Abstract
The rosy apple aphid (Dysaphis plantaginea), the leaf-curling aphid (Dysaphis cf. devecta) and the green apple aphid (Aphis pomi) are widespread pest insects that reduce growth of leaves, fruits and shoots in apple (Malus × domestica). Aphid control in apple orchards is generally achieved by insecticides, but alternative management options like growing resistant cultivars are needed for a more sustainable integrated pest management (IPM). A linkage map available for a segregating F1-cross of the apple cultivars ‘Fiesta’ and ‘Discovery’ was used to investigate the genetic basis of resistance to aphids. Aphid infestation and plant growth characteristics were repeatedly assessed for the same 160 apple genotypes in three different environments and 2 consecutive years. We identified amplified fragment length polymorphism (AFLP) markers linked to quantitative trait loci (QTLs) for resistance to D. plantaginea (‘Fiesta’ linkage group 17, locus 57.7, marker E33M35–0269; heritability: 28.3%), and to D. cf. devecta (‘Fiesta’ linkage group 7, locus 4.5, marker E32M39–0195; heritability: 50.2%). Interactions between aphid species, differences in climatic conditions and the spatial distribution of aphid infestation were identified as possible factors impeding the detection of QTLs. A pedigree analysis of simple sequence repeat (SSR) marker alleles closely associated with the QTL markers revealed the presence of the alleles in other apple cultivars with reported aphid resistance (‘Wagener’, ‘Cox’s Orange Pippin’), highlighting the genetic basis and also the potential for gene pyramiding of aphid resistance in apple. Finally, significant QTLs for shoot length and stem diameter were identified, while there was no relationship between aphid resistance and plant trait QTLs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Apple (Malus × domestica Borkh.) is the most relevant fruit crop in the temperate region, and production quantity has even increased by 27% between 1995 and 2006 (http://faostat.fao.org). Insects from different orders have a negative impact on the quantity and quality of fruit yield and require control (Beers et al. 2003). Development of sustainable insect pest management is particularly promising in perennial crops such as apple, as measures taken to reduce insect pest populations in one growing season are likely to also affect the next growing season (Dorn et al. 1999). Although quantitative trait loci (QTLs)-based approaches to host-plant resistance against insects are common in annual crops (Frei et al. 2005; Fujita et al. 2006), and are intensively studied in apple to suppress diseases (Cheng et al. 1998; James et al. 2004; Calenge and Durel 2006; Khan et al. 2007), they have received little attention so far in apple in connection with aphid suppression (Roche et al. 1997; Bus et al. 2008).
The rosy apple aphid (Dysaphis plantaginea Pass.) is one of the major insect pests on apple, causing stunting and malformation of leaves and fruits (Graf et al. 2006). The economic threshold level is extremely low, with 1% of infested flower buds in spring, because even light infestations produce unmarketable fruits (Blommers 1994). The leaf-curling aphids (Dysaphis cf. devecta Wlk.) belong to a species complex (Stekolshchikov and Lobanov 2004). They are pests of cultivated apple species and cause red-curled leaves leading to high economic damage through malformation of leaves and fruits and inhibited shoot growth (Cevik and King 2002). The green apple aphid (Aphis pomi De Geer), found in most areas where apple is cultivated, negatively affects the shoot and leaf of apple trees by sucking the phloem-sap (Arbab et al. 2006). Population outbursts can lead to serious economic losses as aphids feeding on immature and mature fruits produce deformed apples of reduced quality (e.g., russeted fruits or reduced sugar content) (Hamilton et al. 1986; Arbab et al. 2006).
Besides direct injuries to leaves, shoots, and fruits, aphids may act as vectors for virus transmission, while the release of honeydew fosters sooty mold infestation (Arbab et al. 2006). As a consequence, large amounts of insecticides are needed to manage aphid pests, for example, up to four applications of pirimicarb (250–750 g/ha) per season in the Swiss midlands (Maag 1996; Cevik and King 2002). For economic and environmental reasons, integrated pest management (IPM) programs have been developed (Zehnder et al. 2007). Potential tools within IPM programs in fruit orchards include natural enemies as biological control measures and orchard management. These tools have, however, in recent examinations been found to be of minor importance for the suppression of the mentioned aphid species (Miñarro et al. 2005; Omkar and Pervez 2005; Simon et al. 2006).
Breeding for resistant cultivars is a further possibility. Several authors have found variable susceptibility of different apple cultivars to aphids (Alston and Briggs 1977; Lespinasse et al. 1985; Graf et al. 1998; Habekuss et al. 2000; Qubbaj et al. 2005; Angeli and Simoni 2006). Based on these observations, a genetic basis of aphid resistance in apple was assumed. So far, only the Sd1 gene for D. cf. devecta resistance and the ER1, ER2, and ER3 genes for woolly apple aphid (Eriosoma lanigerum Hausm.) resistance were studied for application in apple breeding (Roche et al. 1997; Bus et al. 2008). The detection of resistance-breaking biotypes of D. plantaginea (Rat-Morris et al. 1999) emphasizes the need for a better understanding of the genetic background of aphid resistance, and application of new molecular tools such as QTL analysis could help.
The purpose of this study was to investigate resistance in apple to the aphid species, D. plantaginea, D. cf. devecta and A. pomi, using linkage map data available for a segregating F1-cross of the apple cultivars ‘Fiesta’ and ‘Discovery’ (Liebhard et al. 2003; Silfverberg-Dilworth et al. 2006). Besides detection of potential QTLs, knowledge on the D. cf. devecta resistance gene (Sd1) (Alston and Briggs 1977; Roche et al. 1997; Cevik and King 2002) should be improved by describing more markers (especially simple sequence repeat [SSR] markers) linked to this resistance gene. These could then be used for gene pyramiding in plant breeding. As plant growth characteristics could influence aphid infestations and erroneously be interpreted as QTLs for physiological resistance, the relationship of plant traits and aphid infestation was assessed by QTL analyses of plant growth characteristics. To account for environmental factors causing instable QTL effects at different study sites, interactions between the aphid species studied, the climatic conditions at the study sites, and the spatial variability of aphid infestations were examined.
Materials and methods
Orchard location and plant material
The resistance of apple trees to the three aphid species was studied in the field at three different study sites in Switzerland, situated in Zurich (Wadenswil; at 47°13′20″ N, 8°40′05″ E, 455 m altitude), Valais (Conthey; at 46°12′30″ N, 7°18′15″ E, 478 m altitude), and Ticino (Cadenazzo; at 46°09′35″ N, 8°56′00″ E, 203 m altitude) in 2005 and 2006 (year 1 and year 2). The climate from March to August was characterized for each site by monthly average temperature and the sum of rainfall obtained from MeteoSwiss (http://www.meteoschweiz.ch) (Table S1). Compared to the standard value (30-year average: 1960–1990), temperature in the study years was 1–2°C higher, and at the Ticino site the measured sum of rainfall was 60–70% of the standard values (Table S1). Orchards were treated with fertilizers and herbicides, but no insecticides and fungicides were applied.
The surveyed apple genotypes were previously described (Liebhard et al. 2003). They are a segregating F1-cross of the apple varieties ‘Fiesta’ (syn. ‘Red Pippin’) and ‘Discovery’. ‘Fiesta’ was named as a sort 1985 in England (HRI East Malling), as a cross of ‘Cox’s Orange Pippin’ and ‘Idared’, and only resistance properties to one aphid species (Dysaphis devecta) were yet reported (Roche et al. 1997). ‘Discovery’ is a cross of ‘Worcester Pearmain’ as mother sort and probably ‘Beauty of Bath’ as father sort. It was discovered around 1949 in England and is attributed a high, polygenic resistance to apple scab (Liebhard et al. 2003). The apple trees were tripled in summer 1998 by bud grafting on M27 rootstocks and planted in winter 1998/1999 at the three sites. The apple trees were planted in rows 3.50 m apart and with a tree-to-tree distance of 1.25 m at the Ticino site and of 0.50 m at the Valais and Zurich site. All tree genotypes that were present at all three sites were surveyed. Sample size occasionally differed from the maximum of 160 apple genotypes, as some trees died since plantation establishment, and as for specific analyses not all genotypes were considered (e.g., neighborhood effect).
Aphid survey
Aphid infestation was assessed on all shoots of each apple genotype. To quantify Dysaphis plantaginea infestation, the number of aphid colonies per tree was counted two times per year in both study years (sampling date: May and June). The number of red-curled leaves per tree was used as a measure of Dysaphis cf. devecta infestation (May, June, and July). The number of green apple aphids was counted three times (May, June and July) for each shoot to quantify infestation with Aphis pomi. In the second study year, a fourth A. pomi survey was carried out in August at the Valais site. In case of large A. pomi colonies, aphid number was estimated by reference counts. Reference counts were obtained by assessing the number of A. pomi aphids on a part of a shoot to obtain a reference for a shoot section hosting 50 aphids. Extrapolation of this information was then used to quantify the size of large A. pomi colonies.
Plant-growth characteristics
Stem diameter was measured 1.2 m above soil. For quantification of tree crown volume, the crown diameter at the widest part, the diameter perpendicular to the latter and the crown height were measured (crown volume = 4π/3 abc; a, b, c = the three sides of an ellipsoid). Tree height was determined as another plant trait, from the soil surface to the top (highest point) of the tree. The term ‘new shoots’ was used for the current year’s actively growing shoots. The total length of shoots (including new shoots) and the length of new shoots, separately, were measured and the sum and the number were calculated. Shoot length was assessed in May, new shoot length end of June. Shoots and new shoots that were very short (shoot length <20 cm; new shoot length <5 cm) were not considered.
QTL mapping
Quantitative trait loci (QTLs) analyses of mean aphid infestation per year and location (Krakowsky et al. 2004) were carried out with MapQTL® 4.0 (van Ooijen et al. 2002). The same software was used for QTL analysis of plant characteristics data (per year and location, one replication). The genetic linkage maps for both ‘Fiesta’ and ‘Discovery’ (single parent maps), used in QTL analysis, were calculated with 251 apple genotypes and were already published (Liebhard et al. 2003). The maps consist of a total of 345 (‘Fiesta’) and 389 (‘Discovery’) markers and include 137 amplified fragment length polymorphism (AFLP), 108 microsatellites (SSR), and 100 RAPD markers in ‘Fiesta’, and 160 AFLP, 103 SSR, one SCAR and 125 RAPD markers in ‘Discovery’. The average linkage group length was 66.96 cM for ‘Fiesta’ and 84.36 cM for ‘Discovery’. Kruskal–Wallis tests and interval mapping (IM) were used for QTL analysis. Logarithm of odds (LOD) threshold values were determined by 100-fold-permutation tests (MapQTL® 4.0) at a significance level of 95% (genome-wide) (King et al. 2000). The proportion of variation in aphid infestation that can be explained by the genetic variation among the apple progenies was analyzed by broad-sense heritability, which was estimated by the formula \(H^2 = {{\sigma _{\text{g}}^2 } \mathord{\left/ {\vphantom {{\sigma _{\text{g}}^2 } {\sigma _{\text{p}}^{\text{2}} }}} \right. \kern-\nulldelimiterspace} {\sigma _{\text{p}}^{\text{2}} }}\) and \(\sigma _{\text{p}}^{\text{2}} = \left( {\sigma _{\text{g}}^{\text{2}} + {{\sigma _{\text{e}}^{\text{2}} } \mathord{\left/ {\vphantom {{\sigma _{\text{e}}^{\text{2}} } n}} \right. \kern-\nulldelimiterspace} n}} \right)\), where \(\sigma _{\text{g}}^2 \) is the genetic variance, \(\sigma _{\text{p}}^2 \) is the phenotypic variance, \(\sigma _{\text{e}}^2 \) is the environmental variance and n is the number of replicates per genotype (Lauter and Doebley 2002). Variance components were based on mean square analysis of variance (ANOVA) results (Frei et al. 2005). To test for interactions between different QTLs, multiple QTL mapping (MQM) was carried out for QTLs with LOD scores exceeding the significant LOD threshold in IM. For MQM, markers with the highest likelihood ratios (i.e., LOD test statistic) positioned on the linkage group containing the QTL were selected as the initial set of possible MQM cofactors. A backward elimination procedure was applied to this initial set of cofactors using a significance level of 0.02 (Larson et al. 2006). The 2-LOD support interval was calculated to estimate the position of significant QTLs with 95% confidence (King et al. 2000). The ‘Fiesta’ × ‘Discovery’ population was divided into subpopulations based on the presence/absence of the marker closest to the detected QTL and phenotypic difference was tested considering the two subpopulations (Mann–Whitney U test).
Pedigree analysis
Polymerase chain reactions (PCR) amplifications with the primers of the SSR markers Hi03a10 and Hi07h02 (Silfverberg-Dilworth et al. 2006) were performed in a 10-μl volume containing 5 μl of a DNA solution (1 ng/µl), 1× reaction buffer (Amersham Pharmacia, Dübendorf, Switzerland), 0.1 mM of each deoxyribonucleotide triphosphate, 0.2 μM of dye-labeled forward primer and 0.2 μM of reverse primer, and 0.7 U of Taq Polymerase (Amersham Pharmacia, Dübendorf, Switzerland) per reaction. Forward primers were labeled with the following dyes: Hi03a10f with NED (Applied Biosystems) and Hi07h02f with HEX (Biomers, Germany). PCRs were performed in a Gene Amp PCR system 9600 (PerkinElmer, Foster City, CA) under the following conditions: 5 min at 96°C, 35 cycles of 30 s at 96°C, 30 s at 60°C, and 30 s at 72°C with a final extension of 10 min at 72°C. For each sample, 1 μl of PCR product was mixed with 10 μl deionized formamide and 0.2 μl 500-LIZ ladder (Applied Biosystems). Microsatellite fragments were separated by electrophoresis on an ABI PRISM 3700 capillary sequencer (Applied Biosystems). Chromatographs were generated using Genescan 3.7 software and microsatellite fragment lengths were scored with Genotyper 3.6 (Applied Biosystems).
Spatial distribution of aphids
The distribution patterns of aphids on individual apple genotypes were characterized by computing the index of dispersion (I D) (Southwood and Henderson 2000), with \(I_{\text{D}} = s^2 {{\left( {\nu - 1} \right)} \mathord{\left/ {\vphantom {{\left( {\nu - 1} \right)} m}} \right. \kern-\nulldelimiterspace} m}\), where m and s 2 are the sample mean and variance, respectively, and ν is the sample size. I D values significantly greater than the χ 2 statistic (0.025 probability level) with (ν-1) degrees of freedom indicate an aggregated distribution of the studied species (significant variation considering the level of infestation of individual trees) (Ludwig and Reynolds 1988). The calculation of the I D values was performed with BiodiversityPro 2.0 (McAleece, Lambshead and Paterson; The Natural History Museum, London). Potential effects of the spatial position of trees in the study sites on aphid infestation were inferred from analyses of spatial autocorrelation, computing Moran’s I (Legendre and Legendre 1998) and corresponding z values (significance levels) using the software CrimeStat III (Levine 2007). Moran’s I varies between −1.0 for negative spatial autocorrelation (nearby trees have dissimilar aphid infestation level) and +1 for positive spatial autocorrelation (nearby trees have similar aphid infestation level). If no spatial autocorrelation exists, the expected value for Moran‘s I is \(eI = {{ - 1} \mathord{\left/ {\vphantom {{ - 1} {\left( {n - 1} \right)}}} \right. \kern-\nulldelimiterspace} {\left( {n - 1} \right)}}\). Values of I greater than the expected I indicate clustering, while values of I less than the expected I indicate dispersion. The significance test of the z-values indicates whether these differences are greater than what would be expected by chance. The z values were compared to a standard normal table, and absolute values greater than 1.96 indicate a spatial autocorrelation at a 5% significance level. In addition, we described the spatial distribution of aphids by fitting trend surfaces on contour plots by kriging (best unbiased generalized least squares estimation) with an exponential covariance function (Venables and Ripley 2002). The distribution of aphids on a small scale level was analyzed by comparing aphid infestation on neighbor trees. The number of aphids on an individual tree was compared to the number of aphids on the neighbor trees (sum) using Spearman’s rank tests to estimate neighborhood effects. Only those genotypes were included in this analysis that had direct neighbor genotypes (a dead or missing genotype was not regarded as neighbor genotype).
Statistical analysis
A log10(x + 1) transformation was applied to normalize error distribution of aphid infestation. To evaluate the effect of genotype, site, and year, a three-factor mixed-model ANOVA, with year as within-subject effect (repeated measure), and genotype and site as between-subjects fixed effects, was applied. Spearman’s rank tests were used to study relationships between different sites, years, sampling dates, aphid species, and plant characteristics. When multiple correlation tests were carried out, the Benjamini-Hochberg procedure was used to correct for false discovery rates (type I errors) (Benjamini and Hochberg 1995; Verhoeven et al. 2005). All statistical analyses were performed with SPSS 16.0 for Mac OS X (SPSS, Chicago, IL) and R 2.6.0 (R Development Core Team, Vienna).
Results
Aphid infestation
Aphid infestation of individual apple trees varied strongly among sites and study years (Table 1). The highest number of Dysaphis plantaginea colonies per tree was found at the Ticino site in both study years (means: 13.5 and 17.8), and the lowest number at the Zurich site (means: 0.9 and 0.0). Incidence ranged between 0% (Zurich Year 2) and 92% (Valais Year 2) infested trees. The mean number of red-curled leaves per tree caused by Dysaphis cf. devecta varied between 1.6 and 2.8 at the Ticino site and between 0.8 and 4.0 at the Valais site. Incidence ranged from 0% (Zurich, years 1 and 2) to 18% (Valais, year 1) and 59% (Valais, year 2) infested trees. Infestation by Aphis pomi was highest for the Valais site in both study years (average: 443.5 and 63.4 aphids per tree) followed by the Ticino (mean: 62.9 and 45.6) and the Zurich site (average: 5.8 and 10.2). Incidence ranged from 26% (Zurich year 1) to 87% (Valais, year 1) infested trees.
Influence of genotype, site, and year on aphid abundance
A significant influence of genotype was found for infestation by D. plantaginea and D. cf. devecta but not by A. pomi. Site and year contributed significantly to infestation patterns of all three aphid species (Table 2). Broad-sense heritability (H 2) was highest for D. cf. devecta resistance (50.2%) and lowest for A. pomi infestation (15.7%) (Table 3). For resistance to D. plantaginea, H 2 was intermediate (28.3%).
No correlation between the three sites was found for D. plantaginea infestation per tree in May and June (P > 0.05, Spearman’s rank test, Benjamini–Hochberg procedure). D. cf. devecta infestation per tree was correlated between the Ticino and Valais site in June of year 1 (n = 130, r s = 0.185, P = 0.035), but it was not correlated between the Ticino and Valais site in May of both years and June of year 2 (P > 0.05, Spearman’s rank test, Benjamini–Hochberg procedure). The number of A. pomi per tree correlated significantly between the Ticino and Valais site in July (year 1) (Spearman’s rank test, Benjamini–Hochberg procedure; n = 129, r s = 0.198, P = 0.025), but no correlation for A. pomi infestation was found in May and June of both study years, and between the Ticino and Valais site, between the Ticino and Zurich site, and between the Valais and Zurich site in July of year 2.
Aphid infestation of the same tree individuals was significantly correlated between the two study years for D. plantaginea at the Ticino site (Spearman’s rank test; May: n = 144, r s = 0.19, P = 0.026; June: n = 144, r s = 0.17, P = 0.046), but not at the Valais site (P > 0.05). For D. cf. devecta, infestation of the same tree individuals was correlated between the two consecutive years at the Valais site (May: n = 142, r s = 0.339, P < 0.001, June: n = 142, r s = 0.368, P < 0.0001), but not at the Ticino site. For A. pomi, infestation of the same tree individuals was correlated between the two study years at the Valais site (Spearman’s rank test, May: n = 142, r s = 0.184, P = 0.028; June: n = 142, r s = 0.286, P = 0.001, July: n = 142, r s = 0.237, P = 0.005) and the Ticino site in June (n = 143, r s = 0.30, P < 0.0001), but not at the Zurich site (P > 0.05).
The within-year infestation of the same tree individuals was significantly correlated in both years for all the three aphid species (P < 0.05, Spearman rank correlation and Benjamini–Hochberg procedure; analysis not shown). Comparing May and June, D. plantaginea abundance was generally higher in June than in May. The same pattern was found for A. pomi. In addition, the potential to rapidly increase population size was detected for A. pomi at the Valais site. The number of red-curled leaves caused by D. cf. devecta was higher in May compared to June.
QTLs for aphid resistance
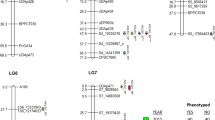
A significant QTL for D. plantaginea resistance in apple was identified at the Ticino site in year 1. Multiple QTL mapping (MQM; data not shown) did not reveal any multiple linked QTLs, and results were therefore based on interval mapping (IM). The QTL was associated with the AFLP marker E33M35–0269 at 57.7 cM on linkage group 17 of the ‘Fiesta’ chromosome. The LOD score at the marker position was 2.85 and explained 8.5% of phenotypic variability (PVE, Table 4, Fig. 1a). The 2-LOD support interval ranged from map positions 45–67 cM (Ticino, year 1). A significant lower D. plantaginea infestation was found for the apple subpopulation amplifying the marker E33M35–0269 compared to apple genotypes not amplifying the marker (Fig. 2). The origin of the QTL associated to aphid resistance was followed in the pedigree of ‘Fiesta’. The allele ‘255 bp’ of the SSR marker Hi07h02 (Silfverberg-Dilworth et al. 2006), which is closely located and in coupling with the AFLP marker E33M35–0269 (10.2 cM distance between Hi07h02 and E33M35–0269), has been inherited from ‘Wagener’ to ‘Idared’ (Fig. 3a).
QTLs for resistance to Dysaphis plantaginea (a), identified on linkage group 17 of ‘Fiesta’, and Dysaphis cf. devecta (b), identified on linkage group 7 of ‘Fiesta’, based on IM results. The x-axis indicates the linkage map of ‘Fiesta’ in cM and marker names; the y-axis shows the LOD scores. The solid black bar indicates the 2-LOD support interval for the position of the QTL (Ticino year 1). Average values of log10(x + 1) transformed data were used for QTL analysis. LOD threshold values at different study sites and in different years ranged between 2.7 and 3.3. The closest marker to the QTL is underlined
Mean Dysaphis plantaginea and D. cf. devecta infestation of the two subpopulations of the ‘Fiesta’ and ‘Discovery’ cross, based on presence and absence of the nearest marker associated to a QTL on the ‘Fiesta’ chromosome. The subpopulations for AFLP markers E33M35–0269 (D. plantaginea) and E32M39–0195 (D. cf. devecta) are presented. Significant differences between subpopulations are indicated by *P < 0.001 and **P < 0.0001 (Mann–Whitney U test)
Analysis of the pedigree of the apple variety ‘Fiesta’ with the two SSRs Hi07h02 and Hi03a10 associated with the QTLs for resistance to Dysaphis plantaginea (a) and to D. cf. devecta (b). The SSR marker alleles associated with resistance are highlighted (allele ‘255 bp’ for Hi07h02 and allele ‘216 bp’ for Hi03a10)
We detected a significant QTL for D. cf. devecta resistance on ‘Fiesta’ linkage group 7 at the Ticino site in both years, and at the Valais site in year 2 (Table 4, Fig. 1b). QTL results based on IM as well as MQM mapping (data not shown) did not identify any multiple linked QTLs. The significant LOD scores for the AFLP marker E32M39–195 at different sites and in different years ranged from 3.36 (Ticino, year 2) to 6.82 (Valais, year 2), and PVE by this QTL varied between 10.3% (Ticino, Year 1) and 20.4% (Valais, year 2). The 2-LOD support interval ranged from map position 0 to 15 cM (Valais Year 2). Apple genotypes containing the marker E32M39–0195 showed significantly lower D. cf. devecta infestation compared to the subpopulation missing the marker (Fig. 2). The origin of the QTL associated to D. cf. devecta resistance was followed in the pedigree of ‘Fiesta’. The allele ‘216 bp’ of the SSR marker Hi03a10, which is in close proximity and in coupling with the AFLP marker E32M39–0195 (9.9 cM distance between Hi03a10 and E32M39–0195) (Silfverberg-Dilworth et al. 2006), was found to derive from ‘Cox’s Orange Pippin’, which is a progeny of ‘Blenheim Orange’ (Fig. 3b). No QTL was identified for A. pomi resistance in apple.
QTLs for different plant growth characteristics
Significant QTLs were identified at the Ticino site in year 1 (shoot length and stem diameter) and at the Valais site in year 2 (stem diameter) (Table 5). MQM mapping (data not shown) did not identify any multiple linked QTLs and results were therefore based on IM. The QTL for shoot length was significant at the Ticino site in year 1 and was positioned on ‘Fiesta’ linkage group 7 (SSR marker CH04e05 on locus 26.7). The allele of CH04e05 conferring increased length is 199 bp long. The highest LOD score for the shoot length QTL obtained at the Ticino site in year 1 was 2.8, while the PVE by this QTL was 8.8%. The proportion of variation in shoot length that can be explained by the genetic variation (broad-sense heritability) was 44.9% (data not shown).
Two markers, linked to QTLs for stem diameter, were identified on ‘Discovery’ linkage groups 1 and 13 (AFLP marker E31M38–0080 on locus 15.2, and AFLP marker E35M41–0575 on locus 29.7) at the Ticino site in year 1. LOD scores were 2.58 and 1.62, respectively. The markers explained 7.1% and 4.5% of the phenotypic variability. Another marker linked to a QTL for larger stem diameter was identified on ‘Discovery’ linkage group 14 at the Valais site in year 2 (RAPD marker D01–1200 on locus 10.3). The LOD score and PVE were 2.1 and 6.2%, respectively. Broad-sense heritability for stem diameter was 15.8% (data not shown).
Spatial distribution of three aphid species
The distribution of the aphid species on individual apple trees was highly aggregated, as was indicated by the index of dispersion (I D) (Table S2). This means that some trees were strongly infested, whereas other trees were not infested at all. This finding was consistent for all sites and years. I D values ranged from 946 (D. cf. devecta) to 137,360 (A. pomi). I D values for D. plantaginea varied between 1,093 and 5,600. A higher aggregation of D. plantaginea and D. cf. devecta was found at the Ticino site compared to the Valais site, whereas for A. pomi a higher aggregation was found at the Valais site compared to the Ticino site.
Spatial distribution patterns on the orchard scale varied among aphid species (Table 6, Fig. S1). For D. plantaginea distribution, a significant positive spatial autocorrelation was found at the Ticino site in both study years and at the Zurich site in year 1. Infestation levels appeared to be higher at the southeastern part at the Ticino site and lower at the middle row at the Zurich site (contour plots, Fig. S1). No spatial autocorrelation was found for D. cf. devecta distribution (Table 6). This is in line with the findings from the contour plots (Fig. S1). The distribution of A. pomi within an orchard seemed to be most strongly autocorrelated. There was a significant positive autocorrelation at the Ticino site in year 1, at the Valais site in year 2, and at the Zurich site in both years (Table 6). At the Ticino site, rows at the southern part showed a higher infestation compared to northern rows, and the border rows were more infested compared to the middle row at the Zurich site (Fig. S1).
Possible effects of neighborhood on aphid infestation of specific apple trees differed among sites and aphid species (Table 7). D. plantaginea infestation of a specific tree and of the two neighbor trees was significantly correlated at the Zurich site in year 1 and the Valais site in year 2 (no D. plantaginea infestation was detected at the Zurich site in year 2). For the same aphid species, no neighborhood effect was found at the Ticino site (P > 0.05; Spearman’s rank test). For D. cf. devecta infestation, no significant correlation between aphid infestation of a specific tree and of the two neighbor trees was found (Table 7). For A. pomi infestation, possible neighborhood effects were found at the Ticino site in year 1, at the Valais site in both study years and the Zurich site in year 2 (Table 7).
Relationship between different aphid species
Dysaphis plantaginea infestation per tree was positively correlated with D. cf. devecta and A. pomi infestation per tree at the Ticino site in year 1 (Spearman’s rank test, Benjamini–Hochberg procedure; n = 143, r s = 0.222, P = 0.008; n = 143, r s = 0.445, P < 0.0001) but not in year 2 (P > 0.05, Table S3). There was no significant relationship between D. cf. devecta and A. pomi (P > 0.05, Table S3). At the Valais and Zurich site no significant relationship between the three aphid species was found (P > 0.05, Table S3). In general, the relationship between the aphid species was positive; however, the correlation was weak and not significant (Table S3).
Relationship between different plant growth characteristics and aphid infestation
Aphid infestation was positively correlated to different plant growth characteristics (Table 8). The influence of plant growth traits on aphid infestation differed among aphid species. Plant growth traits were most important for A. pomi and least important for D. cf. devecta. A significant relationship between D. plantaginea infestation per tree and plant growth characteristics was found for the Ticino site (Spearman’s rank test, Benjamini–Hochberg procedure; Table 8). Stem diameter, shoot length, and new shoot length were significantly correlated to the number of D. plantaginea colonies. At the Valais site in year 2, shoot length and new shoot length were significantly related to D. plantaginea infestation. At the Zurich site, no significant relationship was found.
The number of red-curled leaves per tree induced by D. cf. devecta was significantly related to tree height at the Ticino site in year 2 and at the Valais site in year 2, to shoot length at the Ticino site in year 1 and at the Valais site in year 2 (Table 8). There was no significant relationship between D. cf. devecta infestation and stem diameter and new shoot length, respectively. The length of new shoots had a significant influence on A. pomi infestation per tree (Table 8). A stronger correlation was detected between A. pomi infestation and new shoot length compared to shoot length (Ticino, Valais year 1, and Zurich). At the Ticino site, the relation between the number of A. pomi per tree and shoot length was not significant, whereas a significant correlation between A. pomi infestation per tree and the length of new shoots was found. The number of A. pomi aphids was also significantly correlated to stem diameter at the Ticino site in year 2, at the Valais site and at the Zurich site, to tree height at the Valais site in year 1 and at the Zurich site in year 1 (Table 8).
Discussion
Genetic background of aphid resistance in apple
The present study was aimed at investigating the resistance of apple trees to the aphid species Dysaphis plantaginea, D. cf. devecta and Aphis pomi by QTL analysis. The relationships between aphid infestation and plant growth characteristics, and interactions between the studied aphid species, the climatic conditions at the study sites and the spatial variability of aphid infestation were also assessed. We identified a putative QTL for resistance to D. plantaginea and a QTL for resistance to D. cf. devecta in apple. The markers closely linked to the QTLs were related to significantly lower D. plantaginea infestation at the Ticino site in year 1 (AFLP marker E33M35–0269), and to significantly lower injuries caused by D. cf. devecta at the Ticino site in years 1 and 2 and at the Valais site in year 2 (AFLP marker E32M39–0195). Broad-sense heritability (H 2) was higher for D. cf. devecta (50.2%) compared to D. plantaginea (28.3%).
We found a strong variation in aphid numbers on individual apple trees. While some trees were highly infested, others were not infested at all. Although in some instances aphid infestation patterns appeared to be spatially autocorrelated, no distinct clusters of highly infested trees were detectable. ANOVA analyses indicated that the genotype was a significant factor for this variation in infestation by D. plantaginea and D. cf. devecta, but not by A. pomi.
The identified QTLs and the findings on variable infestation of individual apple genotypes highlight the genetic background of aphid resistance in apple. The existence of some genetically based resistance to aphids in apple is also indicated by studies reporting that susceptibility to D. plantaginea, D. cf. devecta, and A. pomi may differ among apple varieties (Alston and Briggs 1970, 1977; Roche et al. 1997; Graf et al. 1998; Habekuss et al. 2000; Cevik and King 2002; Qubbaj et al. 2005; Angeli and Simoni 2006; see Table S4 for detailed list). Despite this considerable coverage of aphid resistance in apple, information on the genetic background of this resistance is scarce. MAL 59/9, a Malus robusta derivative, was shown to carry a single dominant gene for hypersensitivity to D. plantaginea (Alston and Briggs 1970), but until today the gene has not been located. Qubbaj et al. (2005) investigated gene expression in a resistant (‘Florina’) and a susceptible (‘Topaz’) apple cultivar after infestation with D. plantaginea.
The authors describe gene fragments expressed only in the resistant or susceptible cultivar (e.g., RNase-l-inhibitor-like protein), and mention that the identified genes show homologies to genes related to plant stress. For D. cf. devecta, the existence of a resistance locus at the top of linkage group 7 in ‘Fiesta’ (Sd1) has previously been reported (Roche et al. 1997; Cevik and King 2002). The mentioned studies used aphid inoculations, and the experiments were either carried out in glasshouses (Alston and Briggs 1977; Roche et al. 1997), or in glasshouses and in the field (Cevik and King 2002). In our study, 140–160 apple genotypes were exposed, in comparison, to aphid field infestation under different environmental conditions. This study is the first QTL analysis of D. plantaginea and A. pomi resistance, and it presents the probable position (QTL identified only once and in a single place) of a D. plantaginea resistance gene within the apple genome.
In addition, the present study confirms the results of the mapping of the resistance gene Sd1 (Cevik and King 2002) in a different genetic background (‘Fiesta’ × ‘Discovery’) under natural infestation by different populations of the D. cf. devecta species complex. Cevik and King (2002) mapped the Sd1 resistance gene to D. cf. devecta within an interval of 1.3 cM delimited by the molecular markers SdSSRa and 2B12a at the top of the ‘Fiesta’ chromosome 7. Khan et al. (2007) developed a SSR marker, called CH-Sd1, from the sequence of the BAC clone 49N23 belonging to the contig spanning the Sd1 locus (Cevik and King 2002). CH-Sd1 has been mapped in the ‘Fiesta’ × ‘Discovery’ cross used in this study on F7 between the markers P13–2000 and E35M42–0480 at only 2 cM from E32M39–0195 (Khan et al. 2007). The CH-Sd1 locus is therefore exactly in the confidence interval of the QTL identified in this study on ‘Fiesta’ linkage group 7. This resistance is therefore due to Sd1.
To evaluate the applicability in apple breeding of the SSR markers Hi07h02 and Hi03a10, which were found to be closely positioned to the QTLs for D. plantaginea and D. cf. devecta resistance, apple cultivars described as resistant to aphids could be used. If the alleles of the SSR markers in coupling with these resistances (allele ‘255 bp’ for Hi07h02 and allele ‘216 bp’ for Hi03a10) are present in the resistant apple varieties, these cultivars and the SSR markers could be used for marker-assisted breeding. Based on our pedigree analyses, the alleles in coupling with the resistance occur in ‘Idared’ and ‘Wagener’ (Hi07h02–255: D. plantaginea resistance), and in ‘Cox’s Orange Pippin’ and ‘Blenheim Orange’ (Hi03a10–216: D. cf. devecta resistance). For ‘Wagener’ and ‘Cox’s Orange Pippin’, resistance to D. plantaginea and D. cf. devecta, respectively, was reported (Alston and Briggs 1977), whereas no information on resistance was found for ‘Idared’ and ‘Blenheim Orange’. In combination, these results suggest that apple varieties with known resistance to aphids should be tested for the presence of the markers, and that the markers could also be used to detect so far unknown resistance in other apple cultivars.
Environmental factors related to aphid resistance
Our study shows that for field populations of insect pests and especially for aphids, the genetic background of the host plant could partly explain infestation levels but that environmental and neighborhood effects render their identification very difficult. The identified QTLs were not stable for different sites and years, and aphid abundance on specific apple genotypes was not strongly correlated between different sites and years. Different climatic conditions at the sites may impede the identification of genetically based resistance. Highest aphid infestation was found for the Ticino site, which was characterized by dry and warm climate conditions that are known to favor aphid population growth (Blommers et al. 2004). Drought stress in apple increases emission of volatile secondary plant metabolites (Vallat et al. 2005), and plant volatiles were shown to affect aphid distribution (Bernasconi et al. 1998).
Field observations showed that plant phenology was more progressed at the Ticino site, allowing an earlier infestation with faster population buildup. This may partly explain the lack of correlations of A. pomi infestation between the sites in May and June, and its presence in July. D. cf. devecta was not present at the Zurich site. Insecticides were applied in neighboring orchards, and this may have reduced population buildup due to low aphid immigration.
Besides population fluctuation and climate, various other environmental parameters may affect aphid abundance and blur genetically based resistance in apple. Highly susceptible genotypes may be severely infested by aphids and serve as a source for aphid colonization of neighboring trees (Blommers et al. 2004). A significant spatial pattern and an influence of neighbor tree infestation level were found for D. plantaginea and A. pomi. The positive spatial autocorrelation as well as significant neighborhood effects may partly explain why the QTL for D. plantaginea resistance was not identified in year 2 at the Ticino site and at the Zurich site and why no QTL was identified for A. pomi.
There was a significant effect of neighbor tree infestation mainly at the Valais and Zurich sites. At these sites, trees were planted at a distance of 0.5 m compared to the 1.25 m at the Ticino site, which can increase the tree-to-tree colonization by aphids. On an interesting note, the QTL on the ‘Fiesta’ linkage group 17 for D. plantaginea resistance was identified at the Ticino site, but not at the Valais and Zurich site, where a neighborhood effect was present. Orchard-wide distribution patterns were also identified for the woolly apple aphid (Eriosoma lanigerum) (Alspach and Bus 1999). The authors mention that computation of genetically based resistance may be influenced by significant spatial autocorrelation.
Direct interspecific interactions between aphids or similar resource requirements may also affect the distribution of individual species. At the Ticino site in year 1, distribution of all the aphid species was positively correlated. In general, no negative relationships between the aphid species were found, indicating that competition among aphids was not an important factor affecting their distribution. Similar resource requirements mediated by plant characteristics may, however, help to explain the concordant distribution patterns. If resources commonly used by different species are reflected by specific plant growth characteristics, distribution of different herbivore species should independently correlate with those characteristics.
Indications for such congruent resource use were mainly found for stem diameter and for shoot length and new shoot length, showing a significant positive correlation especially with D. plantaginea and A. pomi infestation. The QTL analysis of different plant growth characteristics revealed that QTLs for shoot length and stem diameter were generally positioned on other linkage groups than the QTLs for aphid resistance. This finding indicates that the identified QTLs for aphid resistance are not likely to be correlative artifacts of plant trait effects. This reasoning may also apply to the putative QTL for shoot length and the QTL for D. cf. devecta resistance. Although both QTLs were positioned on ‘Fiesta’ linkage group 7, they do not appear to be strongly related.
Whereas the QTL for D. cf. devecta resistance was found at different sites in both study years, the putative QTL for shoot length was only found at the Ticino site in year 1. In addition, the correlations between D. cf. devecta infestation and shoot length for the Ticino site were weak or not significant.
Effects of predators on aphid populations are variable (Dixon 2000). In our study, field observations did not reveal relevant predator densities, suggesting that quantification of aphid infestation level was not strongly influenced by predation.
Outlook
The identified SSR markers for aphid resistance could be used in breeding programs where there is a demand for increased fruit quality and plant resistance (Avilla and Riedl 2003). The value of the detected markers linked to a resistance QTL applicable for MAS depends on the ease with which they can be introgressed into commercial cultivars. A combined use of the identified markers associated to resistance to D. plantaginea and D. cf. devecta may facilitate the breeding of new apple cultivars resistant to several aphid species. As markers on linkage group 7 of ‘Fiesta’ are linked to fire blight resistance (Khan et al. 2007), the study also emphasizes the potential for gene pyramiding in broad-spectrum resistance breeding of apple. The same applies to aphid resistance, as the putative QTL for D. plantaginea resistance, identified in the present study, was found on the ‘Fiesta’ linkage group 17, as was a gene most recently found for E. lanigerum resistance (Bus et al. 2008). Furthermore, apple cultivars described as resistant to aphids and amplifying the SSR alleles associated with increased resistance to aphids, provide a useful basis to facilitate traditional apple breeding. The utilization of apple cultivars with the resistance-related marker alleles can minimize failures in apple breeding as resistance should be genetically based and less environmentally dependent.
References
Alspach PA, Bus VGM (1999) Spatial variation of woolly apple aphid (Eriosoma lanigerum Hausmann) in a genetically diverse apple planting. N Z J Ecol 23:39–44
Alston FH, Briggs JB (1970) Inheritance of hypersensitivity to rosy apple aphid (Dysaphis plantaginea) in apple. Can J Genet Cytol 12:257–258
Alston FH, Briggs JB (1977) Resistance genes in apple and biotypes of Dysaphis devecta. Ann Appl Biol 87:75–81
Angeli G, Simoni S (2006) Apple cultivars acceptance by Dysaphis plantaginea Passerini (Homoptera: Aphididae). J Pest Sci 79:175–179
Arbab A, Kontodimas DC, Sahragard A (2006) Estimating development of Aphis pomi (DeGeer) (Homoptera: Aphididae) using linear and nonlinear models. Environ Entomol 35:1208–1215
Avilla J, Riedl H (2003) Integrated fruit production for apples—principles and guidelines. In: Ferree DC, Warrington IJ (eds) Apples: botany, production and uses. CABI Publishing, Wallingford, UK, pp 539–549
Beers EH, Suckling DM, Prokopy RJ, Avilla J (2003) Ecology and management of apple arthropod pests. In: Ferree DC, Warrington IJ (eds) Apples: botany, production and uses. CABI Publishing, Wallingford, UK, pp 489–514
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B (Stat Method) 57:289–300
Bernasconi ML, Turlings TCJ, Ambrosetti L, Bassetti P, Dorn S (1998) Herbivore-induced emissions of maize volatiles repel the corn leaf aphid (Rhopalosiphum maidis). Entomol Exp Appl 87:133–142
Blommers L (1994) Integrated pest-management in European apple orchards. Annu Rev Entomol 39:213–241
Blommers LH, Helsen HHM, Vaal FWNM (2004) Life history data of the rosy apple aphid Dysaphis plantaginea (Pass.) (Homopt., Aphididae) on plantain and as migrant to apple. J Pest Sci 77:155–163
Bus VGM, Chagné D, Bassett HCM, Bowatte D, Calenge F, Celton J-M, Durel C-E, Malone MT, Patocchi A, Ranatunga AC, Rikkerink EHA, Tustin DS, Zhou J, Gardiner SE (2008) Genome mapping of three major resistance genes to woolly apple aphid (Eriosoma lanigerum Hausm.). Tree Genet Genom 4:223–236
Calenge F, Durel C-E (2006) Both stable and unstable QTLs for resistance to powdery mildew are detected in apple after four years of field assessment. Mol Breed 17:329–339
Cevik V, King GJ (2002) High-resolution genetic analysis of the Sd1 aphid resistance locus in Malus spp. Theor Appl Genet 105:346–354
Cheng FS, Weeden NF, Brown SK, Aldwinckle HS, Gardiner SE, Bus VG (1998) Development of a DNA marker for Vm, a gene conferring resistance to apple scab. Genome 41:208–214
Dixon AFG (2000) Insect predator–prey dynamics: ladybird beetles and biological control. Cambridge University, Cambridge, UK, p 257
Dorn S, Schumacher P, Abivardi C, Meyhöfer R (1999) Global and regional pest insects and their antagonists in orchards: spatial dynamics. Agric Ecosyst Environ 73:111–118
Frei A, Blair MW, Cardona C, Beebe SE, Gu H, Dorn S (2005) QTL mapping of resistance to Thrips palmi (Karny) in common bean. Crop Sci 45:379–387
Fujita D, Doi K, Yoshimura A, Yasui H (2006) Molecular mapping of a novel gene, Grh5, conferring resistance to green rice leafhopper (Nephotettix cincticeps Uhler) in rice (Oryza sativa L). Theor Appl Genet 113:567–573
Graf B, Höpfli HU, Höhn H, Kellerhals M, Krebs C (1998) Schorfresistente Apfelsorten: Wie steht‘s mit der Schädlingsanfälligkeit? Schweiz Z Obst-Weinbau 3:71–73
Graf B, Höpli H, Höhn H, Samietz J (2006) Temperature effects on egg development of the rosy apple aphid and forecasting of egg hatch. Entomol Exp Appl 119:207–211
Habekuss A, Proeseler G, Schliephake E, Fischer C (2000) Resistance of apple to spider mites and aphids. Proc Eucarpia Symp 1:271–278
Hamilton GC, Swift FC, Marini RP (1986) Effect of Aphis pomi (Homoptera: Aphididae) density on apples. J Econ Entomol 79:471–478
James CM, Clarke JB, Evans KM (2004) Identification of molecular markers linked to the mildew resistance gene PL-d in apple. Theor Appl Genet 110:175–181
Khan MA, Durel C-E, Duffy B, Drouet D, Kellerhals M, Gessler C, Patocchi A (2007) Development of molecular markers linked to the ‘Fiesta’ linkage group 7 major QTL for fire blight resistance and their application for marker-assisted selection. Genome 50:568–577
King GJ, Maliepaard C, Lynn JR, Alston FH, Durel C-E, Evans KM, Griffon B, Laurens F, Manganaris AG, Schrevens E, Tartarini S, Verhaegh J (2000) Quantitative genetic analysis and comparison of physical and sensory descriptors relating to fruit flesh firmness in apple (Malus pumila Mill.). Theor Appl Genet 100:1074–1084
Krakowsky MD, Lee M, Woodman-Clikeman WL, Long MJ, Sharpova N (2004) QTL mapping of resistance to stalk tunneling by the European corn borer in RILs of maize population B73 × De811. Crop Sci 44:274–282
Larson SR, Wu X, Jones TA, Jensen KB, Chatterton NJ, Waldron BL, Robins JG, Baushman BS, Palazzo AJ (2006) Comparative mapping of growth habit, plant height, and flowering QTLs in two interspecific families of Leymus. Crop Sci 46:2526–2539
Lauter N, Doebley J (2002) Genetic variation for phenotypically invariant traits detected in teosinte: implications for the evolution of novel forms. Genetics 160:333–342
Legendre P, Legendre L (1998) Numerical ecology: developments in environmental modelling 20. Elsevier Science, Amsterdam, The Netherlands, pp 714–721
Lespinasse Y, Olivier JM, Lespinasse JM, LeLezec M (1985) ‘Florina-Quérina’: la résistance du pommier à la Tavelure. Arboric Fruit 378:43–47
Levine N (2007) CrimeStat: A spatial program for the analysis of crime incident locations (v3.1). Ned Levine & Associates; the National Institute of Justice, Houston, TX; Washington, DC
Liebhard R, Koller B, Gianfranceschi L, Gessler C (2003) Creating a saturated reference map for the apple (Malus × domestica Borkh.) genome. Theor Appl Genet 106:1497–1508
Ludwig JA, Reynolds JF (1988) Statistical ecology: a primer on the methods and computing. John Wiley & Sons, New York, USA, pp 26–29
Maag R (1996) Integrierte produktion im Obstbau. Dr. R. Maag AG, Dielsdorf, CH, pp 8–16
Miñarro M, Hemptinne JL, Dapena E (2005) Colonization of apple orchards by predators of Dysaphis plantaginea: sequential arrival, response to prey abundance and consequences for biological control. BioControl 50:403–414
Omkar J, Pervez A (2005) Ecology of two-spotted ladybird (Adalia bipunctata): a review. J Appl Entomol 129:465–474
Qubbaj T, Reineke A, Zebitz CPW (2005) Molecular interactions between rosy apple aphids (Dysaphis plantaginea), and resistant and susceptible cultivars of its primary host Malus domestica. Entomol Exp Appl 115:145152
Rat-Morris E, Crowther S, Guessoum M (1999) Resistance-breaking biotypes of rosy apple aphid (Dysaphis plantaginea), on the resistant cultivar ‘Florina’. IOBC/WPRS Bull 22:71–75
Roche P, Alston FH, Maliepaard C, Evans KM, Vrielink R, Dunemann F, Markussen T, Tartarini S, Brown LM, Ryder C, King GJ (1997) RFLP and RAPD markers linked to the rosy leaf curling aphid resistance gene (Sd1) in apple. Theor Appl Genet 94:528–533
Silfverberg-Dilworth E, Matasci CL, van de Weg WE, van Kaauwen MPW, Walser M, Kodde LP, Soglio V, Gianfranceschi L, Durel C-E, Costa F, Yamamoto T, Koller B, Gessler C, Patocchi A (2006) Microsatellite markers spanning the apple (Malus × domestica Borkh.) genome. Tree Genet Genom 2:202–224
Simon S, Lauri PE, Brun L, Defrance H, Sauphanor B (2006) Does manipulation of fruit-tree architecture affect the development of pests and pathogens? A case study in an organic apple orchard. J Hortic Sci Biotechnol 81:765–773
Southwood TRE, Henderson PA (2000) Ecological Methods. Blackwell Science, Oxford, UK, pp 42–44
Stekolshchikov AV, Lobanov AL (2004) Dysaphis devecta (Walker) species complex (Homoptera, Aphididae): II. Morphological characteristics of the forms. Entomol Obozr 83:115–139
Vallat A, Gu H, Dorn S (2005) How rainfall, relative humidity and temperature influence volatile emissions from apple trees in situ. Phytochemistry 66:1540–1550
van Ooijen JW, Boer MP, Jansen RC, Maliepaard C (2002) MapQTLÒ 4.0, software for the calculation of QTL positions on genetic maps. Plant Research International, Wageningen, NL
Venables WN, Ripley BD (2002) Modern applied statistics with S-Plus. Springer-Verlag, Berlin Heidelberg New York New York, pp 469–485
Verhoeven KJF, Simonsen L, McIntyre LM (2005) Implementing false discovery rate control: increasing your power. Oikos 108:643–647
Zehnder G, Gurr G, Kühne S, Walde MR, Wratten SD, Wyss E (2007) Arthropod pest management in organic crops. Annu Rev Entomol 52:57–80
Acknowledgments
The authors thank Muhammad Awais Khan and Giovanni Broggini (ETH Zurich) and Hans Jansen (Plant Research Wageningen) for helpful support in QTL analysis; Markus Kellerhals and Danilo Christen (Agroscope Changins-Wadenswil Research Station ACW) for access to the study orchards; Davide Gobbin (ETH Zurich) for PCR amplification of SSR markers; Michelle Schmocker, Christoph Rohrer and Sandra Noser for help with fieldwork; Hans-Rudolf Roth, Werner Eugster, Massimo Merlini and Rahel Liesch (ETH Zurich) for statistical consulting; Joel Meier and Hansjoerg Kull (Syngenta Crop Protection Dielsdorf) for useful advise on survey design; Muhammad Awais Khan (ETH Zurich), Kathrin Tschudi-Rein, and two anonymous reviewers for helpful comments on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by P. Arús.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table S1
Climatic characterization of the three study sites during the observation period (March to August) in the 2 years of survey, and comparison to standard valuesa (DOC 33.5 KB)
Table S2
Distribution patterns of aphids (Dysaphis plantaginea, D. cf. devecta, Aphis pomi) on individual apple trees characterized by the index of dispersion (I D). All I D values significantly greater than the χ 2 statistic (0.025 probability level) with (ν-1) degrees of freedom indicate an aggregated distribution of the studied species (P < 0.001 for all observed I D values) (DOC 15.5 KB)
Table S3
Relationship between the aphid species Dysaphis plantaginea, D. cf. devecta, and Aphis pomi assessed by Spearman’s rank test at the Ticino, Valais, and Zurich site. The average aphid infestation at a study site and within a year was used for correlation analysis. Significant correlations after FDR correction are highlighted in bold. The number of colonies (D. plantaginea), the number of red-curled leaves (D. cf. devecta) and the number of aphids (A. pomi) was taken as measure of aphid infestation (DOC 43.0 KB)
Table S4
Apple varieties with reported resistance properties to the three aphid species Dysaphis plantaginea, D. cf. devecta and Aphis pomi (DOC 39.0 KB)
Figure S1
Spatial distribution of aphids (Dysaphis plantaginea, D. cf. devecta and Aphis pomi) described by fitting trend surfaces on contour plots by kriging (best unbiased generalized least squares estimation) with an exponential covariance function. Mean aphid infestation of different observations within a year was considered for contour plots (cf. Table 1). Infestation level of individual trees is reflected by circle size. Yellow circles indicate trees with an infestation greater than zero, whereas uninfested trees are represented by open circles. No contour lines were plotted for the Valais site due to the orchard design with two rows (PDF 250 KB)
Rights and permissions
About this article
Cite this article
Stoeckli, S., Mody, K., Gessler, C. et al. QTL analysis for aphid resistance and growth traits in apple. Tree Genetics & Genomes 4, 833–847 (2008). https://doi.org/10.1007/s11295-008-0156-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-008-0156-y