Abstract
Prosopis species forests in Argentina are increasingly fragmented in the last years mainly by the deforestation activity without any reforestation strategy, the establishment of different crop plantations, and natural fires. The consequence of habitat fragmentation on the genetic potential of Prosopis alba requires a fine-scale analysis of population structure, in particular mating system and pollen dispersal. By means of short sequences repeats, we analyzed a fragmented population of this species in Santiago del Estero (Argentina). Most genetic variation was observed among families within zones (65.5%), whereas the lowest proportion corresponded to the differentiation among zones (2.8%). The fine analysis of structure at family level suggests that this population is complete outcrosser and there is a low but significant biparental inbreeding. Outcrossing rates differ among mother plants and the proportion of full sibs within mother plants ranged from 64% for seeds proceeding from the same pod to 10% for seeds from different pods. The average pollen dispersal distance was estimated to be among 5.36 and 30.92 m by using the KinDist or TwoGener approach. About seven pollen donors are siring each progeny array and the number of seed trees necessary for seed collection aiming to retain an effective population size of 100 was estimated in 16–39 individuals depending on the relatedness estimator used. Pollen and seed dispersal would be limited, what determines the need of conserving short distant patches to avoid the effects of inbreeding and drift within populations as a consequence of intensive use resource for agriculture.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Information on the patterns and the distance of pollen dispersal is fundamental to the development of strategies for conservation in situ and ex situ. Prosopis species (algarrobos) forests in Argentina are increasingly fragmented in the last years. This is especially notorious in the Chaco region, where most of the original Prosopis forests have been already lost (Fernández and Busso 1997; Secretaría de Ambiente y Desarrollo Sustentable de la Nación 2004; FAO 2007). The main causes that should be taken into account are the deforestation activity without any reforestation strategy, the establishment of different crop plantations, and natural fires. The most important damage is caused by irrational deforestation stimulated by the multiple uses of “algarrobo” wood (Roig 1993; Verdes 2007) and the extension of agricultural areas. The consequence of these activities is the desertification of wide areas, endangering these tree species.
Habitat fragmentation reduces areas of continuous forest to small, separate remnants and may decrease the effective size of tree populations (Cascante et al. 2002), disrupt the mating system, and interrupt gene flow (Jump and Penuelas 2006). When populations become genetically isolated, they are at risk of losing the genetic diversity that is critical to their long-term survival (Sork and Smouse 2006). According to the classical view, as a consequence of the isolation, the immediate loss of alleles happens due to the reduction of the population, with the consequence of inbreeding, population divergence increase, and genetic diversity reduction within population patches (Young et al. 1996; Lowe et al. 2005). Relatively recent reviews (Aguilar et al. 2008; Eckert et al. 2008) describe a significant reduction in genetic variability in remnant fragmented populations. However, the longevity of the trees and effective seed and pollen dispersal can enhance their resistance to the negative effect of the forest fragmentation (Hamrick 2004; Jump and Penuelas 2006). Indeed, Kramer et al. (2008) claim that population genetics theory may be misapplied in the case of forest trees and that the actual impact of forest fragmentation on genetic structure depends on a number of ecological factors including the abilities and behavioral characteristics of pollen and seed dispersal vectors.
In Argentina, two groups of “algarrobos” are differentiated by local people in terms of their uses. They are commonly recognized by pod color and shape as white (Prosopis alba and Prosopis chilensis) and black algarrobos (Prosopis flexuosa and Prosopis nigra). Issues relative to uses, conservation status, and actual geographic distribution are different among these groups. Black algarrobos are widely distributed, whereas white algarrobos are found in more humid areas or behave as phreatophyte. The first group is prone to be attacked by borer insects and therefore, they are discarded for furniture, restricting their use to firewood, poles, and stakes. The second group, preferred by the furniture industry because of the highest wood quality (Pometti et al. 2009; 2010), represents an almost exhausted resource, being reduced to a few forests in different provinces of Argentina. In particular, the area Machagai–Quitilipi, Chaco province, which used to be an important timberland center is going to stop sawmill activity in a near future as a consequence of resource extinction. In Santiago del Estero province, forests able to be used for furniture industry are almost absent by now. White algarrobos are restricted to small areas (protected between rivers), road sides, or maintained as small patches in ranches to provide shade to livestock.
The effects of habitat disturbance on the genetic properties of populations are highly dependent on the mating and dispersal mechanism. The distance of pollen movement is a significant determinant of the effective pollination neighborhood for a plant population (Austerlitz and Smouse 2002), which is especially important in calculating the number of seed trees for seed collection as part of ex situ conservation and reforestation programs. Choosing populations that carry most of the species genetic variation is essential in in situ conservation programs, and this choice should be based on population structure analyses. Furthermore, the effective population size can be quiet difficult to predict because it depends not only on the census number but also on the inbreeding coefficient, variation in fertility, nonrandom mating, and fluctuation population sizes (Crow and Kimura 1970).
Available information on population structure of P. alba and other related species (Ferreyra et al. 2007, 2010) indicated that most of genetic variability occurs within populations and regional differentiation is limited. A previous study on the mating system in seven species of Prosopis (Bessega et al. 2000) indicated that they are mostly outcrossers, but information about the genetic structure at a fine scale and pollen dispersal is lacking. Seeds are dispersed endozoically by small mammals (Reynolds 1954; Mares et al. 1977), and pollen is dispersed by insects (Genisse et al. 1990), both conditions usually associated to limited dispersal. However, dispersal patterns may have been highly modified after the European colonization by the introduction of livestock and the honey bee Apis mellifera which might have increased dispersal range. In this paper, we sampled a population of P. alba in Santiago del Estero (Argentina) that represents a remnant of an ancient extended forest in order to evaluate population structure, pollen dispersal distance, and mating system parameters. The hypotheses to be contrasted are (1) the fragmentation prevents gene flow between remnant population patches and the long-term consequence would be a reduction of population diversity and an increase of inbreeding within populations and differentiation among populations; or (2) gene flow might still occur following a stepping stone model with scattered trees or small patches acting as bridges, contributing to the metapopulation viability; this condition added to the long life cycle of P. alba trees would delay the genetic effects of fragmentation.
Material and methods
Study species
P. alba is included in the section Algarobia (Burkart 1976), which comprises ca. 28 species very important economically and ecologically. It is a native tree that grows naturally as secondary tree species in the semiarid region called Chaco, in northern Argentina. It may reach up to 18 m tall and 70–150 cm of trunk diameter. It is an important nitrogen-fixing tree adapted to the semiarid regions. The tree provides fuelwood and dimensionally stable lumber for flooring and furniture; its pods that have high sugar content are consumed by livestock and humans. P. alba has been one of the most heavily harvested species. In the last years, the high use of P. alba wood by the industry yielded a qualitative and quantitative diminution of the populations; the best trees are harvested and the deforestation rate is higher than the grow rate and regeneration.
Flowers are protogynous and are grouped in inflorescences consisting of 7–11 cm long spikes. Pollination in Prosopis is made by insects (Genisse et al. 1990) and seed dispersal is endozoic (Reynolds 1954, Mares et al. 1977). The leaves are pinnately compound with 35 leaflet pairs per pinnae in average (Bessega et al. 2009). The ripe fruits are indehiscent and usually range 12–25 cm long, 12–18 mm wide, and yellow colored. The dried fruits fall from the tree at maturity and are readily eaten by nearly all herbivorous.
The studied population
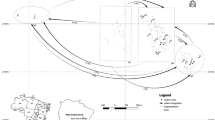
The sampled population of P. alba is located near Fernandez, Santiago del Estero, Argentina (Fig. 1). This population has been a fairly continuous forest of several square kilometers that now has a patchy distribution embedded in an agricultural matrix. Forest cover in this area has been reduced by deforestation and developing of soybean, alfalfa, onion, oat, and cotton, or invaded by Bermuda grass (Cynodon dactylon) as shown in Fig. 1. Taking into account habitat fragmentation, the sampling area was divided into three zones: Ruta 34, Tomey, and Rojas.
Spatial distribution of the sampled Prosopis alba trees (M1–M16). M1, M2, and M16 belong to RUTA 34; M3–M10 belong to ROJAS; and M11–M15 are from TOMEY. D Dismounted, S soy, C cotton, A alfalfa, O oat, On onnion, BG Cynodon dactilon = bermuda grass, EEF Estación Experimental Fernandez (Eucaliptus sp., Casuarina sp., Populus sp., Prosopis sp., Cynodon dactilon, Sorghum halepensis)
Sixteen mother plants were sampled. In all cases, the trees were separated at least 50 m. from each other. About 50 pods were collected from each mother plant. All seeds from each pod were stored in single bags. From each of these bags, seeds were randomly sampled for the short sequence repeats (SSR) analysis of each progeny array. The number of seeds analyzed per mother plant (family array size) was 11.
DNA extraction and genotyping
Leaf material collected in the wild population was dried immediately using silica gel and stored at room temperature until DNA extractions were completed. Genomic DNA was extracted both from the leaves of adult individuals and from 2- to 5-day-old seedling cotyledons using DNA easy Plant mini kit (QIAGEN Inc., Valencia, CA, USA) following the manufacturer’s instructions.
Six SSR that had been developed in P. chilensis exhibits cross-species amplification (Mottura et al. 2005) and shown to be amplified successfully in P. alba (Bessega et al. 2009) were analyzed. Forward primers (website, biomath.trinity.edu/; search term, Prosopis) were labeled with a fluorescent dye (Invitrogen); three loci were labeled with 6-FAM and three with HEX. The PCR amplifications were carried out in a 50 μl reaction volume containing 10–30 ng DNA, 0.6 μM each primer, 0.2 mM dNTPs, 0.3 U Taq DNA polymerase (Invitrogen), and 1.5 mM MgCl2. A MyCycler Thermal Cycler (BioRAD Laboratories) was used for amplifications where the cycling profile was: initial denaturation at 94°C for 5 min followed by 30 cycles at 94° for 45 s denaturation, primer-specific annealing temperature (56°–59°) for 45 s and at 72°C for 45 s extension, and a final extension step at 72° for 10 min. PCR products were electrophoresed in an ABI313XL (HITACHI) automated DNA sequencer and automatically sized using GENEMARKER ver 1.91 (SoftGenetics LLC™ www.softGenetics.com).
Statistical analyses
Analyses of genetic variation and population structure
Possible genotypic associations among the studied loci were tested using the program Multilocus 1. 3 (Agapow and Burt 2001) by the index of association (I A ) and a slightly modified statistic which is independent of the number of loci (r d ). In brief, the "distance" (number of loci at which they differ) between all pairs of individuals is calculated, and the variance of these distances compared to that expected if there is no linkage disequilibrium.
Genetic variation was quantified by means of the number of detected alleles per locus (A), effective number of alleles (N e ), observed (H o ), and expected heterozygosity under Hardy–Weinberg equilibrium (H e ). The distribution of genetic variation was analyzed by nonhierarchical and hierarchical F statistics and analysis of variance components using the software FSTAT (Goudet et al. 1995) and the package HIERFSTAT (Goudet 2005) for R software (R Development Core Team 2011).
The levels considered in the hierarchical analysis were: zone, family, pod, and individual. The fixation index (F IS) was estimated in adult and offspring samples. The significance of the nonhierarchical F ST was estimated using the likelihood ratio G statistic (with 2,000 replicates). Confidence intervals for variance components and F statistics were obtained by bootstrap with 2,000 pseudoreplicates. The significance of each level of the hierarchical F statistics was evaluated by between, between-within, and within F test.
Fine analysis of structure at family level
The sample collected is composed of individuals involving different relationship classes. As mother plants were separated more than 50 m from each other, they, as well as their progeny, are assumed as nonrelated following the Saidman and Vilardi (1993) criterion, which is based on field observations (Palacios, personal communication) that trees isolated by 50 m or larger distance do not produce fruits. As seeds proceed from open pollination, the progeny obtained from each mother tree should involve half sibs, full sibs, self half sibs, and self full sibs (Squillace 1974).
Three approaches were applied in order to asses mating system parameters, pollen dispersal and the structure of each family group.
-
1.
Mating system
Estimates of multilocus (t m ) and mean single locus (t s ) outcrossing rates, correlation of tm within progeny arrays (r t ), the correlation of outcrossed paternity (r p ), and fixation index of maternal parents were calculated using the MLTRwin software (Ritland 2002) This program is based on the multilocus mixed-mating model and the estimation procedure of Ritland and Jain (1981) which assumes that progeny are derived from either random mating (outcrossing) or self-fertilization. The estimation of mating system parameters was made by the expectation–maximization method to assure convergence and 10,000 bootstraps were used to get standard errors for each parameter. Biparental inbreeding was estimated following Ritland (1990) as t m − t s . The effective number of pollen donors over all mothers trees were estimated under sibling pair model (Ritland 1989) by the relation effective number of pollen donors N ep = 1/r p .
In a first analysis, all seeds within each family were considered a single group and all parameters were estimated. As seeds within mother trees are grouped in pods, a second analysis was conducted considering the pod as an additional level within family.
-
2.
Estimation of pollen dispersal parameters based on assumed pollen distribution curves
Pollen dispersal parameters were fit using two indirect methods based on the spatial genetic structure of the pollen pool: TwoGener (Smouse et al. 2001; Austerlitz and Smouse 2002) and KinDist (Robledo-Arnuncio et al. 2006) using the POLDISP 1.0 software (Robledo-Arnuncio et al. 2007). The TwoGener method is based on the relationships between physical distances among a sample of mother trees and pairwise intraclass genetic correlations (ΦFT) for the pollen pools sampled by these mothers. To fit pollen dispersal using the TwoGener approach, an independent estimate (or a joint estimation) of the effective density of pollen donors (de) is needed (Austerlitz and Smouse 2002), which can complicate the accurate estimation of two parameters. In addition, this method assumes uniformity in male fecundity and synchronous flowering. The KinDist method is based on a normalized measure of correlated paternity (ψ) between female pairs (Robledo-Arnuncio et al. 2006). The estimates of pollen dispersal parameters obtained by KinDist are generally more accurate than those from TwoGener because KinDist does not rely on effective density, a parameter, which is very difficult to measure in the field. Joint estimation of de with TwoGener is associated with the usual statistical problems of increasing the dimensionality of the parameter space. The KinDist method is also robust for the levels of male fecundity variation and asynchronous flowering commonly observed in forest tree populations (Robledo-Arnuncio et al. 2006)
The estimate of the pollen structure parameter due only to limited pollen dispersal (Φ FT) was corrected using the formula described in Austerlitz and Smouse (2001):
where, F is the adult (mother tree) inbreeding coefficient. The corrected value was used to estimate N ep under the pollen structure model by the relation:
The estimate of Φ FT was also used to further calculate the derivative measure of the average distance of pollination (δ) using normal and exponential curves with the Subprogram TwoGener.
If we assume a bivariate normal distribution of pollen dispersal, the estimate of pollen structure Φ FT is inversely related the variance of pollen dispersal (σ2) and reproductive adult density (d),
Given that Φ FT is known from genetic data and d from ecological observation, it follows that the variance in pollen dispersal can be computed from the expression above.
Next, σ2 is used to estimate the effective number of pollen donors as:
The effective neighborhood pollination area (A EP) is:
\( {A_{\text{EP}}} = 4\pi {\sigma^2} \) (Austerlitz and Smouse 2001)
-
3.
Average coancestry coefficient (θ) and variance effective population size
We obtained Ritland (1996) and Queller and Goodnight (1989) relatedness estimator (r) for all the individual pairs using GENALEX (Peakall and Smouse 2006). From this estimation, we calculated the coancestry (θ xy ) matrix following the equation \( {r_{{xy}}} = 2{\theta_{{xy}}} \)(Lynch and Walsh 1998). We also estimated pairwise coancestry (θ xy ) coefficients using the Nason (Loiselle et al. 1995) method with the software SPAGeDI (Hardy and Vekemans 2002). We calculated within family θ as the average of θ xy for individuals belonging to the same family, the average of θ within family, and for the whole sample.
The variance effective population size (N ev) for each family was calculated by:
(Cockerham 1969)
where, n is the size family and F o is the inbreeding coefficient in offspring. The total variance effective population size of progeny arrays (N e(t) ) was estimated using the same equation with θ t instead of θ.
The number of seed trees (m) necessary for seed collection was calculated by the method of Sebben (2006) based on the relationship between the proposed effective population size of the conservation program (N e(reference)) and the average N ev:
(Sebben 2006)
Analysis of spatial genetic structure
Correlation between the Ritland (1996), Queller and Goodnight (1989), and Nason (Loiselle et al. 1995) relatedness matrices and the spatial distance matrix was tested using a Mantel (1967) test (with 2,000 permutations) using the package ade4 of R software (R Development Core Team 2011; www.r-project.org).
The association between relatedness and geographic distance was also evaluated using an additive generalized model (gam) using the package mgcv (Wood 2001) of R. Upper and lower confidence limits for the 95% about the null hypothesis of no spatial structure were plotted based on 2,000 permutations of the data set.
Results
Genetic diversity
The genotypes for six loci were determined for 12 individuals of each of the 16 families (making a total of 192). All loci were polymorphic with three to nine (average, 6.5) alleles per locus with a total of 39 alleles. Multilocus linkage disequilibrium coefficients I A = 0.041 and r d = 0.0084 were nonsignificant (P = 0.13), indicating that the studied loci are in linkage equilibrium. The allelic richness varied from 2.48 to 5.77 (3.9), the observed heterozygosity ranged from 0.169 to 0.688 (mean, 0.460; Table 1). Expected heterozygosity within families (H S ) varied between 0.155 and 0.584 (mean = 0.394). Although individual tests of Hardy–Weinberg departures were nonsignificant, multilocus F IS estimate (−0.169) was significant (CI95 = −0.194/−0.133) indicating a significant heterozygote excess in the whole population.
Genetic structure
Heterozygote observed frequencies in the adult (mother) population did not differ from expected (F IS = −0.068, CI95 = −0.205/0.200). Genetic structure of progeny generation was studied through a nonhierarchical and a hierarchical (Table 2) analysis of variance. The first analysis showed a significant heterozygote excess within families (F IS = −0.14, CI95 = −0.162/−0.116) and a highly significant differentiation among families (F ST = 0.172, CI99 = 0.093/0.243).
The hierarchical analysis (Table 2) indicated that the genetic differentiation among zones was nonsignificant, whereas the differentiation among families (within zones) and pods (within families) were highly significant. Expressed as proportion, most of genetic variation was observed among families within zones (65.5%), whereas the lowest proportion corresponded to the differentiation among zones (2.8%; Table 2). A highly significant heterozygote excess was observed within pods.
Mating system and pollen disperal
The analysis of the mating system showed an almost complete outcrossing rate (Table 3). The difference t m − t s was low but significant indicating a low proportion of mating among relatives. The correlation of t m within progeny arrays (r t ) was high, suggesting that outcrossing rates differ among mother plants. The correlation of outcrossed paternity was evaluated at three levels: global, among fruits, and within fruits. In all cases, r p was significantly higher than 0. The r p estimate was maximum for sib sampled from the same fruit (r p = 0.638) but for different fruits on the same plants the value dropped in about 85% (0.098/0.638). These results indicate that the proportion of full sibs is about 64% for seeds proceeding from the same pod, whereas this proportion is only about 10% for seeds from different pods. Effective number of pollen donors, estimated as (1/r p ) indicated that about six pollen donors are siring each progeny array (Table 3). Pollen pool structure analysis with POLDISP gave a Φ FT estimate of about 10%, which translated into an effective N ep represents about five pollen donors per mother plant (Table 3).
Applying KinDist, the average pollen dispersal distance was estimated in 5.36 m when assuming a normal distribution and 11.96 when assuming exponential one of the pollen dispersal distance curve. The model based on exponential distribution curve fits only slightly better than the normal one as described by least square residuals of 26.51 and 26.43, respectively. Therefore, both the normal and exponential curves were used for further analysis using TwoGener.
The estimates made by TwoGener yields estimates of pollen distances a little higher than those obtained with KinDist. Considering a normal distribution, pollen dispersal distances varied depending on the estimation method between 13.74 and 23.74 m. When the same estimates were done using exponential curves, distance estimates varied between 15.5 and 30.92 m.
Effective densities of pollen donor was de = 0.0010 and 0.0007 m2 for both kinds of curves. These estimates are 31% and 24%, respectively, lower than d counted in the field. Finally, the A ep was estimated in 0.150 ha, that is 22 m radiuses circle around each maternal tree.
Number of pollen donors, paternity correlation, coancestry, variance of effective population, and number of seed trees necessary for seed collection
The effective number of N ep, r p , coancestry coefficient θ, and the variance effective size of open pollinated families (N e ) were estimated for each of the 16 families (Table 4). The average paternity correlation was 0.1896 (CI95 = −0.061/0.44), which represents a mean effective number of pollen donors of about seven. The average coancestry coefficient within families estimated from Ritland's (1996) relatedness estimator (θ = 0.053, CI95 = −0.007/0.113) was lower than the expected for panmictic population (θ = 0.125). The estimate obtained by Queller and Goodnight (1989) and Nason methods were higher (respectively, θ QG = 0.168 CI95 = 0.029/0.307 and θ N = 0.165 CI95 = −0.007/0.336) and closer to the expected value. The average variance effective population size estimated from the coancestry coefficients were respectively 8.1, 2.83, and 3.26 for Ritland, Queller and Goodnight, and Nason. The number of seed trees necessary for seed collection aiming to retain an effective population size of 100 was estimated at the minimum of 16–39 individuals depending on the method θ was estimated (Table 4).
Spatial genetic structure
The pairwise Ritland (1996), Queller and Goodnight (1989), and Nason (Loiselle et al. 1995) relatedness matrices were compared with the corresponding geographic distance matrix by mantel test. In all cases, the correlation was significant (R 2 = −0.11, P = 0; R 2 = −0.066, P = 0.013; and R 2 = −0.105, P = 0, respectively, for RI, QG, and N). The plot of relatedness as function of geographic distance shows a deep decrease at short distances, following a gam, suggesting that the relatedness is null for distances over 100 m (Fig. 2).
Discussion
Detrimental effects of habitat fragmentation on species survival are largely dependent on gene flow between spatially isolated populations (Lander et al. 2010). Genetic isolation would result in inbreeding depression, genetic drift, and loss of adaptive potential (Young et al. 1996; Lowe et al. 2005). However, due to their long life cycle, demography, ecology, and reproductive biology, the response of forest tree species to fragmentation may exhibit certain peculiarities in comparison with the expectations according to classical theoretical models (Kramer et al. 2008). Information on the mating systems and parameters of pollen dispersal is paramount to predict the consequence of fragmentation on native forest species.
A fragmented population of the “algarrobo blanco”, P. alba was analyzed in order to study population structure, pollen dispersal distance, and mating system parameters. A significant heterozygote excess (F IS < 0) was found within families suggesting that the pollen cloud differs genetically from the corresponding mother tree. This heterozygote excess suggests high outcrossing rates (see below) and would contribute to reduce the effect of fragmentation on the reduction of genetic variability and increase of inbreeding.
The effect of fragmentation on the distribution of genetic diversity was evaluated by a hierarchical population structure analysis. The differentiation among the three zones recognized in the population (F zone/total) was nonsignificant, whereas the differentiation among families (within zones; F family/zone) and pods (within families; F pod/family) were highly significant. Within pods, a highly significant heterozygote excess was observed (F individual/pod). This result indicates that although pollen dispersal insures that allele frequencies differ between mother plants and pollen donors producing negative F IS and F pod/family estimates, pollen dispersal is not enough as to prevent the differentiation among families within zones. The lack of differentiation among zones is evidence that the genetic variation preserved within each zone is not different up to now from that of the original unfragmented population, most probably due to the fact that fragmentation is a process relatively recent in terms of number of generations, considering that average life span of P. alba is over 100 years.
Further than the evidence from heterozygotes excess within progenies, the relative importance of family structure and outcrossing can be weighed by estimating mating system parameters. The population analyzed is characterized by a complete outcrossing (t m = 1). The low (roughly 5%), but significant, degree of biparental inbreeding (t m − t s ) observed may be caused by the seed dispersion system. Prosopis species have an endozoic seed dispersal system mediated by native small herbivorous and livestock (Mares et al. 1977; Hafez 1962). Although livestock are able to disperse seeds over larger distances (up to 4–6 km) in comparison with small rodents (approximately 50 m; Reynolds 1954, Keys 1993; Keys and Smith 1994), in all cases herbivorous tend to eat as many pods as they can from each single tree, and scarified seeds after passing through the animal gut are deposited in dungs forming groups of related individuals (Keys 1993; Bessega et al. 2000). As an example in Prosopis glandulosa, 75% of the cattle dung pats contained an average of 4.2 seedlings per pat (Brown and Archer 1987). Therefore, a relatively high relatedness is expected for neighbor trees, which are most probably involved in reciprocal matings.
The number of pollen donors siring each mother plant was near six in average (the number varies depending on the estimation method), but there is a great variation occurring among mother plants. Difference in the number of pollen donors per mother plant may be associated to demographic causes. In low-density areas, a lower number of fathers per family is expected respect to more dense regions. This trend was not observed in the present study if the N ep is compared among zones 1–3. Another cause for differences in N ep estimates might be the family structure. If the trees in the neighborhood are highly related, underestimation of the actual number of pollen donors might occur because of the increased difficulty of differentiating pollen donors with similar genotypes.
The joint analysis of outcrossing rates, correlation of outcrossed paternity within r p at different levels (global, among, and within fruits), the correlation of outcrossing rate within progeny arrays (r t ) and average distance of pollination yielded several conclusions.
First, the proportion of full sibs is about 64% for seeds proceeding from the same pod whereas this proportion is only 10% for seeds from different pods. This may be consequence of the pollinator insect behavior and inflorescence anatomy. The pollinator species associated to Prosopis (A. mellifera, Meliponini sp., Xylocopa ordinaria, Xylocopa splendidula, Caupolicana ruficollis, Caupolicana mendocina, Colletini sp.; Genisse et al. 1990) usually focus their foraging efforts on densely flowering plants and limit their interplant movement to nearest neighbors (Levin and Kerster 1974). Consequently, each pollination event involves pollen from only one or a few pollen donors fertilizing each inflorescence, favoring the occurrence of full sibs in each pod. By contrast, different inflorescences would be fertilized by different insects determining that seeds from different pods tend to be half sibs.
Second, the average distance of pollination estimated was from 5 to 30 m depending of the method used. This range agrees with the field observation by Palacios (personal communication) that trees isolated by 50 m from other trees do not produce fruits. Pairwise relatedness was also observed to decrease steeply with distance. According to Dick et al. (2008), a fine-scale spatial genetic structure characterized by a decrease of relatedness between individuals with distance is often interpreted as limited seed dispersal. Under scenarios of long distance seed dispersal with limited pollen flow, there should be no structure because closely related individuals (FS or HS) will be broadly spaced. On the other hand, restricted seed dispersal is always expected to produce a fine-scale genetic structure even if pollen movement is random and long distance because sibs will be aggregated. In P. alba, limited pollen and seed dispersal would account for fine-scale genetic structure.
Third, limited pollen dispersal can produce an increase in the coancestry coefficients within progenies reducing the variance effective population size and a small neighborhood pollination area. The estimated effective neighborhood area of pollination was small, representing a 22 m radius circle around each maternal tree. Coancestry coefficient within families estimated by Ritland (1996; θ xy = 0.05) was much lower than the minimum expected for sibs families (θ xy = 0.125), and the corresponding variance effective population size of open pollinated families was N ev = 8.10, twice as large as the maximum theoretically expected N ev (4). By contrast, Queller and Goodnight (1989) and Nason (Loiselle et al. 1995) methods produced estimates much closer to the expected. The corresponding values of θ xy (0.168 and 0.165) are very similar to each other and agree with the observation of a significant proportion of full sibs within progenies, giving a more conservative N ev (2.83 and 3.26). With our sample, no significant differences were observed between Nason and Queller and Goodnight methods in spite of the fact that the latter assumes HW equilibrium and the former does not.
Fourth, the results here presented have important implications for seed collection strategies for ex situ conservation, tree breeding, and reforestation of P. alba. The spatial genetic structure at short distances indicates that relatedness at distances further than 100 m is negligible. This means that this distance would be enough to avoid duplications in the sampling due to relatedness among harvested seed trees. Seeds for ex situ conservation or tree breeding programs should be collected from trees at least 100 m apart would reduce relatedness among harvested seed trees. An additional caution to be considered is that seeds collected from each mother plant should be taken from different pods to reduce the proportion of full sibs within progeny arrays. Finally, considering that each progeny array represents approximately three non-inbred individuals, the sample aiming to retain an effective population size of 100 should include at least 38 seed trees.
Finally, according to the present results, both pollen and seed dispersal in P. alba would be limited, however, the differentiation among zones was relatively low. Previous works on P. alba geographically distant populations (Ferreyra et al. 2004, 2007, 2010) have also shown that most genetic variation occurs within populations and that the differentiation among populations and regions is rather low. The ability to maintain high genetic coherence in spite of limited dispersal ability may be associated with the long life span of Prosopis species together with large populations’ sizes and continuous distribution that characterized forests in the Chaqueña Region before the habitat fragmentation produced by human activities. Current patchy distribution may still not show the expected effects of drift and inbreeding because of the long life span of P. alba trees. In relict populations, 300-year age trees can be found, which means that, in terms of number of generations, habitat fragmentation is too recent as to have produced statistically significant effects. Also, if patches were separated by shorter distances than the maximum that can be reached either by pollen or seed dispersal vectors, these semi-isolated might serve as stepping stones among groups of individuals which could result in a critical role in determining gene movement within fragmented landscapes (Hamrick 1994; Fuchs and Hamrick 2011). This model is consistent with the hypothesis that gene flow, following a stepping stone model with scattered trees or small patches, would delay the genetic effects of fragmentation in P. alba. Consequently, a strategy to preserve in situ diversity of P. alba compatible with a rational use of resources would imply the conservation of patches connected by pollinators and seed disperser in agro-forestry systems combined with livestock breeding.
References
Agapow PM, Burt A (2001) Indices of multilocus linkage disequilibrium. Mol Ecol Notes 1:101–102
Aguilar R, Quesada M, Ashworth L, Yvonne Herrerias D, Lobo J (2008) Genetic consequences of hábitat fragmentation in plant populations: susceptible signals in plant traits and methodological approaches. Mol Ecol 17:5177–5188
Austerlitz F, Smouse PE (2001) Two-generation analysis of pollen flow across a landscape. II. Relation between FFT, pollen dispersal and interfemale distance. Genetics 157:851–857
Austerlitz F, Smouse PE (2002) Two-generation analysis of pollen flow across a landscape. IV. Estimating the dispersal parameter. Genetics 161:355–363
Bessega C, Ferreyra L, Julio N, Montoya S, Saidman BO, Vilardi JC (2000) Mating system parameters in species of genus Prosopis (Leguminosae). Hereditas 132(1):19–27
Bessega C, Saidman B, Darquier MR, Ewens M, Sánchez L, Rozenberg P, Vilardi JC (2009) Consistency between marker- and genealogy-based heritability estimates in an experimental stand of Prosopis alba (Leguminosae). Am J Botany 96:458–465
Brown JR, Archer S (1987) Woody plants seed dispersal and gap formation in North America subtropical savanna woodland: the role of domestic herbivores. Vegetatio 73:73–80
Burkart A (1976) A monograph of the genus Prosopis (Leguminosae subfam. Mimosoidae). Journal Arnold Arboretum 57:219–249
Cascante A, Quesada M, Lobo JJ, Fuchs EA (2002) Effects of dry forest fragmentation on the reproductive success and genetic structure of the tree Samanea saman. Conservation Biology 16:137–147
Cockerham CC (1969) Variance of gene frequencies. Evolution 23:72–84
Crow JF, Kimura M (1970) An introduction to populations genetics theory. Harper and Row, London
R Development Core Team (2011) R: a language and environment for statistical computing. R Foundation for Statistical Computing: Vienna, Austria. ISBN 3-900051-07-0. Available from: http://www.R-project.org
Dick CW, Hardy OJ, Jones FA, Petit RJ (2008) Spatial scales of pollen and seed-mediated gene flow in tropical rainforest trees. Tropical Plant Biol 1:20–33
Eckert CG, Samis KE, Lougheed SC (2008) Genetic variation across species geographical ranges: the central marginal hypothesis and beyond. Mol Ecol 17:1170–1188
FAO (2007) Situación de los Bosques del Mundo, Roma, Italia. http://www.fao.org/docrep/009/a0773s/a0773s00.htm
Fernández OA, Busso CA (1997) Arid and semi-arid rangelands: two thirds of Argentina. RALA Report: 200.
Ferreyra LI, Bessega C, Vilardi JC, Saidman BO (2004) First report on RAPDS patterns able to differentiate some Argentinean species of Section Algarobia (Prosopis, Leguminosae). Genetica 121(1):33–42
Ferreyra LI, Bessega C, Vilardi JC, Saidman BO (2007) Consistency of population genetics parameters estimated from isozyme and RAPDs dataset in species of genus Prosopis (Leguminosae, Mimosoideae). Genetica 131:217–230
Ferreyra LI, Vilardi JC, Tosto D, Julio N, Saidman BO (2010) Adaptive genetic diversity and population structure of the “algarrobo” [Prosopis chilensis (Molina) Stuntz] analysed by RAPD and isozyme markers. Eur J Forest Res 129:1011–1025. doi:10.1007/s10342-010-0386-z
Fuchs EJ, Hamrick JL (2011) Mating system and pollen flow between remnant populations of the endargered tropical tree, Guaiacum sanctum (Zygophyllaceae). Conserv Genet 12:175–185
Genisse J, Palacios RA, HOC PS, Carrizo R, Moffat L, Mom MP, Agullo MA, Picca (1990) Observaciones sobre la biologia floral de Prosopis (Leguminosae, Mimosoidae). I1 Fases florales y visitantes en el distrito Chaquefio Serrano. Danviniana 30:71–85
Goudet J (2005) Hierfstat, a package for R to compute and test hierarchical F statistics. Mol Ecol Notes 5: 184–186. http://www.unil.ch/popgen/softwares/hierfstat.htm.
Goudet J, Raymond M, DeMeeus T, Rousset F (1995) Testing differentiation in diploid populations. Genetics 144:1933–1940
Hafez ESE (1962) The behavior of domestic animals. Williams and Wilkins, Baltimore, MD
Hamrick JL (1994) Genetic diversity and conservation in tropical forest. In: Drysdale RM, JS Yapa A (eds) Proceedings on International Symposium on Genetic Conservation and Prosuction of Tropical Forest Tree Seed. Asia–Canada Forest Tree Seed Centre, Muack-Lek, Saraubi, Thailand. pp 1–9.
Hamrick JL (2004) Response of forest tree to global environmental changes. Forrest Ecol and Manag 197:323–335
Hardy O, Vekemans X (2002) SPAGeDI: a versatile computer program to analyze spatial genetic structure at the individual or population levels. Mol Ecol Notes 2:618–620
Jump AS, Penuelas J (2006) Genetic effects of chronic habitat fragmentation in a wind-pollinated tree. Proc Natl Acad Sci USA 103:8096–8100
Keys RN (1993) Mating systems and pollination biology of velvet mesquite (Prosopis velutina Wooten). Thesis dissertation. University of Arizona
Keys RN, Smith S (1994) Mating system parameters and population genetic structure in pioneer populations of Prosopis velutina (Leguminosae). Am J Bot 81:1013–1020
Kramer AT, Ison JL, Ashley MV, Howe HF (2008) The paradox of forest fragmentation genetics. Conserv Biol 22:878–885
Lander TA, Boshier DH, Harris SA (2010) Fragmented but not isolated: contribution of single trees, small patches and long-distance pollen flow to genetic connectivity for Gomortega keule, an endangered Chilean tree. Biol Conserv 143(11):2583–2590. doi:10.1016/j.biocon.2010.06.028
Levin DA, Kerster HW (1974) Gene flow in seed plants. Evol Biol 7:139–220
Loiselle BA, Sork VL, Nason J, Graham C (1995) Spatial genetic structure of a tropical understory shrub, Psychotoria officinalis (Rubiaceae). Am J Bot 82:1430–1425
Lowe AJ, Boshier D, Ward M, Bacles CFE, Navarro C (2005) Genetic resource impacts of habitat loss and degradation; reconciling empirical evidence and predicts theory for neotropical trees. Heredity 95:255–273
Lynch M, Walsh B (1998) Genetics and analyses of quantitative traits. Sinauer, Sunderland, MA
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
Mares MA, Enders FA, Kingsolver JM, Neff JL, Simpson BB (1977). In: Simpson BB (ed) Mesquite: its biology in two desert ecosystems, US/IBP synthesis Series. Hutchinson and Ross: Stroudsburg 7: 124–149
Mottura MC, Finkeldey R, Verga AR, Gailing O (2005) Development and characterization of microsatellite markers for Prosopis chilensis and Prosopis flexuosa and cross-species amplification. Mol Ecol Notes 5:487–489
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Pometti CL, Pizzo B, Brunetti M, Macchioni N, Ewens M, Saidman BO (2009) Argentinean native wood species: physical and mechanical characterization of some Prosopis species and Acacia aroma (Leguminosae; Mimosoideae). Bioresour Technol 100(6):1999–2004
Pometti CL, Palanti S, Pizzo B, Charpentier JP, Boizot N, ResioC SBO (2010) Durability of five native Argentine wood species of the genera Prosopis and Acacia decayed by rot fungi and its relationship with extractive content. Biodegradation 21(5):753–760
Queller DC, Goodnight KF (1989) Estimating relatedness using genetic markers. Evolution 43:258–275
Reynolds HG (1954) Some interrelations of the Merriam kangaroo rat to velvet mesquite. J Range Manag 7:176–180
Ritland K (1989) Correlated matings in the partial selfer Mimulus guttatus. Evolution 43:848–859
Ritland K (1990) A series of FORTRAN computer programs for estimating plant mating systems. J Hered 81:235–237
Ritland K (1996) Estimators for pairwise relatedness and individual inbreeding coefficients. Genet Res 67:175–185
Ritland K (2002) Extensions of models for the estimation of mating systems using n independent loci. Heredity 88:221–228
Ritland K, Jain SK (1981) A model for the estimation of outcrossing rate and gene frequencies using n independent loci. Heredity 47:35–52
Robledo-Arnuncio JJ, Austerlitz F, Smouse PE (2006) A new indirect method of estimating the pollen dispersal curve, independently of effective density. Genetics 173:1033–1045
Robledo-Arnuncio JJ, Austerlitz F, Smouse PE (2007) POLDISP: a software package for indirect estimation of contemporary pollen dispersal. Mol Ecol Notes 7:763–766
Roig FA (1993) Aportes a la Etnobotánica del Genero Prosopis. In: Contribuciones Mendocinas a la quinta Reuni6n Regional para América Latina y el Caribe de la Red de Forestación del CIID. Unidades de Botánica y Fisiología vegetal IADIZA. pp 99–121
Saidman BO, Vilardi JC (1993) Genetic variability and germplasm conservation in the genus Prosopis. In: Puri S (ed) Nursery technology of forest tree species of arid and semiarid regions. Winrock–Oxford and IBH, New Delhi, pp 187–198
Sebben AM (2006) Sistema de reproduccion en species arboreas tropicales e suas implicancacoes para la selecao de arvores matrized para reforestamentos ambientais. In: Higa AR, Silva LD (eds) Pomares de sementes en especies forestais nativas. FUPEF, Curitiba, Brasil, pp 93–138
Secretaría de Ambiente y Desarrollo Sustentable de la Nación (2004) Dirección de Bosques. Unidad de Manejo del Sistema de Evaluación Forestal, Informe sobre deforestación en Argentina
Smouse P, Dyer RJ, Westfall RD, Sork VL (2001) Two-generation analysis of pollen flow across a landscape. I. Male gamete heterogeneity among females. Evolution 55:260–271
Sork VL, Smouse PE (2006) Genetic analysis of landscape connectivity in tree populations. Landscape Ecol 21:821–836
Squillace AE (1974) Average genetic correlations among offspring from open-pollinated forest trees. Silvae Genetica 23:149–156
Verdes P (2007) Micropropagación de Prosopis caldenia BURK.: estado actual y perspectives. Revista Científica Agropecuaria 11(1):45–51
Wood SN (2001) mgcv: GAMs and generalized ridge regression for R. R News 1(2):20–25
Young A, Boyle T, Brown T (1996) The population genetic consequences of habitat fragmentation for plants. Trends Ecol Evol 11:413–418
Acknowledgments
This research was supported by funding from Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) PIP 11220090100147 and Universidad de Buenos Aires (EX 201 and 20020100100008) given to JCV and BOS. A special thanks to the Lic. Ariel Guionet from the Servicio de Secuenciación y Genotipificado from Departamento de Ecología Genética y Evolución, FCEyN, UBA.
Ethical standards
In order to fulfill the objectives proposed, the experiments conducted comply with the current Argentine laws and all the trials were made without disturbing the natural ecosystem taking into consideration the current legislation in the country. For the experimental laboratory practice, done at Facultad de Ciencias Exactas y Naturales (Universidad de Buenos Aires), we followed the recommendations from Servicio de Higiene y Seguridad (SHyS, FCEyN, UBA).
Conflict of interest
The authors declare that they have no conflict of interests.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by P. Ingvarsson
Rights and permissions
About this article
Cite this article
Bessega, C., Pometti, C.L., Ewens, M. et al. Strategies for conservation for disturbed Prosopis alba (Leguminosae, Mimosoidae) forests based on mating system and pollen dispersal parameters. Tree Genetics & Genomes 8, 277–288 (2012). https://doi.org/10.1007/s11295-011-0439-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-011-0439-6