of potential orthologous scab resistance genes in the highly collinear apple and pear genomes (Yamamoto et al. 2004). Also, the linkage of Rvi16 with marker NH030a suggests that it maps to a region that is homeologous to a region either at the lower end of LG11 or at the top of LG1 through its linkage with marker CH03g12. Neither region is known to carry genes or QTLs for scab resistance.
While there is a strong suggestion that the gene originated from MIS itself, it is possible that the unknown pollen parent may have contributed the gene, and further research will be required to resolve this question. Rvi16 is named according to the recently proposed nomenclature system for gene-for-gene relationships in the V. inaequalis–Malus pathosystem, based on the fact that it segregates independently of all the other scab resistance genes named to date (Bus et al. 2009).
While the gene conditions an HR in the host to certain isolates, such as J222 in our experiment, the gene conditions partial resistance under different disease pressures and/or environmental conditions involving different pathotypes of the pathogen. This was best demonstrated by the very similar segregation ratios of the F2 families derived from accessions AK617 and AK653, in spite of these resistant seedlings having been classed as 1 and 3A, respectively, after inoculation of the ‘Splendour’ × MIS OP 93.051 G07-098 family with the breeding mix. Some seedlings also sustained low levels of infection in the orchard, but the field screening, together with the glasshouse screening with a single isolate, strongly suggested that only one major gene is involved in the scab resistance of MIS OP and allowed the gene to be mapped more precisely. Previous findings that multiple assessments of accessions carrying the Rvi6 scab resistance gene in the glasshouse and the field increased the reliability of linkage mapping (King et al. 1998) were similar to ours with Rvi16. However, the absence of autofluorescence in one of the differential interactions on the leaves of seedling BD280 showing some sporulation strongly suggests that a race exists in the New Zealand V. inaequalis population that can overcome the gene. Further studies will be performed to investigate this.
The microscopic observations of the resistance reactions showed that they were very superficial as they only affected the epidermis of the leaves. Also, fewer cells were involved in the resistance reactions compared to the HR conditioned by the Rvi5 (Vm) and Rvi4 (Vh4), and the SN conditioned by the Rvi2 (Vh2) and Rvi8 (Vh8) scab resistance genes, which usually involve the palisade mesophyll (Win et al. 2003; Bus et al. 2005a; Bus 2006a). The resistance reactions on hosts carrying the Rvi16 gene therefore have a much more subtle structure and are less intrusive on the leaves than the reactions conditioned by the other genes.
Woolly apple aphid resistance gene Er4
The woolly apple aphid resistance gene Er4 from accession MIS OP 93.051 02-054 whose ‘working name’ was Er-m (Gardiner et al. 2007) mapped to LG7. It segregates independently from the three woolly aphid resistance genes that were recently mapped to LG8 and LG17 (Bus et al. 2008) and therefore is expected to confer a different mechanism of resistance than these genes. The name Er4 follows on from the naming of the Er1, Er2 and Er3 genes. Since the marker alleles linked to the gene in MIS o.p. 93.051 G02-054 of neither CH-Sd1 nor CH04e05 flanking the gene were shared with MIS itself, the gene appears to be derived from an unknown pollen parent. The Er4 gene maps to the upper region of LG7, about 21 cM below the linked Sd-1 and Sd-2 genes for resistance to the rosy leaf-curling aphid (Dysaphis devecta; Cevik and King 2002a, b), which originally were identified in ‘Cox's Orange Pippin’ and ‘Northern Spy’, respectively (Alston and Briggs 1968, 1977). The genes therefore map too far apart to be part of the same resistance gene cluster and to have similar functionalities. The only other resistances mapped to LG7 to date are a minor QTL for leaf scab resistance (Liebhard et al. 2003) and a QTL for fire blight resistance from ‘Fiesta’ (Calenge et al. 2005; Khan et al. 2007). Both are located well below Er4 towards the bottom end of this linkage group.
Powdery mildew resistance gene Pl-m
The Pl-m gene for mildew resistance present in MIS OP 93.051 G02-54 originated, as expected, from MIS and was mapped to LG11. As its name indicates, MIS was originally recognised as a potential source for mildew resistance in apple breeding but has not been used much since the accession was found to be infected by mildew in 1978 (Korban and Dayton 1983). In a glasshouse experiment, it was shown that MIS was strongly infected by mildew inoculum from Illinois and to a lesser extent by inoculum from Pennsylvania, but not other inocula (Lespinasse 1983). The differential interactions strongly suggested that a race of P. leucotricha had developed; however, MIS was not infected in the field in the years following from 1978 (Korban and Dayton 1983). As there are no later reports of the Pl-m gene having been overcome, the gene still has value for resistance breeding, provided it is pyramided with other mildew resistance genes to achieve durable resistance. While it was demonstrated previously that the Pl-m and Pl2 genes are linked (Gardiner et al. 2003) and might be allelic, they however are different genes or alleles as hosts carrying the Pl-m gene are resistant to a P. leucotricha race overcoming the Pl2 gene in New Zealand (Wood and Bus, unpublished data). The gene maps to the same region of LG11 where a scab QTL identified in ‘Fiesta’ has been repeatedly mapped (Durel et al. 2003, 2004; Liebhard et al. 2003) as well as one from ‘Gala’ (Soufflet-Freslon et al. 2008). Although a number of QTLs for powdery mildew resistance have been identified in ‘Discovery’ × TN10-8 family, none of them mapped to LG11 (Calenge and Durel 2006).
Conclusion
In this paper, we have described the identification and mapping of an apple scab, a powdery mildew and a woolly apple aphid resistance gene in two OP derivatives of MIS. The genes will be very useful in the development of new apple cultivars with gene pyramids for multiple, durable resistances. Back-cross lines have been developed to introgress these genes into new apple cultivars with pyramided resistances, with the aid of MAS using the (flanking) markers identified in this study.
References
Aderhold R (1902) Ein Beitrag zur Frage der Empfänglichkeit der Apfelsorten für Fusicladium dendriticum (Wallr.) Fuck. und derem Beziehungen zum Wetter. Arbeiten Biol Abt für Land- und Forstwirtschaft 2:560–566
Alspach PA, Bus VGM (1999) Spatial variation of woolly apple aphid (Eriosoma lanigerum, Hausmann) in a genetically diverse apple planting. NZ J Ecol 23(1):39–44
Alston FH (1977) Practical aspects of breeding for mildew (Podospaera leucotricha) resistance in apples. Proc Eucarpia Fruit Section Symp VII, Top Fruit Breeding, Wageningen, 1976 pp 4–13
Alston FH, Briggs JB (1968) Resistance to Sappaphis devecta (Wlk) in apple. Euphytica 17:468–472
Alston FH, Briggs JB (1977) Resistance genes in apples and biotypes of Dysaphis devecta. Ann appl Biol 87:75–81
Austin P, Norling C, Volz R, Bus V, Gardiner S (2006) Using controlled environments to accelerate flowering of Malus seedlings. Third Rosaceae Genomics Conference, Napier, New Zealand, 19–22 March 2006
Bradley SJ, Murrell VC, Shaw PW, Walker JTS (1997) Effect of orchard pesticides on Aphelinus mali, the woolly apple aphid parasitoid. Proc 50th NZ Plant Protection Conf, pp 218–222
Bruzzese E, Hasan S (1983) A whole leaf clearing and staining technique for host specificity studies of rust fungi. Plant Pathol 32:335–338
Bus V, Bradley S, Hofstee M, Alspach P, Brewer L, Luby J (2000a) Increasing genetic diversity in apple breeding to improve the durability of pest and disease resistance. Acta Hort 538:185–190
Bus V, Plummer K, Rikkerink E, Luby J (2000b) Evaluation of the pathogenicity of two scab isolates derived from the Vf-resistant apple cultivar ‘Baujade’. IOBC WPRS Bull 23(12):231–237
Bus V, Rikkerink E, Aldwinckle HS, Caffier V, Durel C-E, Gardiner S, Gessler C, Groenwold R, Laurens F, Le Cam B, Luby J, Meulenbroek B, Kellerhals M, Parisi L, Patocchi A, Plummer K, Schouten HJ, Tartarini S, Van de Weg E (2009) A proposal for the nomenclature of Venturia inaequalis races. Acta Hort 814:739–746
Bus VGM (2006a) Differential host-pathogen interactions of Venturia inaequalis and Malus. Unpublished thesis. University of Auckland, Auckland
Bus VGM (2006b) Gene-for-gene relationships and durable resistance to apple scab. In: Mercer CF (ed) Proc 13th Australasian Plant Breeding Conf, Breeding for Success: diversity in action, Christchurch, pp 1159–1169
Bus VGM, Alspach PA, Hofstee ME, Brewer LR (2002) Genetic variability and preliminary heritability estimates of resistance to scab (Venturia inaequalis) in an apple genetics population. NZ J Crop Hort Sci 30:83–92
Bus VGM, Laurens FND, van de Weg WE, Rusholme RL, Rikkerink EHA, Gardiner SE, Bassett HCM, Kodde LP, Plummer KM (2005a) The Vh8 locus of a new gene-for-gene interaction between Venturia inaequalis and the wild apple Malus sieversii is closely linked to the Vh2 locus in Malus pumila R12740-7A. New Phytol 166:1035–1049
Bus VGM, Rikkerink EHA, Van de Weg WE, Rusholme RL, Gardiner SE, Bassett HCM, Kodde LP, Parisi L, Laurens F, Meulenbroek EJ, Plummer K (2005b) The Vh2 and Vh4 scab resistance genes in two differential hosts derived from Russian apple R12740-7A map to the same linkage group of apple. Molec Breed 15:103–116
Bus VGM, Chagné D, Bassett HCM, Bowatte D, Calenge F, Celton J-M, Durel C-E, Malone MT, Patocchi A, Ranatunga AC, Rikkerink EHA, Tustin DS, Zhou J, Gardiner SE (2008) Genome mapping of three major resistance genes to woolly aphid (Eriosoma lanigerum Hausm.). Tree Genet Genomes 4:223–236
Caffier V, Laurens F (2005) Breakdown of Pl2, a major gene of resistance to apple powdery mildew, in a French experimental orchard. Plant Pathol 54:116–124
Caffier V, Parisi L (2007) Development of apple powdery mildew on sources of resistance to Podosphaera leucotricha, exposed to an inoculum virulent against resistance gene Pl-2. Plant Breed 126:319–322
Calenge F, Durel CE (2006) Both stable and unstable QTLs for resistance to powdery mildew are detected in apple after four years of field assessments. Molec Breed 17:329–339
Calenge F, Drouet D, Denancé C, Van de Weg WE, Brisset MN, Paulin JP, Durel CE (2005) Identification of a major QTL together with several minor additive or epistatic QTLs for resistance to fire blight in apple in two related progenies. Theor Appl Genet 111:128–135
Celton J-M, Tustin DS, Chagné D, Gardiner SE (2009) Construction of a dense genetic linkage map for apple rootstocks using SSRs developed from Malus ESTs and Pyrus genomic sequences. Tree Genet Genomes 5:93–107
Cevik V, King GJ (2002a) High-resolution genetic analysis of the Sd-1 aphid resistance locus in Malus spp. Theor Appl Genet 105:346–354
Cevik V, King GJ (2002b) Resolving the aphid resistance locus Sd-1 on a BAC contig within a sub-telomeric region of Malus linkage group 7. Genome 45:939–945
Chevalier M, Lespinasse Y, Renaudin S (1991) A microscopic study of the different classes of symptoms coded by the Vf gene in apple for resistance to scab (Venturia inaequalis). Phytopathology 40:249–256
Crane MB (1937) Breeding immune rootsocks. Ann appl Biol 24:188–195
Crosby JA, Janick J, Pecknold PC, Korban SS, O'Connor PA, Ries SM, Goffreda J, Voordeckers A (1992) Breeding apples for scab resistance: 1945-1990. Fruit Var J 46:145–166
Cunningham JL (1972) A miracle mounting fluid for permanent whole-mounts of microfungi. Mycologia 64:906–911
Dayton DF (1977) Genetic immunity to apple mildew incited by Podosphaera leucotricha. HortScience 12:225–226
Durel CE, Parisi L, Laurens F, Van de Weg WE, Liebhard R, Jourjon MF (2003) Genetic dissection of partial resistance to race 6 of Venturia inaequalis in apple. Genome 46:224–234
Durel CE, Calenge F, Parisi L, van de Weg WE, Kodde LP, Liebhard R, Gessler C, Thiermann M, Dunemann F, Gennari F, Tartarini S, Lespinasse Y (2004) An overview of the position and robustness of scab resistance QTLs and major genes by aligning genetic mapsof five apple progenies. Acta Hort 663:135–140
Fernández-Fernández F, Harvey NG, James CM (2006) Isolation and characterization of polymorphic microsatellite markers from European pear (Pyrus communis L.). Molec Ecol Notes 6:1039–1041
Gardiner SE, Bassett HCM, Noiton DAM, Bus VG, Hofstee ME, White AG, Ball RD, Forster RLS, Rikkerink EHA (1996) A detailed linkage map around an apple scab resistance gene demonstrates that two disease resistance classes both carry the Vf gene. Theor Appl Genet 93:485–493
Gardiner S, Murdoch J, Meech S, Rusholme R, Bassett H, Cook M, Bus V, Rikkerink E, Gleave A, Crowhurst R, Ross G, Warrington I (2003) Candidate resistance genes from an EST database prove a rich source of markers for major genes conferring resistance to important apple pests and diseases. Acta Hort 622:141–151
Gardiner SE, Bus VGM, Rusholme RL, Chagné D, Rikkerink EHA (2007) Apple. In: Kole C (ed) Genome mapping and molecular breeding in plants, vol 4, Fruits and Nuts. Springer, Heidelberg, pp 1–62
Gessler C, Patocchi A, Sansavini S, Tartarini S, Gianfranceschi L (2006) Venturia inaequalis resistance in apple. Critical Rev Plant Sci 25:473–503
Giliomee JH, Strydom DK, van Zyl HJ (1968) Northern Spy, Merton and Malling-Merton rootstocks susceptible to woolly aphid, Eriosoma lanigerum, in the Western Cape. Sth Afr J Agric Sci 11:183–186
Grattapaglia D, Sederoff R (1994) Genetic linkage maps of Eucalyptus grandis and Eucalyptus urophylla using a pseudo-testcross: mapping strategy and RAPD markers. Genetics 137:1121–1137
Gygax M, Gianfranceschi L, Liebhard R, Kellerhals M, Gessler C, Patocchi A (2004) Molecular markers linked to the apple scab resistance gene Vbj derived from Malus baccata jackii. Theor Appl Genet 109:1702–1709
Hatton RG (1937) Introduction to the workshop “The problems raised by the woolly aphis of the apple—a case for team research”. Ann appl Biol 24:169–173
Khan MA, Durel C-E, Duffy B, Drouet D, Kellerhals M, Gessler C, Patocchi A (2007) Development of molecular markers linked to the ‘Fiesta’ linkage group 7 major QTL for fire blight resistance and their application for marker-assisted selection. Genome 50:568–577
King GJ, Alston FH, Brown LM, Chevreau E, Evans KM, Dunemann F, Janse J, Laurens F, Lynn JR, Maliepaard C, Manganaris AG, Roche P, Schmidt H, Tartarini S, Verhaegh J, Vrielink R (1998) Multiple field and glasshouse assessments increase the reliability of linkage mapping of the Vf source of scab rsistance in apple. Theor Appl Genet 96:699–708
Korban SS, Dayton DF (1983) Evaluation of Malus germplasm for resistance to powdery mildew. HortScience 18:219–222
Laurens F (1999) Review of the current apple breeding programmes in the world: objectives for scion cultivar improvement. Acta Hort 484:163–170
Lespinasse Y (1983) Amélioration du pommier pour la résistance a l'oidium (Podosphaera leucotricha). Premiers résultats concernant la virulence du champignon. IOBC WPRS Bull 6(4):96–110
Liebhard R, Gianfranceschi L, Koller B, Ryder CD, Tarchini R, Van de Weg WE, Gessler C (2002) Development and characterisation of 140 new microsattellites in apple (Malus x domestica Borkh.). Mol Breed 10:217–241
Liebhard R, Koller B, Patocchi A, Kellerhals M, Pfammatter W, Jermini M, Gessler C (2003) Mapping quantitative field resistance against apple scab in a ‘Fiesta’ x ‘Discovery’ progeny. Phytopathology 93:493–501
Luby JJ, Alspach PA, Bus VGM, Oraguzie NC (2002) Field resistance to fire blight in a diverse apple (Malus sp.) germplasm collection. J Am Soc Hort Sci 127:245–253
Noiton D, Shelbourne CJA (1992) Quantitative genetics in an apple breeding strategy. Euphytica 60:213–219
Parisi L, Lespinasse Y, Guillaumes J, Kruger J (1993) A new race of Venturia inaequalis virulent to apples with resistance due to Vf gene. Phytopathology 83:533–537
Peil A, Garcia T, Richter K, Trognitz FC, Trognitz B, Hanke M-V, Flachowsky H (2007) Strong evidence for a fire blight resistance gene of Malus robusta located on linkage group 3 detected by rapid genome scanning. Plant Breed 126:470–475
Peil A, Hanke M-V, Flachowsky H, Richter K, Garcia-Libreros T, Celton J-M, Gardiner S, Horner M, Bus V (2008) Confirmation of the fire blight QTL of Malus x robusta 5 on linkage group 3. Acta Hort 793:297–303
Pierantoni L, Dondini L, Cho KH, Shin IS, Gennari F, Chiodini R, Tartarini S, Kang SJ, Sanasavini S (2007) Pear scab resistance QTLs via a European pear (Pyrus communis) map. Tree Genet Genomes 3:311–317
Rock GC, Zeiger DC (1974) Woolly apple aphid infests Malling and Malling-Merton rootstocks in propagation beds in North Carolina. J Econ Entomol 67:137–138
Sandanayaka WRM, Bus VGM, Connolly P, Newcomb R (2003) Characteristics associated with woolly apple aphid, Eriosoma lanigerum, resistance of three apple rootstocks. Entomol Exp Appl 109:63–72
Seglias NP, Gessler C (1997) Genetics of apple powdery mildew resistance from Malus zumi (Pl2). IOBC WPRS Bull 20(9):195–208
Sen Gupta GC, Miles PW (1975) Studies on the susceptibility of varieties of apple to the feeding of two strains of woolly aphis (Homoptera) in relation to the chemical content of the tissues of the host. Aus J Agr Res 26:157–168
Shaw PW, Walker JTS (1996) Biological control of woolly apple aphid by Aphelinus mali in an integrated fruit production programme in Nelson. Proc 49th NZ Plant Protection Conf, pp 59–63
Shay JR, Williams EB (1956) Identification of three physiologic races of Venturia inaequalis. Phytopathology 46:190–193
Silfverberg-Dilworth E, Matasci CL, Van de Weg WE, Van Kaauwen MPW, Walser M, Kodde LP, Soglio V, Gianfranceschi L, Durel CE, Costa F, Yamamoto T, Koller B, Gessler C, Patocchi A (2006) Microsatellite markers spanning the apple (Malus x domestica Borkh.) genome. Tree Genet Genomes 2:202–224
Soufflet-Freslon V, Gianfranceschi L, Patocchi A, Durel C-E (2008) Inheritance studies of apple scab resistance and identification of Rvi14, a new major gene that acts together with other broad-spectrum QTL. Genome 51:657–667
Volz R, Rikkerink E, Austin P, Lawrence T, De Silva N, Bus V (2009) “Fast-breeding” in apple: a strategy to accelerate introgression of new traits into elite germplasm. Acta Hort 814:163–168
Way RD, Aldwinckle HS, Lamb RC, Rejman A, Sansavini S, Shen T, Watkins R, Westwood MN, Yoshida Y (1989) Apples (Malus). Acta Hort 290:1–62
Williams EB, Kuć J (1969) Resistance in Malus to Venturia inaequalis. Ann Rev Phytopathol 7:223–246
Win J, Greenwood DR, Plummer KM (2003) Characterisation of a protein from Venturia inaequalis that induces necrosis in Malus carrying the Vm resistance gene. Physiol Molec Plant Pathol 62:193–202
Yamamoto T, Kimura T, Shoda M, Imai T, Saito T, Sawamura Y, Kotobuki K, Hayashi T, Matsuta N (2002) Genetic linkage maps constructed by using an interspecific cross between Japanese and European pears. Theor Appl Genet 106:9–18
Yamamoto T, Kimura T, Saito T, Kotobuki K, Matsuta N, Liebhard R, Gessler C, Van de Weg WE, Hayashii T (2004) Genetic linkage maps of Japanese and European pears aligned to the apple consensus map. Acta Hort 663:51–56
Acknowledgements
We thank Sarah Meech and Ananda Anandan for their technical assistance in the genetic marker research of the project. We thank Dawn Dellefave, Amy Szewc-Mcfadden, Bill Smrack and Chuck Simon from the Plant Genetic Resource Unit of USDA-ARS, Geneva, NY, USA for supplying the leaves of MIS. This research was funded by the New Zealand Foundation for Research, Science and Technology programmes Pipfruit Breeding Consortium (PREV0401) and Genetic Approaches to Sustainable Agricultural Production (C02X0406).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by E. Dirlewanger
Rights and permissions
About this article
Cite this article
Bus, V.G.M., Bassett, H.C.M., Bowatte, D. et al. Genome mapping of an apple scab, a powdery mildew and a woolly apple aphid resistance gene from open-pollinated Mildew Immune Selection. Tree Genetics & Genomes 6, 477–487 (2010). https://doi.org/10.1007/s11295-009-0265-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-009-0265-2