Abstract
The “algarrobo” [Prosopis chilensis (Molina) Stuntz] is a tree species that represents an important natural resource in arid and semi-arid regions of Argentina. In this paper, we analysed and compared the variability of 46 RAPD (Random Amplified Polymorphic DNA) loci with previous estimates obtained from 12 isozyme markers in nine Argentinean populations of P. chilensis representative of the whole range of this species in Argentina. We evaluated the population structure and the existence of genetic variants associated with environmental variables. Expected heterozygosity (H e) estimated from RAPD varied significantly among populations and regions. Hierarchical analysis of genetic variability (AMOVA) showed that most (88.1%) of the total diversity occurs within populations, the component among populations within regions (9.3%) was intermediate, while the between-region component was the lowest (2.6%). All three variance components were highly significant. The MDS plot from pair-wise Φ ST matrix was consistent with the highly significant among-region differentiation indicated by the AMOVA. All 12 variable isozyme loci and 26 out of 46 RAPD loci showed significant or highly significant association with at least one geographic/climatic variable according to the stepwise multiple regression analysis. These results imply that the genetic differentiation among populations is better explained by environmental or biogeographical grounds than by geographical distances, suggesting gametic disequilibrium with loci responsible for the adaptation to particular environmental conditions. The information from RAPD markers would provide a relevant criterion to preserve genetic diversity in programmes for conservation and rationale use of this species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The effect of habitat fragmentation on genetic diversity and biodiversity is of increasing concern since more natural habitats have been modified as a result of human activities. Modern agriculture and inappropriate management of natural resources have produced an accelerated and sustained erosion of variability that leads to desertification in many areas of the world, one of the major environmental problems affecting humanity.
Fragmentation, decline or perturbation of environment can lead to genetic changes that can have adverse implications for the conservation of the species. These changes include loss of differentiated populations, alteration of differentiation between populations, loss of variation among members of the same population and changes in the level of inbreeding (Sherwin and Moritz 2000).
Some species of Prosopis genus, mainly those belonging to section Algarobia (locally known as “algarrobos”), have been subjected to unsustainable exploitation to obtain multiple products: wood, charcoal, forage, gum arabic, tannins, mucilage, ethanol and human food (Karlin et al. 1997; Pasiecznik et al. 2001). The accelerated and uncontrolled use of this important natural resource for fuel and timber has led to increasing deforestation and has caused the loss of many elite trees and the destruction of extensive Prosopis forests (Hunziker et al. 1986).
The genus includes approximately 45 species distributed in arid and semiarid regions of the Americas, Africa and West Asia. Argentina is the country with the greatest species diversity, with more than 27 species, 11 of which are endemic (Burkart 1976). The section Algarobia of this genus includes the most important species from an economic and ecological standpoint.
Prosopis chilensis is an important species within this section that exhibits high variability in morphological characters, and it has been suggested that it should be included in reforestation programmes as it is one of the fastest growing species under arid conditions in Argentina (Burkart 1976; Dutton 1989; Karlin et al. 1997). Although Burkart (1976) described three varieties, P. chilensis var. chilensis, P. chilensis var. catamarcana and P. chilensis var. riojana, different evidence lines suggest that these taxa would not be natural groups. According to Roig (1993), the differences in spine, fruit and leaf morphology are not too general, Dalmasso (1993) described six morphotypes in a single population of San Juan (Argentina), Verga (1995) suggested that P. chilensis var. riojana is actually a hybrid between P. chilensis and P. flexuosa and Vázquez-Garcidueñas et al. (2003) based on RAPD markers suggested that P. chilensis var. chilensis is more closely related to P. flexuosa var. flexuosa than to P. chilensis var. riojana.
According to Burkart (1976), the native range of P. chilensis includes southern Peru, Bolivia, Chile and Argentina. However, Galera (1996) states that this species was introduced into Chile at the beginnings of the XIX Century. In Argentina, it is distributed in the political provinces of Cordoba, Catamarca, La Rioja, Salta and San Juan, which correspond to the biogeographical provinces of Chaco, Monte and Prepuna (for a thorough description of biogeographical regions, see Cabrera and Willink 1980).
This species has wide climatic tolerance (Galera 2000) from an absolute maximum temperature of 48°C to an absolute minimum temperature of −7°C. P. chilensis, like most Prosopis species, can grow in many different soils, commonly in sandy soils; it exhibits a great resistance to salinity and tolerates alkaline soils (pH 7.7–8.9) with a high concentration of sodium. As many other legume species, its ability to develop a symbiotic relationship with Rhizobium, which allows N2 fixation, (Aiazzi et al. 1995) is important to resist and improve soils poor in nutrients. For these reasons, afforestation or reforestation with this species is promising to prevent desertification.
According to Verzino et al. (2003), P. chilensis exhibits throughout its distribution in Argentina an important variation for morphological and physiological quantitative traits. This result implies genetic differences associated with adaptation to particular environments, and these authors pointed out the importance of analysing the relation of adaptive genetic variation to particular environmental conditions.
Genetic markers provide a method for assaying levels of genetic variation and population structure in natural populations of forest trees and for measuring population processes of ecological importance and conservation forest management (Rieseberg 1996a; Shea and Furnier 2002; Lee et al. 2002; Renau-Morata et al. 2005; Belletti et al. 2008). Among several methods, random amplified polymorphic DNA (RAPD) has been commonly used because data can be generated relatively quickly and with less labour than other methods for comparative studies in plant collections. Some authors have suggested that the reliability of RAPD and isozyme data should be comparable at the intraspecific level and moderate and high correspondence between results of these markers has been reported (Nybom and Bartish 2000), while other authors state that RAPD provides a better genome random sample than allozymes do (Fristch and Rieseberg 1996; Nybom 2004). Thus, a complementary approach using both isozyme and RAPD markers may be suitable for generating accurate estimates of genetic diversity and population structure. Moreover, these markers have shown significant correlation with climatic factors (Bulinska-Radomska 2000; Lewinsohn et al. 2001; Semagn et al. 2000) and may be useful in predicting sampling strategies for the conservation of genetic resources.
In Prosopis, isozyme markers have allowed the characterization of many species of section Algarobia (Julio 2000; Saidman et al. 2000; Bessega et al. 2005; Ferreyra et al. 2007) while RAPD markers have provided species-specific bands (Juárez-Muñoz et al. 2002; Vázquez-Garcidueñas et al. 2003; Ferreyra et al. 2004; Landeras et al. 2005). Nevertheless, studies using both isozymes and RAPDs are scarce and have included few populations of each species (Bessega et al. 2000a; Ferreyra 2001; Ferreyra et al. 2007), and the studies so far conducted in species of section Algarobia have failed to show genetic differentiation associated with environmental variables (Julio 2000; Saidman et al. 2000; Ferreyra et al. 2007).
In this study, we analysed the variability of RAPD markers in nine Argentine populations of P. chilensis representative of the three biogeographical provinces: Chaco, Prepuna and Monte. RAPD data were compared with previous isozyme studies (Julio 2000), and the joint information from both markers was used to (1) explore the organization of genetic variability at different hierarchical levels (within populations, among populations within regions and among regions) and (2) investigate the association of genetic variation with climatic and geographic variables throughout the main distribution area of this species in Argentina, in order to apply this information to developing forest conservation strategies.
Methods
Populations sampled
The seeds analysed in this study were provided by the Banco Nacional de Germoplasma de Prosopis (NGBP) of the Facultad de Ciencias Agropecuarias, Universidad de Córdoba (Verzino 2005). Plants collected were taxonomically identified by Joseau and Castro Schule (2005) following the morphological criteria described in Burkart (1976). From each sampled tree, a voucher specimen was obtained and kept in the herbarium of the NGBP. According to R. Palacios, all material used in the present study belong to the same variety (P. chilensis v. chilensis). Seeds for isozyme and RAPD analyses were obtained from the same mother plants.
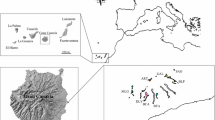
The present study involved nine sample sites representative of the range of P. chilensis in Argentina (Table 1; Fig. 1). Four of them belong to the biogeographical province of Chaco: Villa Dolores (DOL), La Higuerita (LHI), Soto (SOT) and Conlara (CON). Three populations are situated in the Monte biogeographical province: Patquía (PAT), Talampaya (TAL) and Astica (AST). The remaining two samples correspond to Prepuna biogeographical province: Las Talas (LTA) and Belén (BEL) (see Cabrera and Willink 1980).
Map of Argentina indicating the nine sample sites representative of the distribution range of Prosopis chilensis in Argentina. The corresponding population acronyms are indicated in Table 1
As P. chilensis predominantly outcrosses like most species of section Algarobia (Bessega et al. 2000b), most genetic variability is captured with seeds collected from a relatively small number of mother plants (Saidman and Vilardi 1993). In fact, studies in related species of Prosopis with mating systems similar to that of P. chilensis (Saidman et al. 2000; Ferreyra et al. 2004; Ferreyra et al. 2007) showed that more than 85% of the genetic diversity detected was found within population suggesting that the sampling error by applying this strategy is minimal. According to this, the seeds sampled (450 for isozyme and 141 for RAPD analysis) were obtained from seven to nine trees per population. Within each site, the sampled trees were separated at least 100 m from each other in order to reduce the probability of duplication in the samples.
Isoenzyme technique
Six isoenzymatic systems were previously analysed by Julio (2000) in populations of P. chilensis: (EST), (Enzyme Commission) E.C. 3.1.1.1; glutamate oxalacetate transaminase (GOT), E.C. 2.6.1.1; leucilamino peptidase (AMP), E.C. 3.4.1.1.1; alcohol dehydrogenase (ADH), E.C. 1.1.1.1; superoxide dismutase (SOD), E.C. 1.15.1.1; peroxidase (PRX), E.C. 1.11.1.7 using horizontal electrophoresis. A total of 450 individuals (seeds) were analysed, involving 50 seeds per system per population. The information of buffers, electrophoresis conditions and staining methods is given in Online Resource 1. Homogenates were obtained from cotyledons of 7-day-old seedlings for all systems but ADH, for which whole seeds soaked for 24 h were used. Seedlings were obtained under the same conditions of lighting, humidity and temperature (Saidman 1985) in order to assure reproducibility of isozyme banding patterns. Electrophoretic phenotypes of isozymes are available in Online Resource 2 and 3.
Band patterns in P. chilensis were homologous to those observed in related species of section Algarobia (Saidman 1985, Saidman et al. 1997; Bessega et al. 2000a; Ferreyra 2001).
RAPD technique
Genomic DNA was extracted from cotyledons of 8–13-day-old seedlings using the method of Dellaporta et al. (1983). DNA concentration was estimated by comparing electrophoretic patterns on 0.8% agarose (in 1× TAE buffer) gels with standard DNA marker sets (phage λ double-digested with EcoRI and HindIII).
The PCR involving arbitrary primers (Promega) was carried out in a thermal cycler (GeneE, Cambridge, Techne Ltd) that was programmed for one cycle of 94°C for 6 min, 45 cycles of 94°C for 1 min, 36°C for 1 min and 72°C for 2 min followed by a final extension step of 72°C for 6 min. PCR was carried out in 50 μl final volume using 40–60 ng of genomic DNA, 5 μl dNTP mix (1 mM each dNTP), 10 μl of primer (3 ng per μl), 10 μl Taq DNA polymerase buffer (10 mM Tris–HCl, pH 9 at 25°C, 50 mM KCl, 0.1% Triton X-100), 6 μl MgCl2 (25 mM) and 0.5 units of Taq DNA polymerase (Promega). These conditions were maintained in all experiments in order to avoid reproducibility and scoring problems. The amplification products were resolved on 1.4% agarose gels and stained with EtBr. Fragments were observed and photographed on a UV transilluminator (312 nm). The usual cautions needed to prevent contamination of PCR experiments with previously amplified fragments were observed. Reliability of PCR products was tested by several controls that were routinely used, one without primer, a second one with no Taq DNA polymerase and the third one with no genomic DNA. No amplification occurred in any of these negative controls. Re-amplification was performed routinely to ensure reproducibility of banding patterns. Each individual DNA sample was amplified three times and seeded in three different gels, and the same pattern was obtained. In each gel, we analysed individuals from different populations in order to avoid bias in the comparisons among populations attributable to experimental error.
Twenty primers were tested, and five of them were selected on the basis of reproducibility of bands retrieved and their ability to show polymorphisms. The selected primers were B01 (5′-TCG AAG TCC T-3′), B07 (5′-AGA TCG AGC C-3′), B10 (5′-CAG GCA CTA G-3′), A09 (5′-CTA ATG CCG T-3′) and A06 (5′-GAG TCT CAG G-3′). A total of 141 individuals (seeds) were analysed, involving 14–20 per population, with the exception of La Higuerita where the sample was considerable lower (8 individuals) (Table 4).
Homology of RAPD bands
The use of RAPDs for comparative purposes relies on the assumption that similarity of fragment size is a dependable indicator of homology (Rieseberg 1996b). To test the validity of this assumption, homology of the polymorphic RAPD DNA fragments (A09-3 and A09-5) between different populations of P. chilensis was determined and further investigated by DNA sequence alignment. These bands derived from the primer A09 (5′-CTA ATG CCG T-3′) were chosen because they showed association with environmental variables. We cloned and sequenced the bands taken from random selected individuals of different populations. A09-3 bands were sequenced in individuals from TAL and DOL, whereas A09-5 bands were sequenced in individuals from PAT, TAL, DOL and SOT. The bands were excised from 1, 5% agarose gels with a sterile gel slicer, and the DNA was purified using the QIAEX II Gel Extraction Kit (QIAGEN, USA). The eluted fragments were ligated into pGEM T Easy Vector (Promega, USA) following the supplier’s instructions. The recombinant plasmid DNA was used to transform Escherichia coli (DH5a) competent cells. Plasmid DNA was purified from five independent transformed clones as described by Sambrook et al. (1989). The size of cloned DNA inserts was checked by digestion with EcoRI (New England Biolabs). The complete sequences of the cloned fragments were obtained using an ABI Prism 3100 automated DNA sequencer (Unidad de Genómica, Inst. Biotechnology, INTA Castelar, Argentina).
Assembly and editing of sequences was performed with BioEdit version 5.0.9 (Hall 1999). Alignments were performed using CLUSTALW (Thompson et al. 1997).
Genetic variability
Genetic variability and structure parameters of nine populations of Prosopis chilensis were studied from RAPD markers. Binary matrix of band presence/absence was converted into allelic frequencies by a Bayesian method (Zhivotovsky 1999) with non-uniform prior distribution using the software AFLP-SURV 1.0 (Vekemans et al. 2002). Genetic variability was quantified by the unbiased expected heterozygosity (H e) (Nei 1978), percentage of polymorphic loci (P) and mean number of alleles per locus (A).
Expected heterozygosities were compared among populations by means of the asymptotic Friedman test, taking populations and loci as factors. Variability was also compared among regions. In this case, as the model was unbalanced (different numbers of populations between regions), we applied the K-sample permutation test. Pair-wise comparisons of H e between populations were conducted by Wilcoxon test. The correlation among variability from different markers was evaluated by Spearman′s rank correlation test. These non-parametric methods were chosen because they do not involve any assumptions regarding distribution of variability estimates. They were performed using the packages coin (Hothorn et al. 2007) and exactRankTests (Hothorn and Hornik 2007) of the program R (versión 2.9) (R Development Core Team 2009).
Gametic disequilibrium
Linkage between RAPDs loci was estimated by Otha’s (1982) method using the program RAPDLD from the RAPDs package (Black 1996).
Hierarchical analysis of genetic variability
The distribution of RAPD genetic variability at different hierarchical levels was studied by analysis of molecular variance (AMOVA) (Excoffier et al. 1992). Also, non-hierarchical Φ ST was estimated to evaluate overall genetic differentiation. These analyses were conducted using the software GeneticStudio (Dyer 2005–2007).
Clustering methods and linear discriminant analysis
Genetic differentiation between populations was measured by pair-wise Wright’s Φ ST obtained from the RAPD dataset using the software AFLP-SURV 1.0 (Vekemans et al. 2002).
From the genetic distance matrix, a phenogram was obtained by applying the unweighted pair grouping method based on arithmetical means (UPGMA) using the program R. The robustness of the phenogram branching was evaluated by the analysis of consensus of 100 trees obtained from 100 bootstrapped pseudoreplicates of the original data matrix.
Bootstrapping was performed by AFLP-SURV 1.0 (Vekemans et al. 2002), and the consensus analysis was made with PHYLIP program (Felsestein 1993).
The structures of the genetic distance matrices and trees obtained from RAPD loci were compared with those obtained by Julio (2000) from isozymes using Mantel test. The consistence of isozyme and RAPD-based phenograms was evaluated by comparing the corresponding cophenetic matrices by the same statistical method. To conduct Mantel tests, we used the package ape (Paradis et al. 2005–2007) of program R.
The relationships between populations retrieved in the matrix of pair-wise Φ ST were represented graphically by Kruskal’s non-metric multidimensional scaling (MDS) to determine whether populations are grouped according to their corresponding regions.
Linear discriminant analysis (LDA) was conducted, trying to determine how best to separate known groups of individuals and predict to which predefined class an individual belongs. LDA and MDS were made with the package MASS of program R.
Climatic and geographic information
The climatic and geographic variables were obtained using the New locClim program (ver. 1.10) (Grieser 2006). These variables were the following: average minimum temperature, mean annual temperature, average maximum temperature, evapotranspiration potential, mean number of rainy days, sunshine fraction, sunshine hours, mean annual rainfall per month, aridity, effective rain, dryness, latitude, longitude and altitude. From this information, standardized Euclidean pair-wise climatic distances were estimated between populations using the program R.
Isolation by distance
The hypothesis that the genetic differentiation between populations is due to isolation by distance was tested comparing geographic and genetic distance matrices by Mantel test. Distances between sample sites were estimated using the measuring tool of Google Earth ver. 4.1 (http://earth.google.com).
Correlation of genetic variables with climatic and geographic variables
The association of band frequencies with environmental variables cannot be properly analysed with simple regression of band frequencies on environmental variables because this method does not take into account that the environmental and geographical variables are not independent. Taking into account this issue, for each locus, a stepwise multiple regression analysis (backward and forward) was applied to determine direct effects of environmental variables on allelic frequencies. The procedure applied was a generalized linear model defined by the following equation:
where y is response variable (band presence or absence), μ is the overall mean, the explanatory variables x i are the climatic or geographic records corresponding to each population and β i are the partial regression coefficients for each explanatory variable.
As in this case we deal with proportion data (band frequencies), we considered three important properties that affect the way the data should be analysed: (1) the data are strictly bounded (between 0 and 1), (2) the variance is non-constant and (3) errors are non-normal. With proportion data, if band frequency is 0, then there will be no band in repeated trials (sampled individuals), all the data will be zeros and hence the variance will be zero. Likewise, if band frequency is 1, then all sampled individuals will show the band, and again the variance will be 0. For proportion data, therefore, the variance increases with the mean up to a maximum (when band frequency is one half) then declines again towards zero as the mean approaches 1. The variance–mean relationship is humped, rather than constant as assumed in the classical tests. The final assumption is that the errors (the differences between the data and the fitted values estimated by the model) are normally distributed. This cannot be so in proportional data because the data are bounded above (1) and below (0). All these issues (boundedness, non-constant variance and non-normal errors) are dealt with by using a generalized linear model with a binomial error structure. To take into account sample sizes with binomial errors, we must give the number of “failures” (band absence) as well as the numbers of “successes” (band presence) in a two-vector response variable. To do this, we bind together two vectors into a single object, “y ” (the response variable), comprising the numbers of successes and the number of failures. The binomial denominator, “n ”, is the total sample. The procedure we applied carries out weighted regression, using the individual sample sizes as weights, and the logit link function to ensure linearity (see Crawley 2007). The method applies the Akaike (1974) information criterion (AIC) to select predictor variables to be included in the model. The regression analysis was conducted with the package stats of the program R (versión 2.9) (R Development Core Team 2009).
Results
RAPD analysis
With the five (out of 20) primers selected, we detected 46 reproducible and unambiguously scored bands. RAPD patterns obtained with the primers A01, A06, A09, B01 and B10 are given in Online Resource 4 and 5. The molecular weight of the bands ranged from 300 to 1,600 bp. All of them were polymorphic (sensu Ford 1940 and see Ridley 1993 p. 69, 639) showing variation within at least one population (data not shown). Allelic frequencies of RAPD loci in the studied populations of Prosopis chilensis are given in Online Resource 6. After applying Otha′s (1982) method, only 2 out of 1035 comparisons (0.19%) suggested epistasis disequilibrium between RAPD markers.
Analysis of band homology
The sequences of bands A09-3 and A09-5 were deposited in Gene Bank, with the information on accession numbers and molecular weights reported in Table 2. The comparison of the DNA sequences of band A09-5 in individuals from different populations (Table 3) indicated a high sequence homology ranging from 91.0 to 97.4%. The two sequences analysed of band A09-3 in one individual from DOL and one from TAL exhibit a homology of 99.7%. These results were consistent with the hypothesis of homology of comigrating bands observed in individuals from different populations.
Genetic variability
The average number of alleles per locus showed low variation among populations ranging from 1.4 to 1.7 (Table 4). The percentage of polymorphic loci exhibited a large range (from 37 in LHI to 71% in LTA) (Table 4) indicating considerable differences in variability among populations.
He estimated from RAPD (Table 4) showed significant differences among populations according to Friedman test (χ2 = 18.88, DF = 8, P = 0.01). Out of 36 pair-wise comparisons between populations, only five yielded significant or highly significant results when compared by Wilcoxon test (Table 5). According to the K-sample permutation test, He estimates between regions differed significantly (T = 2.85, P = 0.012).
Julio (2000) on the basis on 6 isoenzymatic systems detected a total of 17 loci, 12 of them variable. Isozyme patterns of ADH, PRX, EST, AMP, GOT and SOD are given in Online Resources 2 and 3. The variability estimates from these loci in the same populations studied here were not significantly correlated with those based on RAPD according to Spearman′s rank correlation test (Z = −0.33, P = 0.74).
Hierarchical and non-hierarchical genetic structure analysis
The analysis of the distribution of the genetic variability of RAPD loci by AMOVA (Excoffier et al. 1992) indicated that total differentiation among populations was highly significant (non-hierarchical Φ ST = 0.113, P = 0.001). The variability among populations (Φ ST) was partitioned into the differentiation among populations within regions (Φ SR) and among regions (Φ RT). The highest variance component was observed within populations (Table 6). The results, expressed in terms of gene diversity (Nei 1978), showed that most (88.1%) of the total diversity (1 − Φ ST) occurs within populations, the component among populations within regions (9.3%) (Φ ST–Φ RT) was intermediate, while the between-region component (Φ RT) was the lowest (2.6%) (Table 6). All three variance components were highly significant.
Genetic differentiation between populations was also analysed by Kruskal’s non-metric multidimensional scaling from the matrix of pair-wise Φ ST values. The results of this analysis (Fig. 2) were consistent with the highly significant among-region differentiation indicated by the AMOVA. Axis 1 separates Prepuna (positive values) from Monte populations (negative values). Axis 2 separates Chaco populations (positive values) from the remaining populations (negative values).
Phenetic relationships
The structure of the genetic distance matrices obtained from isozyme (Julio 2000) and RAPD data was not correlated with each other according to Mantel test (r = 0.103, Z = 0.367, P = 0.34, based on 1000 permutations). The RAPD-based phenogram (Fig. 3) showed good cophenetic correlation respect to the corresponding distance matrix (r = 0.77, Z = 0.269, P = 0 based on 1000 permutations). Phenogram nodes showed relatively low (lower than 50%) bootstrap support. Populations from Prepuna province (BEL and LTA) are clustered together. PAT is not grouped with the other two populations (AST and TAL) from the Monte province. Genetic distances between populations from the Chaco (LHI, DOL, SOT and CON) are higher than those between populations within the other biogeographical provinces.
Linear discriminant analysis
The linear discriminant analysis succeeded in differentiating individuals from different populations on the basis of presence or absence of RAPD band. The consistency between prior and post classifications was 100%. The first three linear discriminant axes (LD1-LD3) explained about 77% of total variation, and the tridimensional dispersion plot according to these axes shows most of the individuals in each population grouped (Fig. 4).
Climatic, geographic and genetic distance
The genetic (Φ ST) distance matrix obtained from RAPD was compared with the matrix of geographical distances in order to determine whether they fit to the expectations according to the isolation by distance model. Mantel test (based on 1,000 permutations) indicated that genetic distances were not significantly correlated with geographical distances (r = 0.148, Z = 852.87, P = 0.24).
Band frequencies and geographic and climatic variables
All 12 variable isozyme loci showed significant or highly significant association with at least one geographic/climatic variable according to the stepwise multiple regression analysis (Table 7). We conducted a similar analysis for RAPD data. Out of 46 RAPD loci, 26 showed significant or highly significant association with at least one climatic or geographic variable (Table 8).
Discussion
Despite the ecological importance of Prosopis chilensis and its potential use for reforestation programmes in arid soils, little is known about the distribution of its genetic variability and population structure. This information is essential for establishing programmes of conservation of genetic resources.
Biochemical and molecular markers have been used to study the diversity within and among related forest species and to assist taxonomists in species determination. Among several efficient methods, allozymes have been the most widely used genetic marker in the study of the differentiation and variation at the population and species levels (Crawford 1997). Polymorphic DNA (RAPDs) has also been used as a genetic marker in forest plants (Lee et al. 2002; Shea and Furnier 2002; Renau-Morata et al. 2005). This marker has found the widest application in systematic studies (Bussell et al. 2005) and particularly in the quantification of genetic variability in closely related species (Crawford 1997; Nybom and Bartish 2000; Nybom 2004). The advantages and limitations of co-dominant versus dominant markers have been largely discussed. Although as a general rule highly polymorphic co-dominant loci offer the highest precision for most purposes, tens to hundreds of dominant polymorphic loci are commonly reported (Hardy 2003), which offers opportunities to make inferences at much finer scales of resolution (Ritland 2005). Furthermore, in comparison to co-dominant markers, dominant markers can be developed relatively easily even for species for which no prior genetic information is available and at a relatively low cost (Belletti et al. 2008).
In reference to genus Prosopis, Bessega et al. (2000a) obtained similar estimates of gene flow between populations of the North American species P. velutina and P. glandulosa from isozymes and RAPDs; however, two isoenzyme systems produced diagnostic loci, whereas no diagnostic bands were obtained from RAPDs useful to distinguish these species. Ferreyra et al. (2007) applied both techniques to analyse variability and differentiation among five Argentinean species of Prosopis and natural hybrids closely related to P. chilensis, obtaining similar conclusions from both methods in reference to population structure and phenetic relationships among species. Moreover, Ferreyra et al. (2004) found diagnostic RAPD band combinations useful to distinguish five species of section Algarobia of Prosopis, which in isoenzymatic analysis (Ferreyra et al. 2007) showed only small differences in allele frequencies.
In this paper, the ranges of the different genetic variability measures (A, P and H e) obtained from RAPD were similar to those observed in other related species of section Algarobia (Ferreyra et al. 2004). However, the variability estimates based on RAPD and isoenzymes were not correlated with each other. These results are explained because RAPD-based variability estimates showed significant differences among populations, whereas isozyme-based ones did not. A possible explanation for this different behaviour is that isozyme systems are more or less randomly chosen without taking into account the variability detected. By contrast, the primers for PCR are usually selected to obtain as many informative bands as possible. According to our results, RAPD-based heterozygosity estimates varied significantly among regions, suggesting that genetic variability is associated with some environmental factors.
The analysis of genetic structure from RAPD loci included populations that are representative of almost the entire range of this species in Argentina. The differentiation among populations was highly significant. Although the AMOVA showed that most variability occurs within populations, the diversity among regions and among populations within regions was highly significant. The high variability within populations may be explained by the open pollinated system with a preponderance of an outcrossing mating system as observed in the species so far studied of section Algarobia (Bessega et al. 2000b). The highly significant variability among populations is consistent with the relatively higher differentiation observed for insect-pollinated and animal-dispersed tree species in comparison with wind-pollinated and wind-dispersed trees (Hamrick et al. 1992; Jordano and Godoy 2000; Hilfiker et al. 2004). It is interesting that the differentiation among populations and regions persists in spite of the highly interactive relationship between humans and algarrobos, which probably inadvertently changed the original genetics of population structure by stimulating the mixing of populations: People returning to the trees with sweetest pods, which were later defecated and germinated, or allowing erect thornless trees to grow near their homes while eliminating spiny ones with poor growth. Also, the introduction of livestock by Europeans contributed to the mixing of populations as animals were moved between distant regions (D’ Antoni and Solbrig 1977). The finding of significant variation between regions should be considered in programmes of genetic resources conservation, and this implies that populations from all biogeographical provinces should be maintained to preserve the full genetic variation across the species range.
The structure of the genetic distance matrices obtained from isozyme and RAPD data was not correlated according to Mantel test. These results can be explained by the fact that RAPD markers provide information of a higher number of polymorphic loci and probably represent a more accurate picture of the real distances between populations.
The RAPD phenogram representative of genetic distances (quantified as pair-wise Φ ST) between populations did not agree with the expected one according to geographical distances. Moreover, the multidimensional scaling analysis indicated that populations tend to be grouped according to their corresponding biogeographical province. These results might be interpreted as evidence that the ecological differentiation among biogeographical provinces is reflected in allele frequency variation.
Earlier studies in this species (Verzino et al. 2003) reported a weak association between some macroenvironmental variables and quantitative physiological traits and pointed out the importance of the knowledge of adaptive genetic variation in relation to particular environmental conditions. In contrast, in this study, a deeper insight was obtained by the regression analysis of allele/band frequencies on environmental variables. Twenty-six out of 46 RAPD loci and all 12 isozyme loci showed dependence on climatic or geographic variables. The isozyme systems analysed in this study were previously applied to species belonging to Algarobia section by our group (Saidman 1985, 1990; Saidman et al. 1997; Bessega et al. 2000a; Ferreyra 2001; Ferreyra et al. 2007) and by other authors (Verga 1995). The isozyme banding patterns were consistent, and homology was proved even in different species. Saidman (1990) analysed hybrid segregation between P. caldenia and P. flexuosa and proved homology between isozyme loci of both species. Respect to RAPDs, in this paper, we analysed the homology of some bands that showed highly significant association with several geographic or climatic variables by DNA sequencing. The high homology observed between individuals from different populations reinforces our results. The variables that affected gene frequencies were associated with rainfall, dryness, sun incidence, temperature, altitude, latitude and longitude. The results obtained in the present study give support to the hypothesis that the morphological and physiological differences observed among populations have a genetic basis. Moreover, the discriminant analysis indicated that the individuals can be assigned unequivocally to their corresponding population on the basis of their RAPD pattern. This result together with the highly significant non-hierarchical Φ ST suggests that the gene flow between populations is relatively low, allowing adaptive local differentiation.
According to the neutral theory (Kimura 1983), most variation of molecular markers is caused by random drift and mutant genes selectively equivalent. RAPD and isozymes are usually considered neutral markers. However, several studies suggest that their variation may have adaptive significance (Nevo et al. 1988; Matrajt et al. 1996; Confalonieri et al. 1998; Semagn et al. 2000; Lewinsohn et al. 2001). Furthermore, it has been argued that the environmental heterogeneity is a major factor in maintaining and structuring the genetic variation in natural populations (Bulinska-Radomska 2000) and that the amount of variation may be regarded as an adaptive strategy for increasing population fitness in an heterogeneous environment (Nevo et al. 1982; Baum et al. 1997; Confalonieri et al. 1998; Lewinsohn et al. 2001; Fahima et al. 2002).
In the present paper, variability estimates (H e) differ among biogeographical provinces, and as it was previously stated, allele frequencies of more than half of RAPD loci and all polymorphic isozyme loci are affected by at least one climatic/geographic variable. These results imply that genetic distances between populations are better explained by biogeographical grounds than by physical distances.
The lack of previous molecular support to the morphological ecotypes described for P. chilensis might be explained by the fact that within-population variation explains nearly 90% of total diversity. However, a wider sampling of genetic markers coupled with environmental information might be a useful tool to identify selective variation. We hypothesize that the correlation of isozyme and RAPD loci with climatic and geographic variables reported in this paper as well as the highly significant differentiation among regions is at least partially associated with gametic disequilibrium between the genetic markers here analysed and loci related to local adaptation. These results provide some orientation for the genetic management of this important species.
Conclusions
RAPD and Isozyme markers proved to be a powerful tool to analyse population structure and to assess the importance of environmental and biogeographical factors in the distribution of genetic variability of Prosopis chilensis in Argentina. We detected significant differences of variability estimates among biogeographical provinces. The analysis of population structure indicated that most of genetic diversity (88%) occurs within populations, the diversity among populations within each province is intermediate (9%) and the lowest component corresponds to diversity among provinces diversity (3%). All three components were highly significant. The high variability within populations is consistent with the mostly outcrossing mating system found in this species, while the high differentiation among populations is explained by the insect-pollinated and animal seed dispersion systems. This among population differentiation persists despite anthropogenic interactions that probably increased the dispersion rate. The phenogram and MDS plots did not agree with geographical distances and were best explained on the grounds of biogeographical information. All polymorphic isozyme loci and more than half (26/46) of RAPD band are significantly dependent on at least one climatic or geographic variable. These results allowed us to hypothesize the occurrence of gametic disequilibrium between the genetic markers here analysed and loci responsible for local adaptation. The results of our study contribute to a better understanding of the distribution of genetic variability and population structure and provide orientation for the genetic management and preservation of this important species. The significant variation observed between regions has implications for conservation purposes because populations from all biogeographical provinces should be maintained to preserve the full genetic variation across the species range.
References
Aiazzi MT, Di Rienzo A, Ledesma A, Arguello J (1995) Growth dynamics and nitrogen fixation in young Prosopis chilensis (Mol.). Phyton 57:121–126
Akaike H (1974) A new look at the statistical model identification. IEEE Trans Automat Contr 19(6):716–723
Baum BR, Nevo E, Johnson DA, Beiles A (1997) Genetic diversity in wild barley (Hordeum spontaneum C. Koch) in the Near East: a molecular analysis using random amplified polymorphic DNA (RAPD) markers. Genet Resour Crop Evol 44:147–157. doi:10.1023/A:1008655023906
Belletti P, Monteleone I, Ferrazzini D (2008) A population genetic study in a scattered forest species, wild service tree [Sorbus torminalis (L.) Crantz], using RAPD markers. Eur J Forest Res 127:103–114. doi:10.1007/s10342.007-0187-1
Bessega C, Vilardi JC, Saidman BO (2000a) Isozyme and RAPD studies in Prosopis glandulosa and P. velutina (Leguminosae, Mimosoideae). Genet Mol Biol 23:639–648. doi:10.1590/S1415-47572000000300024
Bessega C, Ferreyra LI, Julio N, Montoya S, Saidman BO, Vilardi JC (2000b) Mating system parameters in species of genus Prosopis (Leguminosae). Hereditas 132:19–27. doi:10.1111/j.1601-5223.2000.00019.x
Bessega C, Saidman BO, Vilardi JC (2005) Genetic relations among American species of Prosopis (Leguminosae) based on enzyme markers. Genet Mol Biol 28:277–286. doi:10.1590/S1415-47572005000200018
Black WC IV (1996) Fortran programs for the analysis of RAPD-PCR markers in populations. Colorado State University, Ft. Collins, CO
Bulinska-Radomska Z (2000) Enzyme polymorphism and adaptation in strawberry clover (Trifolium fragiferum L.). Genet Resour Crop Evol 47:197–205. doi:10.1007/S10722-006-9162-9
Burkart A (1976) A monograph of the genus Prosopis (Leguminosae, Mimosoideae). J Arnold Arb 57 (3, 4): 219–249; 450–525
Bussell JD, Waycott M, Chappill A (2005) Arbitrarily amplified DNA markers as characters for phylogenetic inference. Perspect Plant Ecol Evol Syst 7:3–26. doi:10.1078/1433-8319-00007
Cabrera AL, Willink A (1980) Biogeografía de América Latina. The General Secretarial of the Organization of American States, Washington, DC
Confalonieri V, Sequeira A, Todaro L, Vilardi J (1998) Mitochondrial DNA and phylogeography of the grasshopper Trimerotropis pallidipennis in relation to clinal distribution of chromosome polymorphisms. Heredity 81:444–452
Crawford DJ (1997) Molecular marker for the study of genetic variation within and between populations of rare plants. Opera Bot 132:149–157
Crawley MJ (2007) The R Book. Wiley, The Atrium, Southern Gate, Chichester, West Sussex PO19 8SQ, England, pp 942
D′ Antoni HL, Solbrig OT (1977) Algarrobos in South American cultures: past and present. In: Simpson BB (ed) Mesquite its biology in two desert scrub ecosystems. Stroudsburg, Pensilvania, pp 189–202
Dalmasso A (1993) Selección de formas de una población de Prosopis chilensis stuntz (Leguminosae) para ensayos de forestación. Conservación y mejoramiento de especies del género Prosopis. In: Contribuciones mendocinas a la quinta reunión general para América latina y el Caribe de la red de forestación del CIID. IADIZA-CRICYT-CIID, Mendoza, Argentina
Dellaporta SL, Wood J, Hicks JB (1983) A plant DNA minipreparation: version II. Plant Mol Biol Rep 1:19–21
Dutton RW (1989) Prosopis sp: A justification for their future research and development. In: Dutton R (ed) A Research and development strategy for Prosopis. Center for Overseas Research and Development, Durham, pp 33–40
Dyer R (2005–2007) Genetic Studio. http://dyerlab.bio.vcu.edu
Excoffier L, Smouse P, Quattro J (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Fahima T, Roder MS, Wendehake K, Kirzhner VM, Nevo E (2002) Microsatellite polymorphism in natural populations of wild emmer wheat, Triticum dicoccoides, in Israel. Theor Appl Genet 62:241–254
Felsestein J (1993) PHYLIP (Phylogeny Inference Package), version 3.6. Department of Genoma Sciencies, University of Washington, Seattle
Ferreyra LI (2001) Estudio de la variabilidad y la diferenciación genética por medio de técnicas de isoenzimas y RAPD en poblaciones naturales de especies e híbridos del género Prosopis (Leguminosae). PhD Thesis, Facultad de Ciencias Exactas y Naturales, Universidad de Buenos Aires, Buenos Aires, Argentina
Ferreyra LI, Bessega C, Vilardi JC, Saidman BO (2004) First report on RAPD patterns able to differentiate some Argentinean species of section Algarobia (Prosopis, Leguminosae). Genetica 121:33–42. doi:10.1023/B:GENE.0000019925.06080.c7
Ferreyra LI, Bessega C, Vilardi JC, Saidman BO (2007) Consistency of population genetics parameters estimated from isozyme and RAPDs dataset in species of genus Prosopis (Leguminosae, Mimosoideae). Genetica 131:217–230. doi:10.1007/s10709-006-9132-3
Ford EB (1940) Polymorphism and taxonomy. In: Huxley JS (ed) New systematic. Clarendon Press, Oxford, p 493
Fristch P, Rieseberg LH (1996) The use of random amplified polymorphic DNA (RAPD) in conservation genetics. In: Smith TB, Wayne RK (eds) Molecular genetics approaches in conservation genetics. Oxford University Press, Oxford, pp 54–73
Galera FM (1996) Bioecología de especies del Género Prosopis con perspectiva de uso en la alimentación de rumiantes: potencial y limitaciones. Univer. Food Agricultural Organization (FAO), Rómulo Gallegos
Galera FM (2000) Las especies del género Prosopis (algarrobos) de América Latina con especial énfasis en aquellas de interés económico. Universidad Nacional de Córdoba, FAO
Grieser J (2006) New LocClim, version 1.10, Agrometeorology Group, FAO/SDRN, Via delle Terme di Caracalla, 00100, Rome, Italy
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41:95–98
Hamrick JL, Godt MJW, Sherman-Broyles SL (1992) Factors influencing levels of genetic diversity in woody plant species. New Forest 5:95–124
Hardy OJ (2003) Estimation of pairwise relatedness between individuals and characterization of isolation-by-distance processes using dominant genetic markers. Mol Ecol 12:1577–1588
Hilfiker K, Holderegger R, Rotach P, Gugerli F (2004) Dynamics of genetic variation in Taxus baccata: local versus regional perspectives. Can J Bot 82:219–227
Hothorn T, Hornik K (2007) The exactRankTests Package. http://www.r-project.org
Hothorn T, Hornik K, Van de Wiel M, Zeileis A (2007) COIN: Conditional inference procedures in a permutation test framework. http://www.r-project.org
Hunziker JH, Saidman BO, Naranjo CA, Palacios RA, Poggio L, Burghardt AD (1986) Hybridization and genetic variation of Argentine species of Prosopis. Forest Ecol Manag 16:301–315. doi:10.1016/0378-1127(86)90030-7
Jordano P, Godoy JA (2000) RAPD variation and population genetic structure in Prunus mahaleb (Rosaceae), an animal-dispersed tree. Mol Ecol 9:1293–1305
Joseau MJ, Castro Schule FF (2005) Caracterización del género Prosopis. In: Verzino GE, Joseau MJ (eds) El Banco Nacional de Germoplasma de Prosopis. Facultad de Ciencias Agropecuarias, Universidad Nacional de Córdoba, Argentina, pp 81–86
Juárez-Muñoz J, Carrillo-Castañeda G, Arreguin R, Rubluo A (2002) Inter and Intra-genetic variation of four wild populations of Prosopis using RAPD-PCR fingerprints. Biod Conserv 11:921–930
Julio NB (2000) Estudios alozímicos sobre variabilidad, estructura y diferenciación genética en Prosopis chilensis (Leguminosae, Mimosoideae) y especies relacionadas. PhD Thesis, Facultad de Ciencias Exactas y Naturales, Universidad Nacional de Córdoba, Córdoba, Argentina
Karlin O, Coirini R, Catalán L, Zapata R (1997) Especies arbóreas y arbustivas para zonas áridas y semiáridas de América Latina. Series zonas áridas y semiáridas Nº 12 OEA, Programa de las Naciones Unidas para el medio ambiente, FAO, pp 41–51
Kimura M (1983) The neutral theory of molecular evolution. Cambridge University Press, Cambridge
Landeras G, Alfonso M, Pasiecznik NM, Harris PJC, Ramírez L (2005) Identification of Prosopis juliflora and Prosopis pallida accessions. Biod Conserv 15:1829–1844
Lee SW, Ledig FT, Johnson DR (2002) Genetic variation at allozyme and RAPD markers in Pinus longaeva (Pinaceae) of the White Mountains, California. Am J Bot 89(4):566–577
Lewinsohn D, Nevo E, Wasser S, Hadar Y, Beharav A (2001) Genetic diversity in populations of the Pleurotus eryngii complex in Israel. Mycol Res 105:941–951
Matrajt M, Confalonieri V, Vilardi JC (1996) Parallel adaptive patterns of allozyme and inversion polymorphisms on an ecological gradient. Heredity 76:346–354
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small numbers of individuals. Genetics 89:583–590
Nevo E, Golenberg EM, Beiles A, Brown ADH, Zahorí D (1982) Genetic diversity and environmental association of wild wheat, Triticum dicoccoides in Israel. Theor Appl Genet 62:241–254. doi:10.1007/BF00303903
Nevo E, Beiles A, Krugman T (1988) Natural selection of allozyme polymorphism: a microgeographic climatic differentiations in wild emmer wheat (Triticum dicoccoides). Theor Appl Genet 75:529–538. doi:10.1007/BF00276761
Nybom H (2004) Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol Ecol 13:1143–1155. doi:10.1111/j.1365-294X.2004.02141.x
Nybom H, Bartish IV (2000) Effects of life history traits and sampling strategies on genetic diversity estimates obtained with RAPD markers in plants. Plant Ecol Evol Syst 3:93–114. doi:10.1078/1433-8319-00006
Otha T (1982) Linkage disequilibria due to random genetic drift in finite subdivided populations. Proc Nat Acad Sc 79:1940–1944
Paradis E, Strimmer K, Claude J, Jobb G, Opgen-Rhein R, Dutheil J, Noel Y, Bolker B, Lemon J (2005–2007) The ape Package. http://www.r-project.org
Pasiecznik NM, Felker P, Harris PJC, Harsh LN, Cruz G, Tewari JC, Cadoret K, Maldonado LJ (2001) The Prosopis juliflora-Prosopis pallida complex: a monograph. HDRA, Coventry, UK
Renau-Morata B, Nebauer SG, Sales E, Allainguillaume J, Caligari P, Segura J (2005) Genetic diversity and structure of natural and managed populations of Cedrus atlántica (Pinaceae) assessed using Random Amplified Polymorphic DNA. Am J Bot 92:875–884
Ridley M (1993) Evolution. Blackwell, Oxford
Rieseberg LH (1996a) The use of random amplified polymorphic DNA (RAPD) in conservation genetics. In: Smith TB, Wayne RK (eds) Molecular genetics approaches in conservation genetics. Oxford University Press, Oxford, pp 54–73
Rieseberg LH (1996b) Homology among RAPD fragments in interspecific comparisons. Mol Ecol 5:99–105
Ritland K (2005) Multilocus estimation of pairwise relatedness with dominant markers. Mol Ecol 14:3157–3165
Roig FA (1993) Aportes a la etnobotánica del género Prosopis. Contribuciones Mendocinas a la quinta Reunión Regional para América Latina y el Caribe de la Red de Forestación del CIID. Unidades de Botánica y Fisiología vegetal, IADIZA, Mendoza, pp 99–121
Saidman BO (1985) Estudio de la variación alozímica en el género Prosopis. PhD Thesis, FCEyN, Universidad de Buenos Aires, Buenos Aires, Argentina
Saidman BO (1990) Isozyme studies on hybrid swarms on Prosopis caldenia and sympatric species. Silvae Genet 39:5–8
Saidman BO, Vilardi JC (1993) Genetic variability and germoplasm conservation in the genus Prosopis. In: Puri S (ed) Nursey technology of forest tree species of arid and semiarid regions. Winrock-Oxford & IBH Publishing Co., New Delhi, pp 187–198
Saidman BO, Vilardi JC, Pocoví MI, Acreche N (1996) Isozyme studies in Argentine species of the Section Strombocarpa, Genus Prosopis (Leguminosae). J Genetics 75:139–149
Saidman BO, Vilardi JC, Montoya S, Poggio L (1997) Genetic variability and ploidy level in species of Prosopis (Leguminosae). Bol Soc Argent Bot 32:217–225
Saidman BO, Bessega C, Ferreyra LI, Julio N, Vilardi JC (2000) The use of genetic markers to assess populations structure and relationships among species of the genus Prosopis (Leguminosae). Bol Soc Arg Bot 35:315–324
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor Laboratory Press, New York 1659
Semagn K, Bjornstad A, Stedje B, Bekete E (2000) Comparison of multivariate methods for the analysis of genetic resources and adaptation in Phytolaca dodecandra using RAPD. Theor Appl Genet 101:1145–1154
Shea KL, Furnier GR (2002) Genetic variation and population structure in central and isolated populations of Balsam fir, Abies Balsamea (Pinaceae). Am J Bot 89:783–791
Sherwin WB, Moritz CM (2000) Managing and monitoring genetic erosion. In: Young A, Clarke G (eds) Genetics, demography and the viability of fragmented populations. Cambridge University Press, Cambridge, pp 9–34
R Development Core Team (2009) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0, URL http://www.R-project.org
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl Acids Res 25:4876–4882
Vázquez-Garcidueñas S, Palacios RA, Segovia-Quiroz J, Hernández JT, Olalde Portugal V, Mollard FO, Vázquez-Marrufo G (2003) Morphological and molecular data to determine the origin and taxonomic status of Prosopis chilensis var. riojana (Fabaceae, Mimosoidae). Can J Bot 81:905–917. doi:10.1139/b03-078
Vekemans X, Beauwens T, Lemaire M, Roldan-Ruiz I (2002) Data from amplified fragment length polymorphism (AFLP) markers show indication of size homoplasy and of a relationship between degree of homoplasy and fragment size. Mol Ecol 11:139–151
Verga AR (1995) Geneische untersuchungen an Prosopis chilensis und Prosopis flexuosa (Mimosaceae) im trockenen Chaco Argentiniens. PhD Thesis, Universität Göttingen
Verzino GE (2005) Conservación ex situ de recursos genéticos forestales. In: Verzino GE, Joseau MJ (eds) El Banco Nacional de Germoplasma de Prosopis. Facultad de Ciencias Agropecuarias, Universidad Nacional de Córdoba, Argentina, pp 15–22
Verzino G, Carranza C, Ledesma M, Joseau J, Di Rienzo J (2003) Adaptive genetic variation of Prosopis chilensis (Mol) Stuntz, Preliminary results from one test-site. For Ecol Manag 175:119–129. doi:10.1016/S0378-1127(02)00124-X
Wright S (1978) Evolution and the genetics of populations. University of Chicago Press, Chicago
Zhivotovsky LA (1999) Estimating population structure in diploids with multilocus dominant DNA markers. Mol Ecol 8:907–913
Acknowledgments
We would like to thank the Banco Nacional de Germoplasma de Prosopis of the Facultad de Ciencias Agropecuarias, Universidad Nacional de Córdoba, that provided the seeds analysed in this study. We also wish to express our gratitude to the comments received by the two anonymous reviewers that greatly contributed to improve the manuscript. This study was financed by Universidad de Buenos Aires (Grant X201 to B.O.S.), Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) (Grant PIP No. 5122 to J.C.V.), and Agencia Nacional de Promoción Científica y Tecnológica (Grant PICT 32064 BID 1728 OC/AR to B.O.S).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by R. Matyssek.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ferreyra, L.I., Vilardi, J.C., Tosto, D.S. et al. Adaptive genetic diversity and population structure of the “algarrobo” [Prosopis chilensis (Molina) Stuntz] analysed by RAPD and isozyme markers. Eur J Forest Res 129, 1011–1025 (2010). https://doi.org/10.1007/s10342-010-0386-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10342-010-0386-z